Abstract
When experiencing resource competition or abrupt environmental change, animals often must transition rapidly from an ancestral diet to a novel, derived diet. Yet, little is known about the proximate mechanisms that mediate such rapid evolutionary transitions. Here, we investigated the role of diet-induced, cryptic genetic variation in facilitating the evolution of novel resource-use traits that are associated with a new feeding strategy—carnivory—in tadpoles of spadefoot toads (genus Spea). We specifically asked whether such variation in trophic morphology and fitness is present in Scaphiopus couchii, a species that serves as a proxy for ancestral Spea. We also asked whether corticosterone, a vertebrate hormone produced in response to environmental signals, mediates the expression of this variation. Specifically, we compared broad-sense heritabilities of tadpoles fed different diets or treated with exogenous corticosterone, and found that novel diets can expose cryptic genetic variation to selection, and that diet-induced hormones may play a role in revealing this variation. Our results therefore suggest that cryptic genetic variation may have enabled the evolutionary transition to carnivory in Spea tadpoles, and that such variation might generally facilitate rapid evolutionary transitions to novel diets.
Keywords: phenotypic plasticity, genetic accommodation, corticosterone, novelty, anuran larvae, heritability
1. Introduction
Understanding the origins of novel, complex traits is a central goal of evolutionary biology (West-Eberhard 2003). Of special interest are novel traits associated with resource use, which may be particularly important because many of life's most spectacular bouts of diversification are characterized by the appearance of new resource-use traits (Schluter 2000). Indeed, studies of invasive species suggest that transitions in resource use may occur rapidly (Carroll et al. 1998; Phillips & Shine 2004). Yet, the proximate mechanisms that generate such new traits are poorly understood.
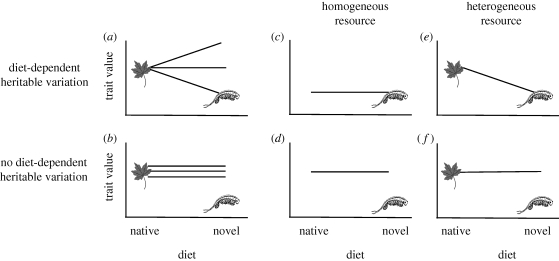
Here, we explore the proximate mechanisms that facilitate the evolution of resource-use traits by considering the role of ‘cryptic’ genetic variation (sensu Gibson & Dworkin 2004; Schlichting 2008) in fuelling evolutionary shifts in resource use. Cryptic genetic variation is genetic variation that is revealed phenotypically under certain circumstances, such as when a population is exposed to a novel diet. Although such variation is not inherently adaptive, it might, by chance, be adaptive for consuming the novel resource. If so, it can mediate phenotypic shifts through genetic accommodation (sensu West-Eberhard 2003), in which selection modifies the expression and functionality of a trait that was initially environmentally induced. This process, acting on cryptic genetic variation, could provide a general explanation for how populations adapt to novel diets (figure 1).
Figure 1.
Adaptive trait evolution in the presence or absence of cryptic genetic variation. In these diagrams, the optimal trait values for native and novel diets are indicated by a leaf and shrimp, respectively. When a population encounters a novel diet, it can either express hidden, diet-dependent heritable variation in a given trait (a) or not (b). If it does, selection can refine this phenotypically expressed genetic variation into an optimal canalized trait (c) if only the novel diet is experienced, or a plastic trait if both diets are experienced (e), a process referred to as ‘genetic accommodation’. In the absence of diet-dependent genetic variation, and the absence of novel mutation, the population cannot evolve towards the new trait optimum (d,f), and may perish if this phenotype–environment mismatch results in low fitness.
Adaptive evolution produced by genetic accommodation has the potential to occur more rapidly than that initiated by novel genetic mutations (West-Eberhard 2003). Given that the rate of adaptive evolution is determined by the frequency of beneficial mutations in a population (Fisher 1930), selection can immediately act on cryptic genetic variation, whereas the generation of novel genetic variation is limited by the mutational rate and population size. In addition, novel mutations initially occur in only one individual, whereas cryptic genetic variation may be exposed in several experiencing the same environmental conditions, and may therefore exist at a relatively high initial frequency. Indeed, these are the same reasons that visible standing genetic variation produces adaptive change more rapidly than novel mutations (Barrett & Schluter 2008). However, visible genetic variation is constantly curtailed by selection, whereas cryptic genetic variation is buffered, allowing it to accumulate. Although cryptic genetic variation in traits associated with the ability to consume and assimilate novel food items could promote rapid dietary transitions, few empirical studies have evaluated the plausibility of this model for the evolution of novel resource-use traits (Ghalambor et al. 2007; Moczek 2007).
To test this model empirically, the expression of cryptic genetic variation must be measured in populations experiencing novel conditions. These measures should be made in a phylogenetic context to determine whether a given trait arose through pre-existing genotypic variation as opposed to novel mutation (Hall 2001). Unfortunately, appropriate substitutes for ancestral populations of taxa with derived feeding strategies are rarely available (but see Parsons & Robinson 2006; Wund et al. 2008). An alternative approach is to study a species that is closely related to the species with the derived feeding strategy, but which has retained the ancestral feeding strategy (Aubret et al. 2007). Finding cryptic genetic variation present in such a species would indicate that cryptic genetic variation might have been present in the ancestral population and may therefore have facilitated the evolution of novel resource-use traits.
We undertook this approach with spadefoot toads. As we describe in detail below, some species of spadefoot toads have evolved a novel larval feeding strategy: in addition to the ancestral larval diet of plants and detritus, they also feed on live animal prey (Bragg 1965). We investigated the mechanisms underlying this dietary transition by measuring cryptic genetic variation in resource-use traits in a species that is ancestral (with respect to feeding strategy) to the clade that has evolved carnivory. This species, therefore, served as a model for the ancestral lineage that first encountered and consumed the novel resource.
In addition to determining whether the ancestral lineage probably harboured cryptic genetic variation that enabled the transition to carnivory, we also sought to investigate the physiological mechanisms that might have revealed this variation to selection. We specifically investigated the role of a single hormone, corticosterone. Hormones such as corticosterone are good candidates for mediating the expression of cryptic genetic variation for at least three reasons. First, hormones are a vital link between environmental change and an organism's morphological and physiological response (Pigliucci 2001; Dufty et al. 2002). Second, hormonally regulated loci are known to possess selectable variation within populations (Suzuki & Nijhout 2006). Third, because many hormones are produced in response to environmental change, they could potentially unmask cryptic genetic variation when it is most needed. Given the concurrent release of cryptic genetic variation and hormones in response to novel environments, it seems likely that hidden variation might be exposed as a result of environmentally induced hormones. Indeed, the results of our analyses suggest that cryptic genetic variation, mediated by an environmentally responsive hormone, may play a substantial role in the rapid evolution of novel feeding strategies.
2. Material and methods
(a). Study system and specific predictions
We used Couch's spadefoot, Scaphiopus couchii, as a model for the ancestral phenotype of the facultatively carnivorous genus Spea. Larvae of most anuran species, including Sc. couchii, feed exclusively on minute organic material (Altig et al. 2007; Ledón-Rettig et al. 2008). In contrast, Spea larvae that develop in the presence of anostracan fairy shrimp often depart from this typical diet and begin to specialize on large animal prey: shrimp and other tadpoles (Pfennig & Murphy 2000, 2002). This diet-induced plasticity is accompanied by several derived traits. In contrast to the long, coiled gut of most anuran larvae, Spea that specialize on invertebrates and tadpoles develop a short, relatively uncoiled gut, which is adaptive for assimilating large particles of a high-quality diet (Barton & Houston 1993; Hume 2005). Carnivorous Spea also develop disproportionately large jaw muscles that help them capture and ingest large prey (Martin & Pfennig 2009). Lastly, unlike Sc. couchii, members of Spea can thrive on and extract nutrients from a shrimp diet, as measured by growth and gut cellular proliferation (Ledón-Rettig et al. 2008, 2009).
Spea very likely evolved from an ancestral population that was similar to Sc. couchii with respect to resource acquisition, as supported by an ancestral character state reconstruction (Ledón-Rettig et al. 2008). The reconstruction was developed using a maximum likelihood model in Mesquite (Maddison & Maddison 2007) and favoured omnivory as the best character state for the ancestral node that gave rise to Scaphiopus (the omnivorous clade) and Spea (the facultatively carnivorous clade). To determine whether cryptic genetic variation might have enabled a dietary transition in spadefoot toad tadpoles, we measured genetic variation in morphological traits (gut length and jaw muscle width) and fitness (growth and developmental speed) when Sc. couchii tadpoles were raised on a surrogate for their native diet of detritus and Spea's derived diet of shrimp.
Further, we hypothesized that cryptic variation can be unmasked by diet-induced changes in glucocorticoid levels. Glucocorticoids cause simultaneous morphological and physiological adjustments in vertebrates (Sapolsky et al. 2000; McEwen & Wingfield 2003) including amphibian tadpoles (Denver 2009). Specifically, corticosterone (CORT, the major glucocorticoid in amphibians) is produced in Sc. couchii tadpoles upon exposure to a novel, carnivorous diet (Ledón-Rettig et al. 2009), although it is unclear whether they experience this increase due to novelty per se, or nutrient restriction resulting from difficulty in assimilating shrimp. Regardless of the mechanism causing the elevated CORT, its production may allow for the unmasking of cryptic variation. Thus, to assess the contribution of diet-induced hormones in the exposure of cryptic variation, we included a CORT treatment in this study (e.g. Hu et al. 2008).
If cryptic genetic variation within a natural population of Sc. couchii is mediated by shrimp-induced elevations in CORT levels, then the effects of shrimp and CORT (regardless of diet) on the degree of variability or direction of expressed variation should be similar. We anticipated that a large proportion of phenotypic variation exposed by dietary or hormonal manipulation would be explained by underlying genetic variation. This would suggest that there is heritable cryptic variation within Sc. couchii and, if similar heritable variation was present in ancestral Spea populations, this variation might have facilitated the evolutionary transition to a shrimp-based diet (figure 1).
(b). Experimental design
In order to detect cryptic genetic variation, we measured heritability in traits associated with both resource use and fitness. We specifically estimated broad-sense heritability using a full sibship analysis. A full sibship analysis is sensitive to three types of variation that can potentially inflate heritability estimates: dominance, maternal and environmental. Environmental effects can be estimated from the phenotypic variation remaining after the effects of treatment (CORT and diet) and family are removed, thus we were able to correct heritabilities for environmental variation. Further, because the treatments and families were randomized and interspersed in the same room, it is unlikely that environmental variation varied systematically among treatments or families (i.e. if heritabilities were inflated by environmental effects, they were inflated equally in all treatments and diets, Hurlbert 1984; Roff 1997). Finally, it might be contended that our estimates of heritabilities were moderately inflated in the laboratory relative to what they would have been in the wild, owing to an underestimate of total phenotypic variation. Even if this were the case, our results and conclusions would not qualitatively change. This is because we are mainly interested in relative levels of heritabilities among diet and hormone treatments (in fact, estimates of trait variance in field-caught and laboratory-reared tadpoles are similar; see electronic supplementary material, appendix S1). Only one condition-dependent maternal effect is known in spadefoot toads, and this effect vanishes after mothers have been equilibrated in the laboratory for 1 year (Pfennig & Martin 2009). The adults bred in this experiment had been housed under similar conditions for 2–3 years, and we found no evidence that maternal condition influenced their offspring's phenotype (electronic supplementary material, appendix S2). Although we were unable to partition out possible environmental or genetic maternal effects (i.e. genetic variation among mothers that can influence their offspring's phenotype) that are uncorrelated with maternal condition (e.g. cytoplasmic factors), or the effect of genetic dominance, the comparisons emphasized in this study are differences in heritabilities between environments, not a derivation of heritability that is accurate enough to predict future trait values in response to selection (Uller et al. 2002). Thus, a full sibship design is probably adequate for our purposes. Nine families of Sc. couchii were used in our analyses. All animals were collected near Portal, Arizona, and had been housed in a colony at the University of North Carolina, Chapel Hill.
To induce breeding, male and female adults were injected with 0.07 ml human luteinizing hormone-releasing hormone (Sigma L-7134) and left for 8 h in nursery tanks. After hatching (3 days after breeding), larvae from each sibship were distributed among 12 × 16 × 8 cm3 plastic boxes filled with 800 ml dechlorinated tap water. For the fitness and morphology aspect of this study, each sibship was subject to a diet or hormone treatment, and each treatment was represented by nine replicate boxes of five tadpoles. Three additional replicate boxes of each family × hormone/diet treatment, each containing five tadpoles, were included for the purpose of determining how family and experimental treatment influenced endogenous CORT levels. All replicates were randomized and interspersed on racks in the same room maintained at 26°C and on a 14 L : 10 D light cycle. Four days after breeding, larvae were fed brine shrimp nauplii, ground fish food (hereafter, detritus) or detritus plus exogenous CORT. Brine shrimp resemble the fairy shrimp that Spea (the genus with the carnivorous feeding strategy) feed on in nature, whereas ground fish food resembles detritus (the native diet of Sc. couchii) in form and nutrition. An amount of 200 µl of 1 mM exogenous CORT (Sigma C2505) was added to the appropriate replicates for a final concentration of 250 nM CORT. This concentration has physiological effects on growth, and mifepristone (a glucocorticoid receptor antagonist) reverses these effects, suggesting that exogenous and endogenous CORT have comparable physiological actions (Ledón-Rettig et al. 2009). Tadpoles in the shrimp treatment were switched from nauplii to adult shrimp 4 days after hatching, simulating the natural development of this resource. On the same day, the exogenous CORT was replenished. Eight days after hatching, tadpoles were sampled for fitness and morphology, or for CORT analysis. Tadpoles sampled for fitness and morphology were euthanized with tricaine methosulphonate (MS 222) and fixed in formalin. Tadpoles sampled for CORT analysis were individually transferred to 12 × 75 mm Falcon tubes, rapidly frozen by immersion in an ethanol and dry ice slurry and transferred to a −80°C freezer.
The developmental stage of tadpoles was determined using Gosner staging (Gosner 1960). Remaining traits of interest were measured with NIH Image J (Rasband 1997–2006) on photographs captured with a Leica (Wetzlar, Germany) DFC480 R2 Camera. Separate images were taken for snout–vent length (SVL) (ventral, magnification ×1.25), (dissected and uncoiled ×0.71) and orbitohyoideous muscle (OH) (lateral ×3.2).
(c). CORT extraction and radioimmunoassay
Weight was recorded for tadpoles before they were transferred to glass tubes for radioimmunoassay (RIA). CORT was extracted from whole tadpoles following the method of Denver (1998) with modifications. Briefly, total lipids were extracted by homogenizing each tadpole in 2 ml ethyl acetate. Samples were centrifuged and the supernatant was reduced by rapid evaporation. All samples were spiked with 3.5 K cpm tritiated CORT ([3H]CORT, Perkin-Elmer) to allow for identification of CORT migration on silica thin-layer chromatography plates (JT Baker Si250F) exposed (twice) to a toluene : cyclohexane (1 : 1) solvent system, followed by a chloroform : methanol (9 : 1) system. We located the CORT migration distance by scraping silica in 1 cm increments from two extra samples on the ends of the plate and determining the radioactive peak using a Beckman LS6500 scintillation counter. We then scraped the region of silica containing CORT for each sample, extracted CORT from silica with 5 ml anhydrous ether and dried samples under nitrogen (repeated to improve efficiency). Each sample was resuspended in 0.5 ml 0.2 M phosphate-buffered saline with 1 per cent gelatin, and a 0.4 ml aliquot of this resuspension was used for RIA. In addition to our experimental samples, two aliquots from a single homogeneous rat plasma sample were subjected to the same extraction procedure and were included in each RIA run for the purpose of calculating the coefficients of variation (CVs) within and between assays. Intra- and inter-assay CVs were 8.7 and 15.4 per cent, respectively. The assay's range of detection was 30–4000 pg CORT per sample. We included samples from each diet and family on each TLC plate and in each RIA.
(d). Statistical analysis: main effects of diet and hormone regime
The effects of diet and exogenous CORT on fitness, morphology and endogenous CORT were determined using linear mixed models with package lme4 of the R Statistical Language, which is designed to model hierarchical, random variation (Baayeen et al. 2008). Model parameters were estimated using restricted maximum likelihood (REML). Response variables included SVL, Gosner stage (GS), gut length, OH width and endogenous CORT (ng g−1 body weight). All parameters except GS were natural log-transformed to reduce skewedness and increase normality. GS is an ordinal variable (discrete with a natural order), and may behave inappropriately in linear models. However, estimates of fixed and random effects for GS obtained using a Bayesian MCMC generalized linear mixed model approach (package MCMCglmm in R; Hadfield 2009), which can accommodate ordinal response variables, were nearly identical to those obtained when GS was treated as a continuous variable in a linear mixed model. Thus, only REML estimates are reported here.
For morphological and fitness traits, the effect of treatment (detritus, shrimp and detritus plus exogenous CORT) on each response variable was determined with models where the fixed effect was treatment and the random effects were family, the family by treatment interaction and replicate. In models where the response variable was a morphological trait (gut length and OH), size (SVL) was included as a fixed covariate. In a similar model with endogenous CORT as the response variable, the fixed effect was treatment and the random effects were family, the family by treatment interaction and assay. To determine treatment effects in each model, we used a Tukey's test from the R package multcomp (Hothorn et al. 2008), which is designed to execute multiple comparisons in linear mixed models.
(e). Statistical analysis: cryptic genetic variation
We employed two methods of quantifying environment-specific genetic variation. First, we measured each trait's environment specific heritability, which is the proportion of phenotypic variation owing to underlying genetic variation (Falconer & Mackay 1996). Genetic variances of fitness and morphological traits within treatments were calculated with linear mixed models including family and replicate as random effects. Genetic variances of endogenous CORT were calculated with models including family and assay as random effects. For each within treatment model, only data pertaining to the respective treatment were used. In models where the response variable was a morphological trait (gut length and OH), size (SVL) was included as a fixed covariate. All parameters except GS were natural log-transformed to reduce skewedness and increase normality. To estimate heritabilities, we followed the protocols of Roff (1997) for a full sibship design,
where H2 is broad-sense heritability, VAF is the variance among families, VAR is the variance among replicates and VWR is the variance within replicates. Significant differences among heritabilities were determined using 500 replicates of non-parametric bootstrapping on the entire statistic. For comparing within-treatment heritabilities, only families represented in both treatments were used (comparisons between detritus- and shrimp-fed tadpoles involved nine families, whereas comparisons between detritus-fed and CORT-exposed tadpoles involved seven families because two families responded to the CORT treatment with excessive mortality).
Second, for accurate comparisons of genetic variation among treatments, and for comparisons with other datasets, we calculated CVs. The CV is a useful measure of variation when two groups differ in their means because, in many cases, the variation of a population measure will increase with its mean (Sokal & Rohlf 1995). Notably, it is independent of environmental variance that might influence heritability estimates (Houle 1992; Charmantier & Garant 2005). Typically, coefficients of additive genetic variation are typically measured. However, because we used a full sibship design, where additive, dominant and maternal effects are indistinguishable, we will refer to our metric as the coefficient of family variation, CVAF.
The genetic variation, VAF, was determined using the same mixed models as for heritability, but using untransformed data (using trait values that can potentially produce negative means results in CVs that are meaningless; Houle 1992). Confidence intervals for estimates of CVAF were created using non-parametric bootstrapping with 500 replicates.
3. Results
(a). Main effects of diet and hormone regime
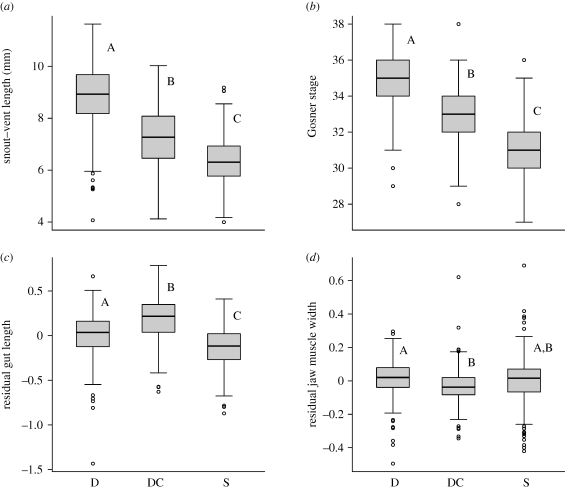
Overall, tadpoles fed shrimp or exposed to exogenous CORT grew significantly less and developed significantly slower than tadpoles fed detritus (for all comparisons, Tukey's test, p < 0.0001, electronic supplementary material, appendix S3 and figure 2). Tadpoles fed shrimp had significantly shorter guts than tadpoles fed detritus after correcting for overall size (p < 0.0001). In contrast, tadpoles exposed to CORT exhibited relatively long guts (p = 0.019). Compared with detritus-fed tadpoles, shrimp-fed and CORT-exposed tadpoles developed relatively small jaw muscles, although this difference was only significant for tadpoles treated with CORT (p = 0.0008). Tadpoles fed shrimp or exposed to CORT exhibited significantly higher levels of endogenous CORT than those fed detritus (for both, p < 0.0001), but were not significantly different from each other (p = 0.42). Therefore, levels of endogenous CORT induced by the exogenous CORT treatment were within a natural, physiological range. Further, the range of endogenous larval CORT produced over all treatments was similar to the range produced by larvae of another spadefoot toad species (Spea hammondii) in an independent study (Denver 1998), suggesting that no treatment used here induced pharmacological levels of CORT.
Figure 2.
The main effects of treatment (D is the native diet, detritus; DC is detritus plus exogenous corticosterone; S is the novel diet, shrimp) on SVL (a), developmental stage (b), gut length (c) and jaw muscle width (d). For the purpose of displaying size-corrected traits, the residuals of natural log-transformed gut length and jaw muscle width on natural log-transformed SVL are shown. Significance levels (indicated with letters) were determined with linear mixed models on the original data with natural log-transformed SVL as a covariate, and natural log-transformed response variables. Bars within boxes indicate median values, boxes indicate interquartile ranges and circles indicate outliers.
(b). Cryptic genetic variation: heritabilities and coefficients of genetic variation
Consistent with our expectations that a novel diet could expose cryptic genetic variation, shrimp-fed Sc. couchii tadpoles possessed greater broad-sense heritability in size, developmental stage and gut length (but not jaw muscle width) as compared with detritus-fed tadpoles (table 1 and figure 3). Likewise, tadpoles exposed to CORT exhibited greater heritability in gut length and developmental stage. However, the heritability of size was not significantly different between CORT-exposed and detritus-fed tadpoles, and tadpoles exposed to CORT in fact exhibited reduced heritability in jaw muscle width. The results produced by comparing coefficients of family variation were qualitatively identical to those produced by comparing heritability estimates (table 1).
Table 1.
Estimated family variance component (VAF) and total estimated variance (VTotal) for fitness, morphological and hormonal parameters. The 95% confidence intervals for broad-sense heritabilities (H2) and coefficients of variation (CVAF, estimates of variation that are independent of sample means) were calculated using 500 bootstrapped replicates of the original data. If H2 or CVAF confidence intervals of alternate diets or hormonal environments were not overlapping for a particular trait, the H2 or CVAF of that trait was considered to be significantly diet or hormone dependent (D is the native diet, detritus; DC is detritus plus exogenous corticosterone; S is the novel diet, shrimp). SVL, snout–vent length; GS, Gosner stage; GL, gut length; OH, orbitohyoideous muscle width; CORT, endogenous CORT (ng g−1 body weight). All parameters except developmental stage were natural log transformed.
| trait | diet | VAF | VTotal | H2 | CI | H2 diet dependent? | CVAF | CI | CVAF diet dependent? |
|---|---|---|---|---|---|---|---|---|---|
| CORT | D | 0.1215 | 0.2580 | 0.12 | (−0.6020,0) | Y | 0 | (−24.63,0) | Y |
| S | 0.1215 | 0.2580 | 0.94 | (0.42,1.16) | 16.39 | (8.38,20.24) | |||
| SVL | D | 0.0005 | 0.0250 | 0.05 | (−0.12,0.09) | Y | 2.73 | (0.75, 5.47) | Y |
| S | 0.0054 | 0.0216 | 0.5 | (0.34,0.65) | 7.35 | (6.11, 8.60) | |||
| GS | D | 0 | 1.8876 | 0 | (−0.13,0) | Y | 0 | (−0.89, 0) | Y |
| S | 0.4856 | 2.8572 | 0.33 | (0.20,0.49) | 2.22 | (1.73,2.75) | |||
| GL | D | 0.0028 | 0.0456 | 0.12 | (−0.04,0.25) | Y | 6.70 | (4.61, 8.75) | Y |
| S | 0.0060 | 0.0431 | 0.28 | (0.13,0.40) | 8.95 | (6.46, 11.38) | |||
| OH | D | 0.0009 | 0.0094 | 0.19 | (0.02,0.33) | N | 2.81 | (1.52, 4.45) | N |
| S | 0.0017 | 0.0179 | 0.19 | (0.03,0.33) | 4.15 | (2.50, 6.26) | |||
| trait | hormonal environ. | VAF | VTotal | H2 | CI | H2 hormone dependent? | CVAF | CI | CVAF hormone dependent? |
| CORT | D | 0 | 0.2045 | 0 | (−0.79,0) | Y | 0 | (−26.20,0) | Y |
| DC | 0.09891 | 0.2061 | 0.10 | (0.36,1.51) | 16.43 | (4.59,24.29) | |||
| SVL | D | 0.0007 | 0.0234 | 0.06 | (−0.14,0.12) | N | 3.21 | (1.20, 6.44) | N |
| DC | 0.0010 | 0.0280 | 0.07 | (−0.09,0.14) | 3.45 | (0.81, 6.89) | |||
| GS | D | 0.0331 | 1.9287 | 0.03 | (−0.15,0.07) | Y | 0.52 | (−0.28, 1.05) | Y |
| DC | 0.3650 | 2.9544 | 0.25 | (0.08,0.45) | 1.82 | (1.26, 3.02) | |||
| GL | D | 0 | 0.0449 | 0 | (−0.09,0) | Y | 1.15 | (−3.59, 2.30) | N |
| DC | 0.0074 | 0.0623 | 0.24 | (0.02,0.41) | 4.99 | (1.59, 9.97) | |||
| OH | D | 0.0011 | 0.0100 | 0.22 | (0.01,0.38) | Y | 3.11 | (1.64, 4.97) | Y |
| DC | 0.0001 | 0.0127 | 0.02 | (−0.31,0.03) | 0 | (−3.98, 0) |
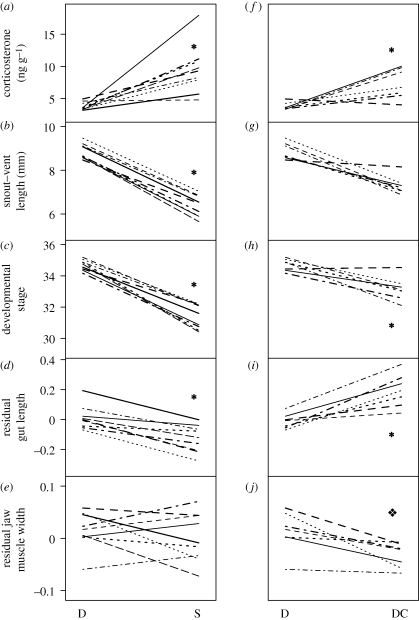
Figure 3.
Reaction norms of endogenous CORT (a and f), SVL (b and g), developmental stage (c and h), gut length (d and i) and jaw muscle width (e and j; for graphical representation, natural log-transformed gut length and jaw muscle width were regressed against natural log-transformed SVL, and then residual values were used as size-independent values; SVL was used as a covariate in the actual statistical analysis). Treatment groups are diets (D for native diet, detritus; S for novel diet, shrimp) and hormonal regimes (DC for exogenous CORT). Only seven families were included in the D versus DC comparison because two families responded to the CORT treatment with excessive mortality. Asterisks and diamonds indicate heritable variation in alternate environments that is significantly higher and lower, respectively. Each family has a unique line pattern and thickness that is retained between graphs.
4. Discussion
We investigated the proximate mechanisms that might have promoted the evolution of a novel resource-use phenotype. We specifically sought to determine whether cryptic genetic variation might have played a role in the evolutionary transition from omnivory to carnivory in spadefoot toad tadpoles. In order to do so, we evaluated whether a novel diet could reveal cryptic genetic variation in an ancestral population, and whether diet-induced hormones could mediate this expression. Although cryptic genetic variation may play a key role in the origins of novel phenotypes, few studies have attempted to measure such variation in a phylogenetic context (although see Etges 1993) or provide a mechanism for its exposure (although see Rutherford & Lindquist 1998; Queitsch et al. 2002; Suzuki & Nijhout 2006; Badyaev 2009).
Here, we assessed the effects of a novel diet and the hormone corticosterone (CORT) on the expression of cryptic genetic variation in fitness and morphological traits in Sc. couchii larvae. Previous research indicated that Sc. couchii is an appropriate model for the ancestral feeding condition of Spea (Ledón-Rettig et al. 2008), the genus that evolved carnivory. Furthermore, Sc. couchii tadpoles produce CORT in response to the derived diet (Ledón-Rettig et al. 2009). We found that shrimp, a novel diet for both Sc. couchii and ancestral Spea, revealed diet-dependent heritable variation in overall growth, developmental speed and gut length (table 1 and figure 3b–d). We were able to recapitulate this effect on developmental speed and gut length by exposing tadpoles to CORT (table 1 and figure 3h,i), suggesting a role for diet-induced hormones in the expression of cryptic genetic variation. Such variation can be immediately refined by natural selection to produce rapid phenotypic evolution in natural populations.
It may seem counterintuitive that the cryptic genetic variation in size and development revealed by shrimp and CORT in Sc. couchii tadpoles could be adaptive, given that the overall effect of these agents was to decrease fitness. However, the relevant comparison here is not between the fitness in the novel environment versus the native environment, but a comparison between situations where expressed genetic variation is present or absent in the novel environment (figure 1). When ancestral populations of Spea made a dietary transition to carnivory, they probably experienced decreased fitness until the costs associated with consuming shrimp were overcome by adaptation (Lande 2009; Ledón-Rettig et al. 2009). On one hand, they might have lacked diet-dependent variation in trophic traits (e.g. if these traits were developmentally constrained or possessed strong cross-environment correlations; Falconer & Mackay 1996; Gomez-Mestre et al. 2008). However, if ancestral Spea possessed cryptic genetic variation in these traits when they encountered their novel resource, this would have made it more likely that adaptive genetic variation was present for consuming shrimp and thus more likely that the population would endure the dietary transition.
Although shrimp and exogenous CORT had similar effects on the heritability of gut length in Sc. couchii larvae, they exerted remarkably different effects with respect to its direction of response. Shrimp-fed tadpoles produced relatively short guts (figures 2c and 3d), which is an adaptive morphology for a carnivorous diet (Hume 2005). By contrast, CORT-exposed tadpoles produced relatively long guts (figures 2c and 3i), despite the fact that shrimp-fed tadpoles from the same families experienced high levels of endogenous CORT and produced shorter guts. Yet, this finding does not militate against CORT having a role in the phenotypic effects generated by a novel diet. For instance, while CORT may amplify family differences in gut length, the interaction between CORT and other aspects of the shrimp diet (e.g. chemical, physical or nutritional) may be necessary to produce the complete natural response (McGuigan & Sgró 2009).
Our relatively large estimates of shrimp- and CORT-dependent, broad-sense heritabilities in size, development and gut length could be due to increased additive genetic variation, decreased environmental variation or increased maternal influence (Charmantier & Garant 2005). However, it is unlikely that increased heritability was caused by decreased environmental variation: our CVs, which are independent of environmental variation, were consistent with our estimates of heritability. It is also unlikely that our heritability estimates were inflated by differences in maternal condition: females used in this study were raised under similar laboratory conditions for 2–3 years, and maternal condition was not statistically correlated with any of the traits measured in this study (electronic supplementary material, appendix S2). Thus, our estimates of heritability probably indicate, to a substantial degree, additive genetic variation.
In contrast to gut length, the magnitude of genetic variation in jaw muscle width was not diet dependent (figure 3e and table 1). Moreover, exogenous CORT decreased the amount of expressed genetic variation in this trait (figure 3j and table 1). Thus, novel diets and CORT may have different effects on cryptic genetic variation in different traits. Although the heritability of jaw muscle width in Sc. couchii larvae was not diet dependent, it was not negligible either (in both shrimp- and detritus-fed tadpoles, heritability for this trait was significantly greater than zero). Genetic variation for consuming the native diet may persist in populations if there is some mechanism for maintaining phenotypic variation, but most probably reflects weak selection on jaw muscle width: i.e. having a particular jaw muscle size when consuming detritus does not confer a significant cost or benefit. Under such a scenario, variation has the opportunity to accumulate without being diminished by selection.
From the cryptic genetic variation revealed by their novel diet, ancestral populations of Spea might have rapidly evolved improved performance (more robust growth and faster development) and adaptive morphologies (shorter guts) when consuming shrimp. However, modern Spea larvae are facultative carnivores that perform well on, and express alternate resource-use phenotypes that are adaptive for either detritus or shrimp (Pfennig & Murphy 2002). Can we infer from Sc. couchii whether this plasticity (i.e. the ability to produce environmentally induced morphologies) evolved from such cryptic genetic variation? Although the heritability of plasticity can be measured (Laurila et al. 2002; Relyea 2005; Gomez-Mestre et al. 2008; Talloen et al. 2009; electronic supplementary material, appendix S4), it is unclear whether selection acts on reaction norms per se. Indeed, empirical studies have found that plasticity may fail to respond to artificial selection in the presence of genetic variation for the reaction norm (van Kluenen et al. 2002) and, conversely, evolve in spite of a strong genetic correlation between traits in alternate environments (Beldade et al. 2002; Czesak et al. 2006). Some have argued that, as long as genetic variation exists for a trait in each environment, reaction norms will arise as an emergent property of selection favouring different trait optima in different environments (Via & Lande 1985; Via 1993). If true, cryptic genetic variation in gut morphology revealed by ancestral Spea tadpoles might have promoted the evolution of gut plasticity in descendent lineages.
In summary, we have found that a novel diet for anuran larvae can unlock cryptic genetic variation that can potentially fuel the evolution of a novel feeding strategy. Although a multigenerational study is not feasible in our system, a selection experiment would be the next critical step in validating the evolvability of traits that emerge from cryptic genetic variation. In situations where populations are faced with a rapid dietary transition (e.g. when confronting intense resource competition or depletion of their usual resource by an environmental disturbance), phenotypes exposed by the novel diet, itself, may be an important source of variation on which natural selection can act, enabling these populations to adapt more rapidly to their new resource. By evaluating the mechanisms underlying the expression of cryptic genetic variation, and the extent to which it facilitates evolution in natural populations, we can illuminate the origins of the complex phenotypes that accompany diverse feeding strategies. More generally, such studies can help explain the origins of novel traits, which is among the central challenges confronting evolutionary biologists.
Acknowledgements
We thank K. Butler for her assistance with collecting and processing these data. We also thank K. Pfennig, C. Burch, R. Warne, R. Martin, S. Dhole, M. Ferris, A. Leichty, J. Santos, L. Bono, A. Moczek and one anonymous reviewer for their thoughtful comments on this manuscript. All procedures were carried out in compliance with the Institutional Animal Care and Use Committee at the University of North Carolina, Chapel Hill, under application no. 03-0110. Funding was provided by the National Science Foundation under grants IOS-0818212, DEB-0640026 and the Graduate Research Fellowship Programme.
References
- Altig R., Whiles M. R., Taylor C. L.2007What do tadpoles really eat? Assessing the trophic status of an understudied and imperiled group of consumers in freshwater habitats. Freshw. Biol. 52, 386–395 (doi:10.1111/j.1365-2427.2006.01694.x) [Google Scholar]
- Aubret F., Bonnet X., Shine R.2007The role of adaptive plasticity in a major evolutionary transition: early aquatic experience affects locomotor performance of terrestrial snakes. Funct. Ecol. 21, 11 541–11 161 [Google Scholar]
- Baayeen R. H., Davidson D. J., Bates D. M.2008Mixed-effects modeling with crossed random effects for subjects and items. J. Mem. Lang. 5, 390–412 [Google Scholar]
- Badyaev A. V.2009Evolutionary significance of phenotypic accommodation in novel environments: an empirical test of the Baldwin effect. Phil. Trans. R. Soc. B 364, 1125–1141 (doi:10.1098/rstb.2008.0285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R. D. H., Schluter D.2008Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44 (doi:10.1016/j.tree.2007.09.008) [DOI] [PubMed] [Google Scholar]
- Barton N. W. H., Houston D. C.1993A comparison of digestive efficiency in birds of prey. Ibis 135, 363–371 (doi:10.1111/j.1474-919X.1993.tb02107.x) [Google Scholar]
- Beldade P., Koops K., Brakefield P. M.2002Modularity, individuality and evo-devo in butterfly wings. Proc. Natl Acad. Sci. USA 99, 14 262–14 267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragg A. N.1965Gnomes of the night: the spadefoot toads. Philadelphia, PA: University of Pennsylvania Press [Google Scholar]
- Carroll S. P., Klassen S. P., Dingle H.1998Rapidly evolving adaptations to host ecology and nutrition in the soapberry bug. Evol. Ecol. 12, 955–968 (doi:10.1023/A:1006568206413) [Google Scholar]
- Charmantier A., Garant D.2005Environmental quality and evolutionary potential: lessons from wild populations. Proc. R. Soc. B 272, 1415–1425 (doi:10.1098/rspb.2005.3117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czesak M. E., Fox C. W., Wolf J. B.2006Experimental evolution of phenotypic plasticity: how predictive are cross-environment correlations? Am. Nat. 168, 323–335 (doi:10.1086/506919) [DOI] [PubMed] [Google Scholar]
- Denver R. J.1998Hormonal correlates of environmentally induced metamorphosis in the Western spadefoot toad, Scaphiopus hammondii. Gen. Comp. Endocrinol. 110, 326–336 (doi:10.1006/gcen.1998.7082) [DOI] [PubMed] [Google Scholar]
- Denver R. J.2009Stress hormones mediate environment–genotype interactions during amphibian development. Gen. Comp. Endocrinol. 164, 20–31 (doi:10.1016/j.ygcen.2009.04.016) [DOI] [PubMed] [Google Scholar]
- Dufty A. M., Colbert J., Moller A. P.2002Hormones, developmental plasticity and adaptation. Trends Evol. Ecol. 17, 190–196 (doi:10.1016/S0169-5347(02)02498-9) [Google Scholar]
- Etges W. J.1993Genetics of host–cactus response and life-history evolution among ancestral and derived populations of cactophilic Drosophila mojavensis. Evolution 47, 750–767 (doi:10.2307/2410181) [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C.1996Introduction to quantitative genetics. Harlow, UK: Pearson Education Limited [Google Scholar]
- Fisher R. A.1930The genetical theory of natural selection. Oxford, UK: Clarendon Press [Google Scholar]
- Ghalambor C. K., McKay J. K., Carroll S. P., Reznick D. N.2007Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Funct. Ecol. 21, 394–407 (doi:10.1111/j.1365-2435.2007.01283.x) [Google Scholar]
- Gibson G., Dworkin I.2004Uncovering cryptic genetic variation. Nat. Rev. Genet. 5, 1199–1212 [DOI] [PubMed] [Google Scholar]
- Gomez-Mestre I., Touchon J. C., Saccoccio V. L., Warkentin K. M.2008Genetic variation in pathogen-induced early hatching of toad embryos. J. Evol. Biol. 21, 791–800 (doi:10.1111/j.1420-9101.2008.01509.x) [DOI] [PubMed] [Google Scholar]
- Gosner K. L.1960A simplified table for staging anuran embryos and larvae with notes on identification. Herpetologica 16, 183–190 [Google Scholar]
- Hadfield J. D.2009. MCMCglmm: Markov chain Monte Carlo methods for generalised linear mixed models. See http://cran.r-project.org/web/packages/MCMCglmm/vignettes/Tutorial.pdf
- Hall B. K.2001Organic selection: proximate environmental effects on the evolution of morphology and behavior. Biol. Phil. 289, 153–161 [Google Scholar]
- Hothorn T., Bretz F., Westfall P.2008Simultaneous inference in general parametric models. Biometrical J. 50, 346–363 (doi:10.1002/bimj.200810425) [DOI] [PubMed] [Google Scholar]
- Houle D.1992Comparing evolvability and variability of quantitative traits. Genetics 130, 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Crespi E. J., Denver R. J.2008Programming neuroendocrine stress axis activity by exposure to glucocorticoids during postembryonic development of the frog Xenopus laevis. Endocrinology 149, 5470–5481 [DOI] [PubMed] [Google Scholar]
- Hume I. D.2005Concepts of digestive efficiency. In Physiological and ecological adaptations to feeding in vertebrates (eds Starck J. M., Wang T.), pp. 43–58 Enfield, UK: Science Publishers [Google Scholar]
- Hurlbert S. H.1984Pseudoreplication and the design of ecological field experiments. Ecol. Monogr. 54, 187–211 (doi:10.2307/1942661) [Google Scholar]
- Lande R.2009Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. J. Exp. Evol. 22, 1435–1446 [DOI] [PubMed] [Google Scholar]
- Laurila A., Karttunen S., Merilä J.2002Adaptive phenotypic plasticity and genetics of larval life histories in two Rana temporaria populations. Evolution 56, 617–627 [DOI] [PubMed] [Google Scholar]
- Ledón-Rettig C. C., Pfennig D. W., Nascone-Yoder N.2008Ancestral variation and the potential for genetic accommodation in larval amphibians: implications for the evolution of novel feeding strategies. Evol. Dev. 10, 316–325 [DOI] [PubMed] [Google Scholar]
- Ledón-Rettig C. C., Pfennig D. W., Crespi E. J.2009Stress hormones and the fitness consequences associated with the transition to a novel diet in larval amphibians. J. Exp. Biol. 212, 3743–3750 (doi:10.1242/jeb.034066) [DOI] [PubMed] [Google Scholar]
- Maddison W. P., Maddison D. R. Mesquite: a modular system for evolutionary analysis, v. 2.01. 2007. See http://mesquiteproject.org .
- Martin R. A., Pfennig D. W.2009Disruptive selection in natural populations: the roles of ecological specialization and resource competition. Am. Nat. 174, 268–281 (doi:10.1086/600090) [DOI] [PubMed] [Google Scholar]
- McEwen B. S., Wingfield J. C.2003The concept of allostasis in biology and biomedicine. Horm. Behav. 43, 2–15 (doi:10.1016/S0018-506X(02)00024-7) [DOI] [PubMed] [Google Scholar]
- McGuigan K., Sgró C. M.2009Evolutionary consequences of cryptic genetic variation. Trends Ecol. Evol. 24, 305–311 (doi:10.106/j.tree.2009.02.001) [DOI] [PubMed] [Google Scholar]
- Moczek A. P.2007Developmental capacitance, genetic accommodation, and adaptive evolution. Evol. Dev. 9, 299–305 [DOI] [PubMed] [Google Scholar]
- Parsons K. J., Robinson B. W.2006Replicated evolution of integrated plastic responses during early adaptive divergence. Evolution 60, 801–813 [PubMed] [Google Scholar]
- Pfennig D. W., Murphy P. J.2000Character displacement in polyphenic tadpoles. Evolution 54, 1738–1749 [DOI] [PubMed] [Google Scholar]
- Pfennig D. W., Murphy P. J.2002How fluctuating competition and phenotypic plasticity mediate species divergence. Evolution 56, 1217–1228 [DOI] [PubMed] [Google Scholar]
- Pfennig D. W., Martin R. A.2009A maternal effect mediates rapid population divergence and character displacement in spadefoot toads. Evolution 63, 898–909 (doi:10.1111/j.1558-5646.2008.00544.x) [DOI] [PubMed] [Google Scholar]
- Phillips B. L., Shine R.2004Adapting to an invasive species: toxic cane toads induce morphological change in Australian snakes. Proc. Natl Acad. Sci. USA 49, 17 150–17 155 (doi:10.1073/pnas.0406440101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M.2001Phenotypic plasticity: beyond nature and nurture. Baltimore, MD: John Hopkins University Press [Google Scholar]
- Queitsch C., Sangster T. A., Lindquist S.2002Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624 (doi:10.1038/nature749) [DOI] [PubMed] [Google Scholar]
- Rasband W. S. ImageJ. Bethesda, MD: National Institute of Health: 1997–2006. See http://rsb.info.nih.gov/ij/ [Google Scholar]
- Relyea R. A.2005The heritability of inducible defenses in tadpoles. J. Evol. Biol. 18, 856–866 (doi:10.1111/j.1420-9101.2005.00882.x) [DOI] [PubMed] [Google Scholar]
- Roff D. A.1997Evolutionary quantitative genetics. New York, NY: Chapman and Hall [Google Scholar]
- Rutherford S. L., Lindquist S.1998Hsp90 as a capacitor for morphological evolution. Nature 396, 336–342 (doi:10.1038/24550) [DOI] [PubMed] [Google Scholar]
- Sapolsky R. M., Romer L. M., Munck A. U.2000How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocrin. Rev. 21, 55–89 (doi:10.1210/er.21.1.55) [DOI] [PubMed] [Google Scholar]
- Schlichting C. D.2008Hidden reaction norms, cryptic genetic variation, and evolvability. Ann. N.Y. Acad. Sci. 113, 187–203 [DOI] [PubMed] [Google Scholar]
- Schluter D.2000The ecology of adaptive radiation. Oxford, UK: Oxford University Press [Google Scholar]
- Sokal R. R., Rohlf F. J.1995Biometry: the principles and practice of statistics in biological research, 3rd edn.New York, NY: W. H. Freeman and Co [Google Scholar]
- Suzuki Y., Nijhout H. F.2006Evolution of a polyphenism by genetic accommodation. Science 311, 650–652 (doi:10.1126/science.1118888) [DOI] [PubMed] [Google Scholar]
- Talloen W., Van Dongen S., Van Dyck H., Lens L.2009Environmental stress and quantitative genetic variation in butterfly wing characteristics. Evol. Ecol. 23, 473–485 (doi:10.1007/s10682-008-9246-4) [Google Scholar]
- Uller T., Olsson M., Ståhlberg F.2002Variation in heritability of tadpole growth: an experimental analysis. Heredity 88, 480–484 (doi:10.1038/sj.hdy.6800088) [DOI] [PubMed] [Google Scholar]
- van Kluenen M., Fischer M., Schmid B.2002Experimental life-history evolution: selection on the allocation to sexual reproduction and its plasticity in a clonal plant. Evolution 56, 2168–2177 [PubMed] [Google Scholar]
- Via S.1993Adaptive phenotypic plasticity: target or by-product of selection in a variable environment? Am. Nat. 142, 352–365 (doi:10.1086/285542) [DOI] [PubMed] [Google Scholar]
- Via S., Lande R.1985Genotype–environment interaction and the evolution of phenotypic plasticity. Evolution 39, 505–522 (doi:10.2307/2408649) [DOI] [PubMed] [Google Scholar]
- West-Eberhard M. J.2003Developmental plasticity and evolution. New York, NY: Oxford University Press [Google Scholar]
- Wund M. A., Baker J. A., Clancy B., Golub J. L., Foster S. A.2008A test of the ‘flexible stem’ model of evolution: ancestral plasticity, genetic accommodation, and morphological divergence in the threespine stickleback radiation. Am. Nat. 172, 449–462 (doi:10.1086/590966) [DOI] [PubMed] [Google Scholar]