Abstract
The evolution of hybrid polyploid vertebrates, their viability and their perpetuation over evolutionary time have always been questions of great interest. However, little is known about the impact of hybridization and polyploidization on the regulatory networks that guarantee the appropriate quantitative and qualitative gene expression programme. The Squalius alburnoides complex of hybrid fish is an attractive system to address these questions, as it includes a wide variety of diploid and polyploid forms, and intricate systems of genetic exchange. Through the study of genome-specific allele expression of seven housekeeping and tissue-specific genes, we found that a gene copy silencing mechanism of dosage compensation exists throughout the distribution range of the complex. Here we show that the allele-specific patterns of silencing vary within the complex, according to the geographical origin and the type of genome involved in the hybridization process. In southern populations, triploids of S. alburnoides show an overall tendency for silencing the allele from the minority genome, while northern population polyploids exhibit preferential biallelic gene expression patterns, irrespective of genomic composition. The present findings further suggest that gene copy silencing and variable expression of specific allele combinations may be important processes in vertebrate polyploid evolution.
Keywords: allopolyploid, gene expression, allele silencing, hybrid lineage evolution
1. Introduction
Polyploidy has been proposed as an important driving force of evolution, and the success of polyploid lineages confirms it as a successful evolutionary transition and a potentially relevant factor in evolutionary diversification (Otto & Whitton 2000). Ploidy rise brought upon by a hybridization event creates an epigenetic instability state that can only be overcome if regulation mechanisms, which contribute to gene copy perpetuation and heterosis, outrun disadvantages and potentiate species adaptation and viability. Therefore, it is important to investigate the functional basis of how polyploids overcome an initial period of instability, and establish processes that allow evolutionary flexibility and efficient competition with their diploid counterparts.
Hybrid polyploidy success has been documented in numerous plant species (Otto & Whitton 2000) and also in animals (Dowling & Secor 1997), but the regulatory changes that contribute to genome stabilization and regulation in the presence of distinct chromosome sets are still elusive. Successful polyploid vertebrates including Rana esculenta (Hotz et al. 1999) and the Bufo viridis complex (Stöck et al. 2002), and asexual hybrid lineages such as the Amazon molly, Poecilia formosa (reviewed in Lampert & Schartl 2008), have been extensively studied regarding different molecular and reproductive aspects that contribute to their viability (Lamatsch & Stöck 2009). In all these systems, several aspects (such as the relevance of heterosis and the role of gene duplicates in lineage-specific evolution) have been comprehensively addressed (Comai 2005; Pignatta & Comai 2009), but the impact of polyploidization on the mechanisms regulating gene expression has not yet been clarified. Several deviations in the expected gene expression patterns have been reported in polyploid plants (Auger et al. 2005), including organ-specific silencing (Adams et al. 2003), epigenetic regulation of duplicates (Comai et al. 2000; Shaked et al. 2001), parent-of-origin-specific control of gene expression (Alleman & Doctor 2000) and a still unclear effect designated as ‘odd ploidy response’ (Guo et al. 1996). In vertebrates, little has been done to understand the effects of ploidy rise on gene regulation and their impact on the evolutionary potential of populations. The Squalius alburnoides complex was the first system in which dosage compensation by gene copy silencing was reported in a polyploid vertebrate context (Pala et al. 2008).
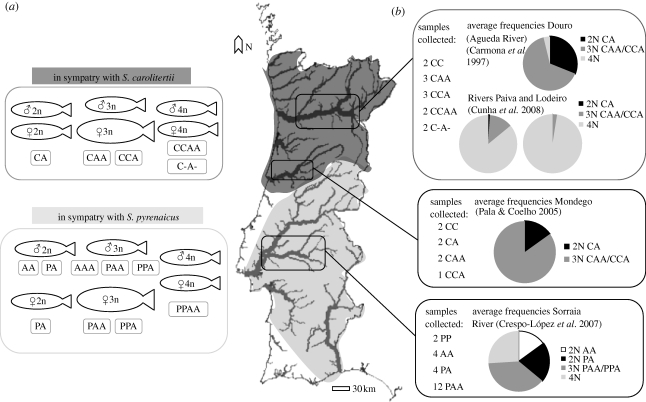
This complex of hybrid fish presents several unique features that make it an ideal system to address the question of gene expression regulation and its impact on the persistence of hybrid lineages over evolutionary time (reviewed in Alves et al. 2001). It is an allopolyploid complex, resulting from interspecific hybridization between two Iberian cyprinid species: Squalius pyrenaicus as the maternal ancestor (contributing with the so-called P genome), and a still undetermined species (closely related to Anaecypris hispanica) as the paternal ancestor (A genome; reviewed in Alves et al. 2001). Presently, the complex is widely distributed in the Iberian Peninsula, where it occurs in sympatry with two species of the genus Squalius: S. pyrenaicus (P genome) in southern basins and S. carolitertii (C genome) in northern basins (figure 1a). These species form a reproductive complex with the hybrids, leading to diploid and polyploid offspring (figure S1, electronic supplementary material). The two bisexual species act as sources of new genetic material, and contribute to the maintenance of gene flow and the continuous cycling of genomes between forms (reviewed in Alves et al. 2001). The complex is composed of animals with different ploidy degrees and genomic constitutions, including diploids (PA, CA), triploids (PAA, PPA, CAA and CCA) and tetraploids, with C or P genome inclusion according to the geographical location (figure 1a). All forms are apparently fertile and interact through diverse reproductive modes (figure S1, electronic supplementary material) that include rare gynogenesis and processes of hybridogenesis (Carmona et al. 1997; Alves et al. 2001; Pala & Coelho 2005). The escape from strictly asexual reproduction and the complexity of the genetic exchange routes definitively contribute to the success of S. alburnoides. In southern populations, an additional form designated as ‘nuclear non-hybrid’ (AA; carrying mtDNA of S. pyrenaicus) occurs and actively contributes to the maintenance of the genetic diversity of the complex (Alves et al. 1999).
Figure 1.
Distribution of S. alburnoides in the areas of sympatry with S. carolitertii (dark grey) and S. pyrenaicus (light grey). (a) The global composition of the populations of the complex in the two areas of sympatry are indicated, although local abundances can vary. (b) Samples collected in each location and reported frequencies of diploid (2N), triploid (3N) and tetraploid (4N) forms in Douro, Mondego and Tejo River basins.
We used the S. alburnoides system to approach the question of gene expression regulation and evolution of polyploid taxa. As an allopolyploid, the S. alburnoides complex offers the advantage of allowing the distinction between the different genome-specific gene copies and how they contribute to overall expression. The presence of lineages within the complex, established differentially in time, including different forms and showing distinct dynamics of genetic exchange and different evolutionary potential, further allows us to explore the question of whether the mechanisms of gene silencing observed in triploids of the southern populations of S. alburnoides (Pala et al. 2008) would also be present in other independent populations and in different genomic compositions.
We have studied the expression pattern of a total of seven widely expressed and tissue-restricted genes in different organs of diploid, triploid and tetraploid forms from southern and northern populations of the complex. We have found differential expression patterns of genome-specific alleles according to geographical location and have attempted to integrate the observed disparities with the distinct genomic and evolutionary features of the different populations of S. alburnoides. We put forward the hypothesis that the type of genomes brought together upon the hybridization process might be an important factor in the establishment of specific mechanisms of gene expression regulation in vertebrate allopolyploids.
2. Material and Methods
(a). Samples, DNA and RNA extraction, and genotyping
Samples of S. alburnoides and of the sympatric Squalius species were collected from three locations corresponding to the northern and southern distribution ranges of the complex (figure 1b). Individuals were sacrificed with an overdose of the anaesthetic MS222. Organs and fin clips were collected for RNA and DNA extraction, which were performed as described in Pala et al. (2008). Blood samples were drawn from the caudal vein, stabilized in buffer (40 mM citric acid trisodium salt, 0.25 M sucrose and 5% dimethyl sulphoxide) and immediately frozen at −80°C. The determination of genotype identity of individual samples was performed by a conjoined approach of flow-cytometry measurements (Dawley & Goddard 1988) and the analysis of microsatellite variation.
Genotypes were determined by cross-species amplification of three microsatellite loci (LCO3, LCO4 and LCO5) using the Multiplex PCR Kit (Qiagen) and identification of alleles specific to the respective genomes (P, C and A), as previously described (Pala & Coelho 2005; Crespo-López et al. 2007; Cunha et al. 2008). In the cases for which it was not possible to identify the three or four distinct alleles in triploids and tetraploids, an additional locus (LCO1) was used (table S1, electronic supplementary material). Amplification products were analysed with an automated sequencer (ABI 310 Genetic Analyser).
(b). Sequence determination and genome expression
Sequences of known teleost orthologues of a total of seven genes—ubiquitously expressed (β-actin, rpl8, ef1a and gapdh), gonad-specific (amh, dmrt1) and eye-specific (rhodopsin)—were used as templates for the design of gene-specific primers (table S2, electronic supplementary material). Primers were initially tested on cDNA samples of S. pyrenaicus (PP), S. carolitertii (CC) and S. alburnoides (AA). Amplifications were performed according to the following PCR conditions: pre-heating at 94°C for 2 min 30 s, 35 cycles at 94°C for 45 s, 52°C (amh, dmrt1)/55°C (rhodopsin, β-actin, rpl8, ef1a and gapdh) for 40 s and 72°C for 1 min 15 s, and a final extension at 72°C for 10 min. Polymorphic sites for the three genomes (P, C and A) were identified for the seven genes by sequence alignment using Sequencher v. 4.0 (Gene Codes Corporation). In hybrid samples, the presence of cDNAs derived from single genome copies or from both genomes was determined through sequence comparison and based on the identified polymorphic sites between genomes (P, C and A). Genome control sequences, representing the P, C and A genomes and obtained from S. pyrenaicus, S. carolitertii and nuclear non-hybrid S. alburnoides, were also analysed. Two to four forward and reverse sequences of each gene were obtained per individual/per organ, using independently synthesized cDNA samples and PCR amplifications. PCR amplifications for replicate reactions were also performed independently, following the procedures described in Pala et al. (2008).
(c). Quantitative real-time PCR analysis
Primers and specific TaqMan probes for β-actin, rpl8, gapdh (Pala et al. 2008) and ef1a (EFreal-F5′-CCGTCTGCCACTTCAGGATG-3′; EFreal-R5′-CATACCAGGCTT GAGGACACC-3′; EF/6VIC 5′-TCCACACGACCCACGGGCACAGT-3′) were used for amplification of muscle, liver and gonad samples of representative specimens of the CAA and CCA triploid (3–5) and CCAA tetraploid (1–2) forms from the Douro and Mondego River basins. Reactions and analysis were conducted as described in Pala et al. (2008).
3. Results
(a). Gene expression patterns differ according to organ and geographical location
The sequence analysis of genome-specific polymorphisms for the different genes in the various organs revealed contrasting results between southern and northern populations of S. alburnoides (table 1).
Table 1.
Relative frequency of allele-specific transcripts of P, C and A genomes in different organs of individuals from southern (Tejo) and northern (Mondego and Douro) populations of the complex.
|
Tejo (south) |
muscle |
liver |
gonad |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ploidy | species | genotype | P | A | PA | P | A | PA | P | A | PA | |
| 2n | S. pyrenaicus | PP | across all genes | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| S. alburnoides | AA | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | ||
| S. alburnoides | PA | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1a | ||
| 3n | S. alburnoides | PAA | β-actin | 0 | 0.67 | 0.33 | 0 | 1 | 0 | 0 | 1 | 0 |
| rpl8 | 0 | 0.67 | 0.33 | 0 | 0.75 | 0.25 | 0 | 1 | 0 | |||
| ef1a | 0 | 0.33 | 0.67 | 0 | 0.83 | 0.17 | 0 | 1 | 0 | |||
| gapdh | 0 | 0 | 1 | 0 | 0.64 | 0.36 | 0 | 1 | 0 | |||
| amh | — | — | — | — | — | — | 0 | 0 | 1 | |||
| dmrt1 | — | — | — | — | — | — | 0 | 0.75 | 0.25 | |||
| Mondego/Douro (north) | ||||||||||||
| ploidy | species | genotype | C | A | CA | C | A | CA | C | A | CA | |
| 2n | S. carolitertii | CC | across all genes | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| S. alburnoides | CA | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1a | ||
| 3n | S. alburnoides | CAA | β-actin | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 |
| rpl8 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | |||
| ef1a | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | |||
| gapdh | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | |||
| amh | — | — | — | — | — | — | 0 | 0 | 1 | |||
| dmrt1 | — | — | — | — | — | — | 0 | 1 | 0 | |||
| 3n | S. alburnoides | CCA | across all genes | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 |
| 4n | S. alburnoides | CCAA | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | |
| C-A- | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | |||
aOnly amh, dmrt1 and β-actin were analysed.
In the sample from the southern population (Tejo Basin), the four housekeeping genes (β-actin, rpl8, ef1a and gapdh) showed differential expression patterns according to organ. In muscle and liver, diploid PA individuals showed the expected simultaneous expression of A and P alleles (referred from now on as biallelic expression). Triploids, on the other hand, exhibited a variety of expression profiles: for β-actin and rpl8 most muscle and liver samples exhibited expression exclusively from the A genome, although biallelic expression was also observed. For ef1a, two triploid samples in muscle and one in liver showed biallelic expression while in the remaining one only A genome transcripts were identified. For gapdh, biallelic expression of P and A alleles was identified in all triploid muscle samples, while in liver samples all genes exhibited a predominantly monoallelic expression of A genome transcripts, with a small number of individuals exhibiting biallelic expression. The most paradigmatic example of this was the β-actin gene, of which transcripts in triploids corresponded exclusively to the A genome.
In individuals from northern populations (Mondego and Douro Basins), in liver and muscle samples, biallelic expression was observed with no exceptions for all gene/organ combinations irrespective of ploidy level and genomic constitution, including CA, CCA, CAA and CCAA S. alburnoides. The same was true for β-actin and rpl8 transcripts in eye samples of triploid (CCA and CAA) and tetraploid individuals: both C and A alleles were expressed (figure S2, electronic supplementary material). A tendency towards a preferential biallelic expression was also observed in eye samples of southern triploid (PAA) specimens: four out of seven samples showed expression of P and A alleles of the β-actin gene, and rhodopsin transcripts resulted from biallelic expression in all samples analysed.
Brain samples from triploid and tetraploid individuals from Mondego and Douro showed an exception to the overall tendency of biallelic expression of C and A genomes in these basins: although C and A β-actin expression was observed in all samples, the gapdh gene showed monoallelic A-genome-exclusive expression in CAA individuals (figure S2, electronic supplementary material).
In adult gonads (table 1) of southern triploid individuals, all four housekeeping genes (β-actin, rpl8, ef1a and gapdh) showed exclusive expression of A genome alleles. The amh gene showed biallelic expression in triploid PAA, while dmrt1 exhibited both monoallelic A expression (nine samples) and biallelic (three samples) genome expression. Diploid PA exhibited a biallelic expression pattern in all genes analysed (amh, dmrt1 and β-actin). In northern samples, patterns were divergent. The amh gene was the only gene for which expression of both C and A alleles was observed in all diploid, triploid and tetraploid forms of S. alburnoides. The remaining five genes exhibited a distinctive expression profile between the two forms of triploids: while in CCA samples the observed expression pattern was biallelic (as in diploid and tetraploid samples from the same locations), in CAA individuals dmrt1, β-actin, rpl8, ef1a and gapdh expression was exclusively from the A genome. Exclusive expression of alleles from the minority genome was never observed in triploids.
(b). Quantitative expression
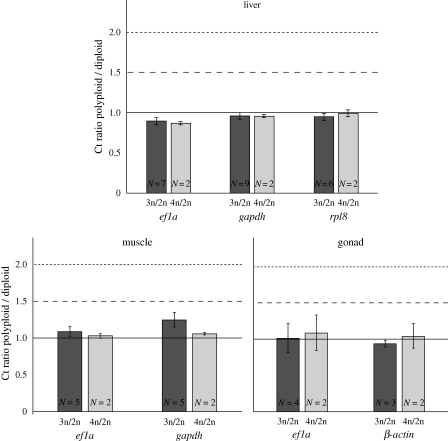
The relative expression ratios obtained from the comparison between average ct values of triploid and tetraploid samples and the diploid controls were always approximately 1 (figure 2), implying dosage compensation by regulation of gene expression to the diploid level in polyploids. Despite qualitative differences in allele representation, a similar effect of dosage compensation was observed in these populations compared with the one described in the southern Tejo basin (Pala et al. 2008). The method was further validated by an independent quantitative real-time PCR analysis comparing definite cell numbers (I. Matos & M. M. Coelho 2009, unpublished data).
Figure 2.
Relative expression (Ct) ratios between polyploid and diploid organ samples of individuals of the northern distribution of the complex, for the ef1a, gapdh, rpl8 and β-actin genes. Expected ratio of 1.5 in the case of no silencing 3n/2n (dashed line), expected ratio of 2, in the case of no silencing 4n/2n (dotted line) and expected ratio of 1 in the case of silencing (solid line). Observed 3n to 2n ratio (dark grey columns), observed 4n to 2n ratio (light grey columns), number of triploid and tetraploid triplicates analysed for each ratio calculation (N).
4. Discussion
In the present work, we have addressed the question of whether the mechanisms of gene silencing and regulation of transcripts to the diploid level, described in triploids of the southern populations of S. alburnoides (Pala et al. 2008), would also be present in other populations and in different genomic contexts. We have studied the expression patterns of housekeeping and tissue-specific genes in individuals of different genomic composition and ploidy degrees, from different locations within the distribution range of the S. alburnoides complex. Based on a representative sample of the forms that constitute the complex in different locations, we have attempted to understand the global effects of hybridization and ploidy rise on gene expression regulation in this system and evaluate their evolutionary implications.
The data obtained in the present work suggest a substantial difference in the genome-specific allele usage, depending on the geographical location of individuals. While a preferential expression of A genome and silencing of P genome alleles was observed in most triploids of southern populations of the complex, the vast majority of samples of the two northern river basins exhibited simultaneous expression of both C and A genome alleles, irrespective of ploidy level or genomic composition.
(a). Gene expression and dosage compensation
In southern populations of the Tejo River basin, the problem of keeping balance of expression regulatory networks in an uneven-numbered genome is apparently overcome by allele silencing (Pala et al. 2008), similar to what has been observed in plant species following polyploidization (Adams et al. 2003, 2004). In plants, it has been proposed that adapted polyploids would avoid extinction by reducing gene redundancy and thus undergoing an evolutionary pathway leading to functional diploidization (Paterson et al. 2004; Wang et al. 2005). The need for correct gene balance in a polyploid context as a way to tolerate the presence of extra genome copies and to maintain stable aneuploids has been further demonstrated in plants (Birchler & Veita 2007) and discussed also as a possibility in vertebrates (Mable 2007). Recently, dosage effects on transcription levels have been revealed in salmon (Ching et al. 2010), and similar levels of gene expression have been reported in triploid and diploid individuals. In the S. alburnoides complex, the necessity for balanced gene expression is apparent in both southern (Pala et al. 2008) and northern populations, with higher ploidy forms exhibiting gene expression regulated to the diploid levels. The similar quantitative outcome, apparently ensured in both geographical locations, is nevertheless accompanied by a distinct usage of specific alleles. In plant allopolyploids, further support for the possibility of different genome-specific allele usage producing similar transcript levels comes from the study of genome-wide expression dominance. The expression of one or the other genome was found to be dependent on the specific genomic combinations that were brought together in the hybrid, irrespective of the level of gene expression (Rapp et al. 2009).
Thus, the preferential monoallelic expression observed in the southern triploids of S. alburnoides, and the usage of both C and A allele copies in northern populations, are both in line with gene expression patterns observed in other polyploid taxa. Variable patterns of expression according to organ, silencing of genome specific alleles and non-additive gene expression have all been observed in plant polyploids (Adams et al. 2004; Auger et al. 2005; Paterson 2005; Salmon et al. 2005) and pointed out as consequences of the integration of divergent regulatory hierarchies.
Population differences in gene expression patterns of polyploids of the same species and ploidy level have also been reported in allopolyploid plants of the genus Tragopodon (Tate et al. 2006), and independent polyploidy events have been pointed out as a likely factor contributing to the observed variation. A similar effect apparently occurs in the S. alburnoides fish complex: patterns of gene expression diverge even within the same hybrid system, as shown by inequality of allele usage according to geographical origin and genome combination. Distinctive patterns of gene expression were also observed in gonads of the two triploid forms (CAA, CCA) in northern populations, contrasting with the overall identity in expression patterns in somatic organs. The patterns of gene expression in CAA gonads were similar to the ones observed in southern PAA samples. This may be related to the type of reproduction and mechanism of gamete production of the different forms, namely the exclusion of the minority genome (Carmona et al. 1997; Alves et al. 1998; Pala & Coelho 2005). Conversely, the overall biallelic expression observed in CCA females is not so easy to integrate. Owing to their low abundance in most populations, no extensive information about the reproductive modes of CCA females has been gathered, and the present results might indicate that they do not follow the same process of hybridogenesis as other triploid females (Crespo-López et al. 2006).
(b). Hybridization events and genome usage
The variation in allele expression dynamics observed here could be related to the difference in the origin and the timing of constitution of the three lineages of the complex (ranging from less than 0.7 to 0.01 Myr, according to Sousa-Santos et al. 2007). Regulation of gene expression can be variable in ancient and newly formed polyploids (Wang et al. 2004; Adams & Wendel 2005), and the disparity in the timing of the onset of hybridization events could be a factor to be taken into account.
An alternative possibility could be the difference in the composition of the genomes involved in the maintenance of the complex in each of the three basins. The C genome, introduced through the bisexual species S. carolitertii in northern populations, was a later addition to the complex, after initial hybridization events involving only S. pyrenaicus (Alves et al. 1997; Cunha et al. 2004). Genomic stress induced by two distinct genomes being brought together in the same cell nucleus often leads to different epigenetic modifications of homeologous (Riddle & Birchler 2003) and differential capacity of regulatory interactions (Comai 2000; Adams & Wendel 2004; Rapp et al. 2009). Both the P and C genome apparently have a good functional ‘affinity’ with the A genome, but it is possible that regulatory elements that have to interact with both heterologous genomes in hybrids respond differentially or have distinct interaction capacity.
Another important difference between the northern and southern populations of the complex, which has been pointed out as a key factor in the maintenance of its diversity and contributing actively to the evolutionary success of S. alburnoides, is the presence of nuclear non-hybrid males of AA genotype (Alves et al. 2002; Crespo-López et al. 2006, 2007). The preferential expression of A genome alleles in southern populations could be related to the establishment of this nuclear ‘non-hybrid’ lineage. A different preference for expressed alleles might have been established in northern populations in which the process of A allele recombination and shifting, mediated by the nuclear non-hybrid form, is not present.
The heterogeneity of the genomes brought together in plant allopolyploids has been proposed as a major factor underlying patterns of non-additive gene expression, as suggested by studies in different ploidy levels of cotton (Flagel et al. 2008) and Senecio (Hegarty et al. 2006). In these species, the effects of genome merger apparently supplant the impact of genome doubling. The differential patterns of gene expression according to genomic composition, reported here for the S. alburnoides complex, also point towards a strong influence of the type of genomes involved in the hybridization events occurring in each geographical location. However, this might not be the sole explanation for the observed patterns. Dosage constrictions and ploidy level might be additional key players in the regulation of gene expression in this system, as indicated by the similar behaviour of diploids of different geographical origins in terms of qualitative and quantitative gene expression, and the switch to differential patterns of genome specific allele usage in triploids of the northern and southern distribution of S. alburnoides.
The elucidation of the mechanism underlying dosage compensation and preferential allele usage, as well as the characterization of the factors that modulate differential gene expression, will hopefully contribute to a better understanding of how vertebrate polyploid genomes are regulated and how the different adopted strategies can influence their odds on the race for adaptability and evolutionary success.
Acknowledgements
The authors thank Carina Cunha for providing samples of the Douro River basin, Judite Alves for providing the means for RNA sample quantification and Maria Ana Aboim for help in the field.
This work was supported by the Project PTDC/BIA-BDE/65586/2006 to M.M.C. and by a grant (SFRH/BD/19286/2004) to I.P., both from the Fundação para a Ciência e a Tecnologia (FCT; part of the Ministry of Science of Portugal); M.S. is supported by the Deutsche Forschungsgemeinschaft (SFB 567).
References
- Adams K. L., Wendel J. F.2004Exploring the genomic mysteries of polyploidy in cotton. Biol. J. Linn. Soc 82, 573–582 (doi:10.1111/j.1095-8312.2004.00342.x) [Google Scholar]
- Adams K. L., Wendel J. F.2005Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8, 135–141 (doi:10.1016/j.pbi.2005.01.001) [DOI] [PubMed] [Google Scholar]
- Adams K. L., Cronn R., Percifield R., Wendel J. F.2003Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl Acad Sci. USA 100, 4649–4654 (doi:10.1073/pnas.0630618100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K. L., Percifield R., Wendel J. F.2004Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168, 2217–2226 (doi:10.1534/genetics.104.033522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alleman M., Doctor J.2000Genomic imprinting in plants: observations and evolutionary implications. Plant Mol. Biol. 43, 147–161 (doi:10.1023/A:1006419025155) [DOI] [PubMed] [Google Scholar]
- Alves M. J., Coelho M. M., Collares-Pereira M. J., Dowling T. E.1997Maternal ancestry of the Rutilus alburnoides complex (Teleostei, Cyprinidae) as determined by analysis of cytochrome b sequences. Evolution 51, 1584–1592 [DOI] [PubMed] [Google Scholar]
- Alves M. J., Coelho M. M., Collares-Pereira M. J.1998Diversity in the reproductive modes of females of the Rutilus alburnoides complex (Teleostei, Cyprinidae): a way to avoid the genetic constraints of uniparentalism. Mol. Biol. Evol. 15, 1233–1242 [Google Scholar]
- Alves M. J., Coelho M. M., Próspero M. I., Collares-Pereira M. J.1999Production of fertile unreduced sperm by hybrid males of the Rutilus alburnoides complex (Teleostei, Cyprinidae): an alternative route to genome tetraploidization in unisexuals. Genetics 151, 277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves M. J., Coelho M. M., Collares-Pereira M. J.2001Evolution in action through hybridisation and polyploidy in an Iberian freshwater fish: a genetic review. Genetica 111, 375–385 (doi:10.1023/A:1013783029921) [DOI] [PubMed] [Google Scholar]
- Alves M. J., Collares-Pereira M. J., Dowling T. E., Coelho M. M.2002The genetics of maintenance of an all-male lineage in the Squalius alburnoides complex. J. Fish Biol. 60, 649–662 (doi:10.1111/j.1095-8649.2002.tb01691.x) [Google Scholar]
- Auger D. L., Gray A. D., Ream T. S., Kato A., Coe E. H., Birchler J. A.2005Nonadditive gene expression in diploid and triploid hybrids of maize. Genetics 169, 389–397 (doi:10.1534/genetics.104.032987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., Veitia R. A.2007The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19, 395–402 (doi:10.1105/tpc.106.049338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona J. A., Sanjur O. I., Doadrio I., Machordom A., Vrijenhoek R. C.1997Hybridogenetic reproduction and maternal ancestry of polyploid Iberian fish: the Tropidophoxinellus alburnoides complex. Genetics 146, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching B., Jamieson S., Heath J. W., Heath D. D., Hubberstey A.2010Transcriptional differences between triploid and diploid Chinook salmon (Oncorhynchus tshawytscha) during live Vibrio anguillarum challenge. Heredity 104, 224–234 (doi:10.1038/hdy.2009) [DOI] [PubMed] [Google Scholar]
- Comai L.2000Genetic and epigenetic interactions in allopolyploid plants. Plant Mol. Biol. 43, 387–399 (doi:10.1023/A:1006480722854) [DOI] [PubMed] [Google Scholar]
- Comai L.2005The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846 (doi:10.1038/nrg1711) [DOI] [PubMed] [Google Scholar]
- Comai L., Tyagi A. P., Winter K., Holmes-Davis R., Reynolds S. H., Stevens Y., Byers B.2000Phenotypic instability and rapid gene silencing in newly formed Arabidopsis allotetraploids. Plant Cell 12, 1551–1568 (doi:10.1105/tpc.12.9.1551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo-López M. E., Duarte T., Dowling T., Coelho M. M.2006Modes of reproduction of the hybridogenetic fish Squalius alburnoides in the Tejo and Guadiana rivers: an approach with microsatellites. Zoology (Jena) 109, 277–286 (doi:10.1016/j.zool.2006.03.008) [DOI] [PubMed] [Google Scholar]
- Crespo-López M. E., Pala I., Duarte T., Dowling T. E., Coelho M. M.2007Genetic structure of the diploid–polyploid fish Squalius alburnoides in southern Iberian basins Tejo and Guadiana, based on microsatellites. J. Fish Biol. 71, 423–436 (doi:10.1111/j.1095-8649.2007.01688.x) [Google Scholar]
- Cunha C., Coelho M. M., Carmona J. A., Doadrio I.2004Phylogeographical insights into the origins of the Squalius alburnoides complex via multiple hybridisation events. Mol. Ecol. 13, 2807–2817 (doi:10.1111/j.1365-294X.2004.02283.x) [DOI] [PubMed] [Google Scholar]
- Cunha C., Doadrio I., Coelho M. M.2008Speciation towards tetraploidization after intermediate processes of non-sexual reproduction. Phil. Trans R. Soc. B 363, 2921–2929 (doi:10.1098/rstb.2008.0048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawley R. M., Goddard K. A.1988Diploid–triploid mosaics among unisexual hybrids of the minnow Phoxinus neogaeus. Evolution 42, 649–659 [DOI] [PubMed] [Google Scholar]
- Dowling T. E., Secor C. L.1997The role of hybridisation and introgression in the diversification of animals. Annu. Rev. Ecol. Syst. 28, 593–620 (doi:10.1146/annurev.ecolsys.28.1.593) [Google Scholar]
- Flagel L., Udall J., Nettleton D., Wendel J.2008Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 6, 16 (doi:10.1186/1741-7007-6-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Davis D., Birchler J. A.1996Dosage effects on gene expression in a maize ploidy series. Genetics 142, 1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty M. J., Barker G. L., Wilson I. D., Abbott R. J., Edwards K. J., Hiscock S. J.2006Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr. Biol. 16, 1652–1659 (doi:10.1016/j.cub.2006.06.071) [DOI] [PubMed] [Google Scholar]
- Hotz H., Semlitsch R. D., Gutmann E., Guex G. D., Beerli P.1999Spontaneous heterosis in larval life-history traits of hemiclonal frog hybrids. Proc. Natl Acad. Sci. USA 96, 2171–2176 (doi:10.1073/pnas.96.5.2171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamatsch D. K., Stöck M.2009Sperm-dependent parthenogenesis and hybridogenesis in teleost fishes. In Lost sex! The evolutionary biology of parthenogenesis (eds Schön I., Martens K., Van Dijk P.), pp. 399–432 Berlin, Germany: Springer [Google Scholar]
- Lampert K. P., Schartl M.2008The origin and evolution of a unisexual hybrid: Poecilia formosa. Phil. Trans. R. Soc. B 363, 2901–2909 (doi:10.1098/rstb.2008.0040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable B. K.2007Sex in the postgenomic era. Trends Ecol. Evol. 22, 559–561 (doi:10.1016/j.tree.2007.07.006) [DOI] [PubMed] [Google Scholar]
- Otto S. P., Whitton J.2000Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437 (doi:10.1146/annurev.genet.34.1.401) [DOI] [PubMed] [Google Scholar]
- Pala I., Coelho M. M.2005Contrasting views over a hybrid complex: between speciation and evolutionary ‘dead-end’. Gene 347, 283–294 (doi:10.1016/j.gene.2004.12.010) [DOI] [PubMed] [Google Scholar]
- Pala I., Coelho M. M., Schartl M.2008Dosage compensation by gene copy silencing in a triploid hybrid fish. Curr. Biol. 18, 1344–1348 (doi:10.1016/j.cub.2008.07.096) [DOI] [PubMed] [Google Scholar]
- Paterson A. H.2005Polyploidy, evolutionary opportunity, and crop adaptation. Genetica 123, 191–196 (doi:10.1007/s10709-003-2742-0) [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Chapman B. A.2004Ancient polyploidisation predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA 101, 9903–9908 (doi:10.1073/pnas.0307901101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatta D., Comai L.2009Parental squabbles and genome expression: lessons from the polyploids. J. Biol. 8, 43 (doi:10.1186/jbiol140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp R. A., Udall J. A., Wendel J. F.2009Genomic expression dominance in allopolyploids. BMC Biol. 7, 18 (doi:10.1186/1741-7007-7-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle N. C., Birchler J. A.2003Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet. 19, 597–600 (doi:10.1016/j.tig.2003.09.005) [DOI] [PubMed] [Google Scholar]
- Salmon A., Ainouche M. L., Wendel J. F.2005Genetic and epigenetic consequences of recent hybridisation and polyploidy in Spartina (Poaceae). Mol. Ecol. 14, 1163–1175 (doi:10.1111/j.1365-294X.2005.02488.x) [DOI] [PubMed] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A. A.2001Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridisation and allopolyploidy in wheat. Plant Cell 13, 1749–1759 (doi:10.1105/tpc.13.8.1749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Santos C., Collares-Pereira M. J., Almada V.2007Reading the history of a hybrid fish complex from its molecular record. Mol. Phylogenet. Evol. 45, 981–996 (doi:10.1016/j.ympev.2007.05.011) [DOI] [PubMed] [Google Scholar]
- Stöck M., et al. 2002A bisexually reproducing all-triploid vertebrate. Nat. Genet. 30, 325–328 (doi:10.1038/ng839) [DOI] [PubMed] [Google Scholar]
- Tate J. A., Ni Z., Scheen A. C., Koh J., Gilbert C. A., Lefkowitz D., Chen Z. J., Soltis P. S., Soltis D. E.2006Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 173, 1599–1611 (doi:10.1534/genetics.106.057646) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., et al. 2004Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167, 1961–1973 (doi:10.1534/genetics.104.027896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Shi X., Hao B., Ge S., Luo J.2005Duplication and DNA segmental loss in the rice genome: implications for diploidisation. New Phytol. 165, 937–946 (doi:10.1111/j.1469-8137.2004.01293.x) [DOI] [PubMed] [Google Scholar]