Abstract
Post-mating reduction in immune defence is common in female insects, and a trade-off between mating and immunity could affect the evolution of immunity. In this work, we tested the capacity of virgin and mated female Drosophila melanogaster to defend against infection by four bacterial pathogens. We found that female D. melanogaster suffer post-mating immunosuppression in a pathogen-dependent manner. The effect of mating was seen after infection with two bacterial pathogens (Providencia rettgeri and Providencia alcalifaciens), though not after infection with two other bacteria (Enterococcus faecalis and Pseudomonas aeruginosa). We then asked whether the evolution of post-mating immunosuppression is primarily a ‘female’ or ‘male’ trait by assaying for genetic variation among females for the degree of post-mating immune suppression they experience and among males for the level of post-mating immunosuppression they elicit in their mates. We also assayed for an interaction between male and female genotypes to test the specific hypothesis that the evolution of a trade-off between mating and immune defence in females might be being driven by sexual conflict. We found that females, but not males, harbour significant genetic variation for post-mating immunosuppression, and we did not detect an interaction between female and male genotypes. We thus conclude that post-mating immune depression is predominantly a ‘female’ trait, and find no evidence that it is evolving under sexual conflict.
Keywords: immune defence, mating and reproduction, Drosophila melanogaster, genetic variation, trade-off, sexual conflict
1. Introduction
Immune defence (defined as the combined ability of an organism to both actively fight and to tolerate an infection (Ayres & Schneider 2008)) is generally considered to be costly in that its maintenance and deployment often results in physiological and evolutionary trade-offs against other traits important for fitness, including longevity (Moret & Schmid-Hempel 2000; DeVeale et al. 2004; Ye et al. 2009), larval ability to compete for food (Kraaijeveld & Godfray 1997), body size (Fellowes et al. 1999), fertility (Ye et al. 2009) and fecundity (Fellowes et al. 1999; McKean et al. 2008). It has been proposed that mating may have immunosuppressive effects in females in order to allow limited resources to be shunted from immunological requirements to reproductive needs (Sheldon & Verhulst 1996). Examples of post-mating immune depression include a reduction in phenoloxidase activity after mating in the beetle Tenebrio molitor (Rolff & Siva-Jothy 2002), and decreased encapsulation ability correlated with increased oviposition in damselflies (Siva-Jothy et al. 1998). Additionally, mating causes decreased survival after infection with a pathogen in female D. melanogaster (Fedorka et al. 2007) but see (McKean & Nunney 2005; Wigby et al. 2008), and increased mating effort leads to decreased hemocyte number, lytic activity and encapsulation ability in the cricket Allonemobius socius (Fedorka et al. 2004). Post-mating immunosuppression may not be universal, as it was not detected in yellow dung flies Scathophaga stercoraria (Schwarzenbach et al. 2005), and phenoloxidase activity and parasite resistance are even increased after mating in female A. socius and Gryllus texensis, respectively (Fedorka et al. 2004; Shoemaker et al. 2006). Determining how and why immune defence is altered by mating (or other fitness-related activities) is crucial to our understanding of how immune defence evolves, as well as how it functions at the whole-organism level.
The immune system of D. melanogaster is well understood, and extensive genetic analysis has revealed many of the genes involved in the function of the humoral and cellular immune response (reviewed in Lemaitre & Hoffmann 2007). This work has mainly focused on the function of the canonical immune system; but pleiotropic connections to mating (or other costly processes) have the potential to dramatically alter or limit the function and evolution of overall levels of defence (reviewed in Lawniczak et al. 2007). In the present study, we first determined whether mating affects the function of female immune defence. We used multiple pathogens in order to establish the generality of the phenomenon and to elucidate the potential importance of pathogen diversity on changes in defence owing to mating. We were also interested in determining the role that this trade-off could play in shaping the evolution of immune defence. We therefore assessed the level of genetic variation among females for the reduction in defence they experience after mating and among males for the level of post-mating immunosuppression they elicit in their mates. We also determined whether the change in defence is dependent on the particular combination of male and female genotypes engaging in copulation. We measured genetic variation in both sexes because we were particularly interested in assessing the potential for ongoing sexual conflict in this system, as it has been suggested in the literature that the fitness of males and females may be affected differently depending on the level of immunosuppression females experience after mating, and that this could lead to sexual conflict (Fedorka et al. 2007; Lawniczak et al. 2007). This hypothesis could be provisionally supported by data we have collected, which suggest that the male ejaculate plays a role in reducing female defence (S. M. Short, M. F. Wolfner & B. P. Lazzaro 2010, unpublished data). Furthermore, components of the seminal fluid have been demonstrated to be involved in dynamic evolutionary interactions such as sexual conflict (Rice 1996), and multiple proteins in the male ejaculate of D. melanogaster have been shown to be rapidly evolving (Swanson et al. 2001; Mueller et al. 2005). If extant genetic variation in female immunosuppression was maintained by an ongoing intersexual interaction (e.g. sexual conflict) in our sampled population, we could expect to observe that the magnitude of post-mating immunosuppression is determined by the specific male and female genotypes participating in a mating (Gillespie & Turelli 1989). D. melanogaster possess significant genetic variation for immune defence (e.g. Lazzaro et al. 2004; Tinsley et al. 2006), and variation in the immunological cost incurred by mating is one potential source of that genetic variation.
2. Material and methods
(a). Fly stocks and maintenance
We used the following lines of D. melanogaster: Canton S (a wild-type inbred strain), and 18 lines chosen randomly from the Drosophila Genetic Reference Panel (DGRP), a collection of inbred isofemale lines collected in Raleigh, NC (Ayroles et al. 2009). Each line is genetically distinct and the total set represents a ‘snapshot’ of naturally occurring genetic variation in this population at the time of sample. Nine DGRP lines (coded 1F–9F) were used to assay female variation. These were RAL-324, RAL-362, RAL-820, RAL-639, RAL-375, RAL-315, RAL-437, RAL-786 and RAL-486. Nine different DGRP lines (coded 1M–9M) were used to assay male variation. These were RAL-391, RAL-774, RAL-358, RAL-303, RAL-380, RAL-712, RAL-732, RAL-208 and RAL-360. Flies for all experiments were reared at 24°C on a 12 h light–dark cycle on standard glucose medium (12 g agar, 100 g glucose and 100 g Brewer's yeast per 1.2 l of water, plus 0.04% phosphoric acid and 0.4% propionic acid (final concentration) added to inhibit microbial growth in the food).
(b). Experimental design
To test the effect of mating on bacterial load and survival after infection with multiple pathogens, we conducted multiple experiments (one per pathogen) each in a complete block design, where both virgin and mated females were assayed for either bacterial load or survival in each replicate of the experiment. To test for genetic variation across lines from the DGRP, we used females from nine lines (coded 1F–9F) and males from nine additional lines (coded 1M–9M). The experiment was conducted in a manner similar to a lattice square design, with minor departures from classical set-up owing to experimental contingencies. Bacterial load data for virgin and mated females were collected for all 81 pairwise crosses between all nine ‘F’ lines and all nine ‘M’ lines, with the entire experiment conducted in duplicate. Owing to the labour involved in assaying infection phenotypes in a crossing scheme of this scale, we opted to measure only the bacterial load phenotype in this part of the experiment. We feel that this is justified in that mated females sustain significantly higher bacterial loads and significantly higher mortality than virgins do after infection with Providencia rettgeri (figures 1a and 2a), so either phenotype is a reliable indicator of overall defence. Additionally, because of the magnitude of the experiment, data for all of the 81 pairwise crosses (comprising a single replicate of the entire experiment) were collected over 9 days. On each day, nine of the 81 pairwise combinations were observed, with females from each ‘F’ line mated to males from a single, randomly assigned ‘M’ line, such that all ‘F’ lines and all ‘M’ lines were used each day. At the end of the 9-day experiment, all ‘F’ lines had been paired to all ‘M’ lines once, with data for virgin and mated females from each of these 81 combinations recorded. The randomization scheme for this experiment was generated using the ‘plan’ procedure in SAS (SAS Institute, Cary, NC, USA). On any given day, we shuffled the order in which each ‘F’ line and each ‘M’ line was mated and infected using the ‘sample’ function in R (R Foundation for Statistical Computing, Vienna, Austria).
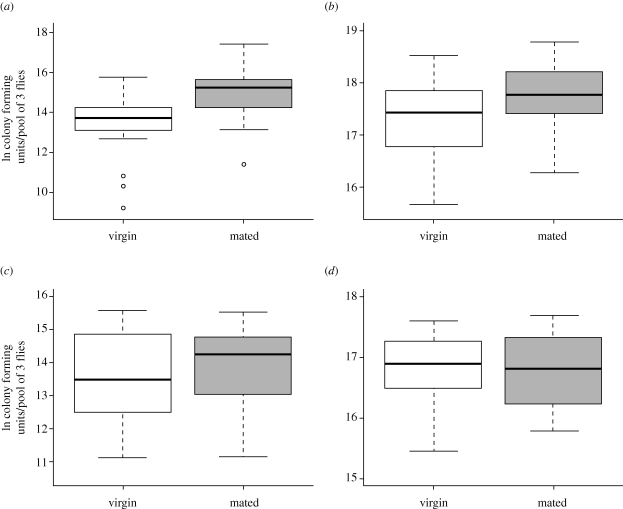
Figure 1.
The effect of mating on female bacterial load after infection with four bacterial pathogens. Bacterial loads of wild-type (Canton S) females mated to wild-type (Canton S) males were significantly higher than those of virgin wild-type (Canton S) females after infection with (a) P. rettgeri (F1,67 = 28.77, p < 0.0001) and (b) P. alcalifaciens (F1,77 = 9.86, p = 0.0024), but not after infection with (c) E. faecalis (F1,52 = 1.20, p = 0.279) or (d) Ps. aeruginosa (F1,32 = 0.17, p = 0.6804). We infected virgin and mated females in parallel 2–3 h after mated females completed copulation. Total sample sizes were as follows: for P. rettgeri, nmated = 36 and nvirgin = 35 for P. alcalifaciens, nmated = 43 and nvirgin = 38, for E. faecalis, nmated = 28 and nvirgin = 28, and for Ps. aeruginosa, nmated = 17 and nvirgin = 18. Each data point consists of three pooled females, and data were collected over three replicates for each bacterial species with the exception of Ps. aeruginosa, for which only two replicates were collected. Uninfected controls (not shown) were sham-infected with a sterile needle and always yielded zero bacteria.
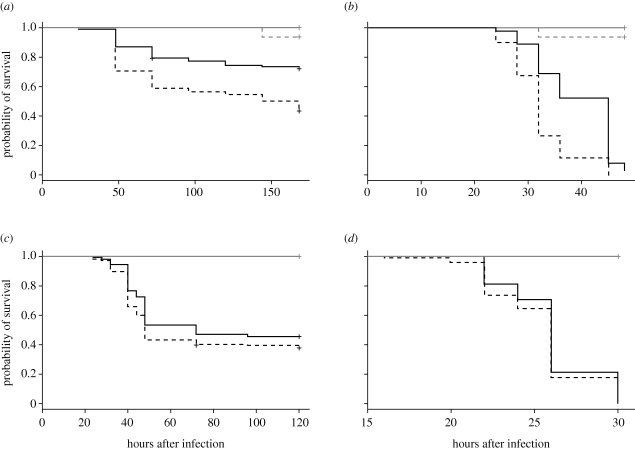
Figure 2.
The effect of mating on female survival after infection with four bacterial pathogens. Survival over time of wild-type (Canton S) females mated to wild-type (Canton S) males was significantly lower than that of virgin wild-type (Canton S) females after infection with P. rettgeri (panel (a), p < 0.0001) and P. alcalifaciens (panel (b), p < 0.0001), but not after infection with E. faecalis (panel (c), p = 0.0811) or P. aeruginosa (panel (d), p = 0.3466). Survival curves were estimated using the Kaplan–Meier method. Significance values are for the effect of mating treatment in infected females and were determined by Cox regression analysis. We infected both mated and virgin females in parallel 2–3 h after mated females complete copulation. N = 44–75 infected females per mating status per replicate, and two to four replicates were performed for each survival experiment. Uninfected controls pierced with a sterile needle (lines shown in grey) showed negligible mortality for both mated (dashed grey line) and virgin (solid grey line) treatments.
(c). Mating set-up
All matings were set up individually between a single virgin female and a single virgin male. All flies (males and females) were collected as virgins and aged 3 days post-eclosion with ad libitum access to food in groups of approximately 30. The day before matings were to be set up, virgin females were anaesthetized with CO2 and placed in individual vials containing abundant media. They were then randomly allocated to ‘virgin’ or ‘mated’ treatment. Females were allowed to recover overnight. The next morning (within 3 h of incubator ‘dawn’), unanaesthetized virgin males were aspirated into each vial assigned to the ‘mated’ treatment and each mating was individually observed. Matings lasting less than 15 min were not used for the experiments testing multiple pathogens, but this lower bound was reduced to a minimum of 10 min in the experiment to assess genetic variation. This was done because many mating pairs in this experiment copulated for shorter times than Canton S flies, possibly owing to natural variation in mating times. Lowering this boundary enabled inclusion of approximately 25 per cent of our final dataset, and therefore significantly increased our sample size. The number of 10 min matings were not equally distributed across genotypes (χ2-test for the null hypothesis of equal distribution across lines for males: χ2 = 228.73, d.f. = 8, p < 2.2 × 1016 and females: χ2 = 68.5, d.f. = 8, p < 9.7 × 1012), but the average length of mating did not correlate with change in bacterial load (for all 81 genotype combinations: r = −0.024, p = 0.83), so we are confident that the inclusion of these shorter matings did not bias the results of our study. After mating, mated females were removed from the presence of males.
(d). Infection procedure
Two to three hours after mating cessation, mated females and their virgin counterparts were alternately anaesthetized in groups of 15 or fewer on CO2 and pricked in the thorax with a needle dipped in dilute bacterial culture (see below). Females were then placed in a vial containing media to recover. A subset of flies from each mating treatment was pricked with a sterile needle as a wounding control. Bacterial species used for infection were as follows: P. rettgeri (isolated from wild-caught D. melanogaster by B. Lazzaro in State College, PA, USA), Providencia alcalifaciens (isolated from wild-caught D. melanogaster by P. Juneja and S. Short in Ithaca, NY, USA), Enterococcus faecalis (isolated from wild-caught D. melanogaster by B. Lazzaro) and Pseudomonas aeruginosa (species type strain, PAO1) (table 1). P. rettgeri, P. alcalifaciens and Ps. aeruginosa are all Gram-negative bacteria, while E. faecalis is Gram-positive. All of these species are opportunistic pathogens with broad host ranges, and all have the ability to infect humans (Devriese et al. 2006; Manos & Belas 2006; Yaher & Parsek 2006). All bacterial cultures were grown overnight in Luria broth (LB) at 37°C from a single bacterial colony. Each overnight culture was then diluted with sterile LB to O.D.600 = 1.0, with the exception of E. faecalis, which was diluted to O.D.600 = 0.5. This resulted in delivery of approximately 3 × 103 bacterial cells to each infected female with P. rettgeri and P. alcalifaciens, approximately 1 × 104 with Ps. aeruginosa and approximately 5 × 102 with E. faecalis.
Table 1.
Pathogens that vary in biology and virulence were used for infection of virgin and mated female D. melanogaster. Per cent mortality is averaged across virgin and mated females, and natural pathogens are those that have been isolated from the hemolymph and/or thoracic muscle of wild-caught D. melanogaster (see §2 for details).
| pathogen | virulence level | natural pathogen |
|---|---|---|
| P. rettgeri | moderate (approx. 40% mortality) | yes |
| P. alcalifaciens | high (approx. 98% mortality) | yes |
| E. faecalis | moderate (approx. 60% mortality) | yes |
| Ps. aeruginosa | high (100% mortality) | no (strain PAO1) |
(e). Bacterial load assay
To assay bacterial load, females were aged after infection with ad libitum access to food for either 24 ± 0.5 h (for the experiments represented in figures 1 and 2) or 26–28 h (for the genetic variation experiment). At this time, females were anaesthetized on CO2 and homogenized in 500 µl LB in pools of three (figures 1 and 2) or five (genetic variation experiment). The homogenate was diluted 1 : 100 for E. faecalis, 1 : 1000 for P. rettgeri, and 1 : 10 000 for P. alcalifaciens and Ps. aeruginosa prior to plating. Fifty microlitres of each diluted homogenate was deposited in a spiral pattern on LB agar using a WASP II spiral plater (Microbiology International, Bethesda, MD, USA), and plates were incubated overnight at 37°C. We verified that the colonies on the plate were of the species used for infection by visual inspection and periodic analysis of 16S rDNA. 16S rDNA was amplified from randomly selected colonies throughout the experiment using the primers fD1 and rP2, which amplify the rDNA of most eubacteria (Weisburg et al. 1991). PCR product from these colonies as well as from positive control colonies taken from a pure freezer stock of each bacterial species was digested with StuI and/or MspI, and the digested products were run on a 1 per cent agarose gel. Digest patterns of colonies taken from infected females always matched those of the pure freezer stock. Bacterial colonies were counted using the ProtoCOL plate counting system (Microbiology International) associated with the spiral plater, allowing estimation of the number of viable bacteria present in each pool of homogenized females. These were the primary data used for analysis. Seven to 16 data points were collected per replicate per treatment for each experiment in figure 1. For the genetic variation experiment, two to five data points were collected per treatment per replicate for each pairwise combination, yielding a total of 4–10 data points per treatment for each pairwise combination, with the exception of 6F × 2M, for which only a single replicate was obtained. Six plates with zero colonies were excluded from analysis for the genetic variation experiment, as these zero counts could represent either an absence of bacteria in the flies or a technical error in the plating process. Since we cannot definitively say the flies contained zero bacterial cells, we chose to exclude these data points. The excluded data represent less than 0.5 per cent of the dataset and eliminating these six data points has a negligible effect on the outcome of the analysis.
(f). Survival assay
To assay survival, females were placed in groups of approximately 10 after infection and observed either daily (for slower-acting pathogens like P. rettgeri), or at shorter intervals for the first 48 h after infection (for fast-acting pathogens like Ps. aeruginosa). Females from both virgin and mated treatments were put onto fresh food every other day. Survival was observed for 7 days after infection with P. rettgeri owing to its gradually induced mortality, but only for 5 days for E. faecalis since most mortality occurs in the first 48 h after infection with this bacterium. Survival for P. alcalifaciens and Ps. aeruginosa was observed for 48 h or until all flies were dead.
(g). Statistical analysis
To assess the effect of mating on bacterial load, we first performed a natural log transformation on bacterial load values for each bacterial pathogen to produce data that more closely fit a normal distribution. We then performed an ANOVA for each bacterial pathogen to determine the effect of mating status on bacterial load. Assumptions for the ANOVA were evaluated by running diagnostic plots (fitted values versus residuals, residual normal quantile–quantile plot) and visually assessing heteroskadicity and normality of residuals. In cases where residuals were found to be non-normal (verified by Shapiro–Wilk test), deviation from normality was due to a few outlier points. Removal of outlier points did not change the significance of mating status, and ANOVA results were therefore considered to be robust. These analyses were performed in SAS (SAS Institute).
To analyse our survival data, we used Cox regression analysis in SAS (SAS Institute) to determine the effect of mating status on survival over time. Event data were recorded for each fly (where an ‘event’ = death), and flies not dead by the last time point recorded were treated as censored data. Survival curves were generated using the Kaplan Meier method in R (R Foundation for Statistical Computing).
To determine the level of genetic variation between lines from the DGRP, we first performed a natural log transformation on the bacterial load data collected from females from each pairwise mating combination. We then performed an analysis of variance with Proc Mixed in SAS (SAS Institute) using the following mixed model:
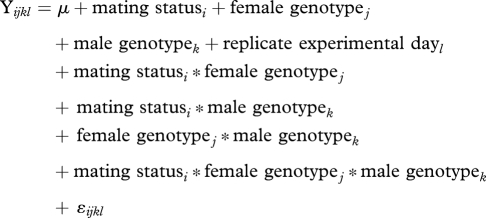 |
where Y = ln(bacterial load) data taken from all females, Mating statusi (i = 1,2) represents whether females were virgin or mated, female genotypej (j = 1,9) represents the DGRP lines contributing females to crosses, male genotypek (k = 1,9) represents the DGRP lines contributing males to crosses and replicate experimental dayl (l = 1,20) is a factor including all days over which the experiment was conducted. Each replicate required 9 days, and two replicates were performed for a total of 18 days. Missing data were subsequently filled in over 2 additional days, resulting in d.f. = 19 for day in the model. The factor mating statusi * female genotypej tests for genetic variation among females for post-mating immunosuppression, while mating statusi * male genotypek tests for male genetic variation for the level of immunosuppression they elicit in their mates. Mating statusi * female genotypej * male genotypek tests whether the effect of a particular male or female genotype on post-mating immunosuppression depends on the genotype of their mate.
Mean bacterial loads for each female and male genotype were obtained by finding the arithmetic mean of the log-transformed bacterial load data and back transforming it to obtain the geometric mean. We then calculated 95% confidence intervals (Sokal & Rohlf 1995).
3. Results and discussion
(a). Mating reduces female immune defence against two natural bacterial pathogens
It is yet unclear how ubiquitous post-mating reduction in immune defence is in insects. Many experiments testing potential trade-offs between immunity and defence have relied on indirect measures of immune quality (e.g. encapsulation ability, phenoloxidase activity or antimicrobial peptide gene expression) in the absence of actual infection (Siva-Jothy et al. 1998; Rolff & Siva-Jothy 2002; Fedorka et al. 2004; Lawniczak & Begun 2004; McGraw et al. 2004; Peng et al. 2005). While informative with regard to the potential mechanisms linking mating and the immune system, these assays do not directly measure changes in the ability of an organism to resist or tolerate an infection, and must be interpreted with care (Adamo 2004). Other studies have measured overall defence as a function of mating in the context of experimental infection (McKean & Nunney 2001, 2005; Shoemaker et al. 2006; Fedorka et al. 2007; Wigby et al. 2008). Three of the cited studies have been performed using female D. melanogaster (McKean & Nunney 2005; Fedorka et al. 2007; Wigby et al. 2008), but no clear consensus has emerged even from those as to whether females suffer a meaningful reduction in immune defence after mating. Two of these studies (McKean & Nunney 2005; Wigby et al. 2008) show no change owing to mating in the ability of females to clear non-pathogenic bacteria, while Fedorka et al. (2007) demonstrated that females infected with a pathogenic bacterium suffer higher mortality if they have mated. We hypothesized that the lack of consensus in this body of literature could be due to the effect of mating being dependent on the assay used to measure defence and/or the microbe used to test changes in defence (for example, pathogenic versus non-pathogenic infection). We, therefore, tested the effect of mating on female immune defence using two different assays (survival and systemic bacterial load) and four pathogens that differ in biology and pathogenicity (table 1).
We infected female D. melanogaster of the strain Canton S with each of the four bacterial pathogens in table 1, 2–3 h after mating cessation. We also infected virgin females in parallel to serve as a control comparison. We then assayed bacterial load (i.e. the number of colony forming units present in a fly) and survival in mated and virgin females after infection with each bacterial species. Females pierced with a sterile needle yielded zero bacterial colonies and had negligible mortality. At 24 h after infection with P. rettgeri (figure 1a) or P. alcalifaciens (figure 1b), we observed significantly higher bacterial loads in mated females compared with virgin females (P. rettgeri: p < 0.0001, P. alcalifaciens: p = 0.0024). We also observed significantly reduced survival in mated females compared with their virgin counterparts after infection with either P. rettgeri (figure 2a, p < 0.0001) or P. alcalifaciens (figure 2b, p < 0.0001). In contrast, we observed no difference in bacterial load owing to mating after infection with either E. faecalis (figure 1c, p = 0.279) or Ps. aeruginosa (figure 1d, p = 0.6804), and no effect of mating on survival after infection with E. faecalis (figure 2c, p = 0.0811) or Ps. aeruginosa (figure 2d, p = 0.3466). Virgins infected with E. faecalis had a slightly (but not significantly) higher probability of survival at multiple time points (e.g. mean per cent survival at 5 days post infection for virgin = 46.5% and for mated = 37.6%), but this effect was apparent in only two of the four replicates in the experiment (difference between treatments in two of the four replicates considered alone: p = 0.0164; in the other two replicates alone: p = 0.8587).
Our data show that mating results in reduced defence for females after infection with at least two pathogenic species of bacteria, both of which are pathogens of wild D. melanogaster. These results, coupled with previous findings showing no effect of mating in females after infection with a non-pathogenic bacterium (McKean & Nunney 2005; Wigby et al. 2008), suggest that, while general immune maintenance and immunocompetence are not impaired after mating, the ability of females to defend against pathogenic infection is hindered. Interestingly, and in contrast to our study, Fedorka et al. (2007) also used Ps. aeruginosa and did detect post-mating immunosuppression, suggesting that the magnitude of the effect may vary over bacterial strains, host genotypes or experimental conditions. Nevertheless, the total data suggest that the quality of immune defence in female D. melanogaster is frequently modulated by mating, an activity that is itself clearly essential to fitness.
(b). Female, but not male, D. melanogaster harbour significant genetic variation for the effect of mating on immune resistance
In order to gain insight into the evolution of the trade-off between mating and immunity, we assayed genetic variation among females for their resistance to infection after mating, genetic variation among males for their ability to alter female resistance and the degree to which the magnitude of post-mating immune depression depends on the specific male and female genotypes in mating pairs. We mated females from nine genetic lines of D. melanogaster to males from nine distinct genetic lines and, for all 81 pairwise crosses, assayed bacterial load after infection with P. rettgeri in both virgin and mated females. The entire experiment of 81 crosses was performed in duplicate, with two to five data points collected per cross in each replicate, where each data point is obtained from a pool of five females. We then performed an analysis of variance on the bacterial load data using the model in table 2.
Table 2.
Analysis of variance for effects of male and female genotype on bacterial load in mated versus virgin females. The experiment was conducted over the course of multiple days. ‘Experimental replicate day’ refers to all days over which the experiment was conducted. ‘Mating status’ refers to mated females versus virgins.
| factor | effect type | d.f. | F value | p-value |
|---|---|---|---|---|
| mating status | fixed | 1 | 36.23 | <0.0001 |
| female genotype | fixed | 8 | 26.42 | <0.0001 |
| male genotype | fixed | 8 | 2.30 | 0.0193 |
| experimental replicate day | random | 19 | ||
| mating status * female genotype | fixed | 8 | 4.32 | <0.0001 |
| mating status * male genotype | fixed | 8 | 0.61 | 0.7730 |
| female genotype * male genotype | fixed | 64 | 0.078 | 0.0777 |
| mating status * female genotype * male genotype | fixed | 64 | 1.16 | 0.1905 |
| residual error | 1129 |
Females are highly significantly genetically variable for the degree of post-mating immune depression that they experience (mating status * female genotype, p < 0.0001, table 2 and figure 3). The bacterial load of mated females relative to virgins ranged across female genotypes from a 4.1-fold increase to essentially no change (figure 3). To our surprise, however, we did not observe significant genetic variation in the ability of males to suppress female immune defence (mating status * male genotype, p = 0.7730, table 2 and figure 4). The bacterial load of mated females relative to virgins was relatively invariant across the male genotypes to which they mated, with the smallest change being a 1.4-fold increase and the largest a twofold increase. Such low levels of genetic variation among males were unexpected because female immune depression occurs only when the male ejaculate is intact with respect to sperm and accessory gland proteins (S. M. Short, M. F. Wolfner & B. P. Lazzaro 2010, unpublished data). Despite that, and despite the known adaptive evolution of some male ejaculate proteins, we fail to reject the null hypothesis that males are not variable for the magnitude of female immune modulation they elicit. We also found no evidence that female post-mating immune depression is determined by an interaction between the specific male and female genotypes engaged in a mating (mating status * female genotype * male genotype, p = 0.1905, table 2), casting further doubt on any hypothesis that this trait is evolving under sexually antagonistic coevolution.
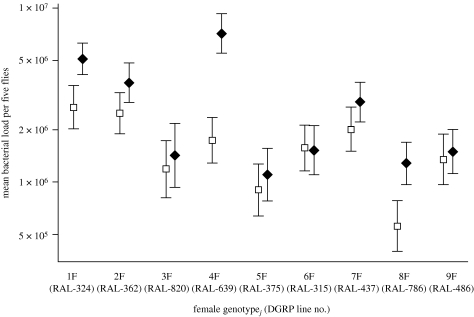
Figure 3.
Variation in the effect of mating across female genotypes. We calculated corrected mean bacterial load for mated females (black diamonds) and virgin females (open squares) of each female genotype pooled across all male genotypes. For example, the black diamond for female 1F corresponds to the mean load sustained by 1F females after mating to males from genotypes 1M–9M, and the open square corresponds to loads sustained by virgin 1F females infected and plated alongside mated 1F females. Mean refers to geometric mean, and error bars represent a 95% confidence interval. The parenthetical numbers on the x-axis are the DGRP stock identity number.
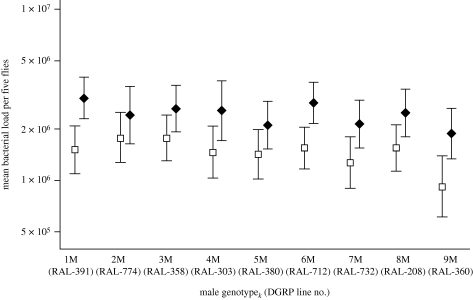
Figure 4.
Variation in the effect of mating across male genotypes. We calculated corrected mean bacterial load for all females mated to each individual male genotype (black diamonds) and the mean bacterial load for the corresponding virgin controls from all female genotypes (open squares). For example, the black diamond for 1M corresponds to the mean of females from genotypes 1F–9F mated to 1M males, and the open square corresponds to the mean of virgin control females from lines 1F–9F infected and plated alongside the females mated to 1M males. Mean refers to geometric mean, and error bars represent a 95% confidence interval. The parenthetical numbers on the x-axis are the DGRP stock identity number.
Our observation that females are highly genetically variable for the degree of post-mating immunosuppression they experience is consistent with a potential evolutionary trade-off between mating (and/or consequent reproduction) and immune defence. However, since we did not directly assay fitness in this experiment, we cannot definitively assess the possibility of an evolutionary trade-off. If such a trade-off does exist, the genetic variability we observe could reflect antagonistic pleiotropy coupled with spatial or temporal environmental variation. In this scenario, conflicting selective pressures related to immunity and reproduction could lead to maintenance of genetic variation (Gillespie & Turelli 1989; Lazzaro & Little 2009). The observation reported here and in McKean & Nunney (2005), Fedorka et al. (2007), Wigby et al. (2008) that mating induces susceptibility to some infections more than to others suggests that microbial heterogeneity might be one such example of environmental variation.
It has been hypothesized that ongoing sexual conflict could manifest in manipulation of female immune defence, such that males could potentially increase their fitness by reducing female immune defence in favour of reproduction (e.g. Lawniczak et al. 2007; Fedorka et al. 2007). However, the fact that we did not observe significant male genetic variation for post-mating female immunosuppression renders this hypothesis unlikely. Our data are not consistent with evolution of this trait being driven by ongoing sexual conflict.
4. Conclusions
In this work, we showed that female D. melanogaster become more susceptible to infection with two different natural bacterial pathogens after mating. Mated females sustained higher bacterial loads and lower survival compared with virgins. However, infection with two other pathogens was not more severe in mated females relative to virgins, revealing the mating effect to be pathogen-dependent. Wild females harbour substantial genetic variation for the magnitude of post-mating susceptibility they experience, but males harbour little if any genetic variability for the degree of immunosuppression they can drive. This effectively eliminates ongoing interlocus sexual conflict as a possible evolutionary scenario under which this trait could be evolving in the sampled population. It is much more likely that there is a physiological and perhaps evolutionary trade-off in females between reproduction and immune defence.
Acknowledgements
We are thankful to Maria Driscoll for invaluable laboratory assistance, and to Andrew Clark, Mariana Wolfner, Daniel Barbash, Punita Juneja, Madeline Galac, Jacob Crawford, Jennifer Comstock, Susan Rottschaefer and Stuart Wigby for helpful discussion and/or comments on the manuscript. We are also thankful to Francoise Vermeylen for statistical advice and Simona Despa and Steven Schwager for assistance with experimental design. This work was supported by NIH grant AI083932.
References
- Adamo S. A.2004How should behavioural ecologists interpret measurements of immunity? Anim. Behav. 68, 1443–1449 (doi:10.1016/j.anbehav.2004.05.005) [Google Scholar]
- Ayres J. S., Schneider D. S.2008A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS Biol. 6, 2764–2773 (doi:10.1371/journal.pbio.0060305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles J. F., et al. 2009Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41, 299–307 (doi:10.1038/ng.332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeale B., Brummel T., Seroude L.2004Immunity and aging: the enemy within? Aging Cell 3, 195–208 (doi:10.1111/j.1474-9728.2004.00106.x) [DOI] [PubMed] [Google Scholar]
- Devriese L., Baele M., Butaye P.2006The genus Enterococcus: taxonomy. In The prokaryotes, vol. 4 (ed. Dworkin M.), pp. 163–174 New York, NY: Springer [Google Scholar]
- Fedorka K. M., Zuk M., Mousseau T. A.2004Immune suppression and the cost of reproduction in the ground cricket, Allonemobius socius. Evolution 58, 2478–2485 (doi:10.1554/04-399) [DOI] [PubMed] [Google Scholar]
- Fedorka K. M., Linder J. E., Winterhalter W., Promislow D.2007Post-mating disparity between potential and realized immune response in Drosophila melanogaster. Proc. R. Soc. B 274, 1211–1217 (doi:10.1098/rspb.2006.0394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellowes M. D. E., Kraaijeveld A. R., Godfray H. C. J.1999The relative fitness of Drosophila melanogaster (Diptera, Drosophilidae) that have successfully defended themselves against the parasitoid Asobara tabida (Hymenoptera, Braconidae). J. Evol. Biol. 12, 123–128 (doi:10.1046/j.1420-9101.1999.00018.x) [Google Scholar]
- Gillespie J. H., Turelli M.1989Genotype-environment interactions and the maintenance of polygenic variation. Genetics 121, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraaijeveld A. R., Godfray H. C.1997Trade-off between parasitoid resistance and larval competitive ability in Drosophila melanogaster. Nature 389, 278–280 (doi:10.1038/38483) [DOI] [PubMed] [Google Scholar]
- Lawniczak M. K., Begun D. J.2004A genome-wide analysis of courting and mating responses in Drosophila melanogaster females. Genome 47, 900–910 (doi:10.1139/g04-050) [DOI] [PubMed] [Google Scholar]
- Lawniczak M. K., Barnes A. I., Linklater J. R., Boone J. M., Wigby S., Chapman T.2007Mating and immunity in invertebrates. Trends Ecol. Evol. 22, 48–55 (doi:10.1016/j.tree.2006.09.012) [DOI] [PubMed] [Google Scholar]
- Lazzaro B. P., Little T. J.2009Immunity in a variable world. Phil. Trans. R. Soc. B 364, 15–26 (doi:10.1098/rstb.2008.0141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B. P., Sceurman B. K., Clark A. G.2004Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science 303, 1873–1876 (doi:10.1126/science.1092447) [DOI] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J.2007The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (doi:10.1146/annurev.immunol.25.022106.141615) [DOI] [PubMed] [Google Scholar]
- Manos J., Belas R.2006The genera Proteus, Providencia, and Morganella. In The prokaryotes, vol. 6 (ed. Dworkin M.), pp. 245–269 New York, NY: Springer [Google Scholar]
- McGraw L. A., Gibson G., Clark A. G., Wolfner M. F.2004Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 14, 1509–1514 (doi:10.1016/j.cub.2004.08.028) [DOI] [PubMed] [Google Scholar]
- McKean K. A., Nunney L.2001Increased sexual activity reduces male immune function in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 98, 7904–7909 (doi:10.1073/pnas.131216398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKean K. A., Nunney L.2005Bateman's principle and immunity: phenotypically plastic reproductive strategies predict changes in immunological sex differences. Evolution 59, 1510–1517 (doi:10.1554/04-657) [PubMed] [Google Scholar]
- McKean K. A., Yourth C. P., Lazzaro B. P., Clark A. G.2008The evolutionary costs of immunological maintenance and deployment. BMC Evol. Biol. 8, 76–95 (doi:10.1186/1471-2148-8-76) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moret Y., Schmid-Hempel P.2000Survival for immunity: the price of immune system activation for bumblebee workers. Science 290, 1166–1168 (doi:10.1126/science.290.5494.1166) [DOI] [PubMed] [Google Scholar]
- Mueller J. L., Ravi Ram K., McGraw L. A., Bloch Qazi M. C., Siggia E. D., Clark A. G., Aquadro C. F., Wolfner M. F.2005Cross-species comparison of Drosophila male accessory gland protein genes. Genetics 171, 131–143 (doi:10.1534/genetics.105.043844) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Zipperlen P., Kubli E.2005Drosophila sex-peptide stimulates female immune system after mating via the toll and imd pathways. Curr. Biol. 15, 1690–1694 (doi:10.1016/j.cub.2005.08.048) [DOI] [PubMed] [Google Scholar]
- Rice W. R.1996Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381, 232–234 (doi:10.1038/381232a0) [DOI] [PubMed] [Google Scholar]
- Rolff J., Siva-Jothy M. T.2002Copulation corrupts immunity: a mechanism for a cost of mating in insects. Proc. Natl Acad. Sci. USA 99, 9916–9918 (doi:10.1073/pnas.152271999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzenbach G. A., Hosken D. J., Ward P. I.2005Sex and immunity in the yellow dung fly Scathophaga stercoraria. J. Evol. Biol. 18, 455–463 (doi:10.1111/j.1420-9101.2004.00820.x) [DOI] [PubMed] [Google Scholar]
- Sheldon B. C., Verhulst S.1996Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol. Evol. 11, 317–321 (doi:10.1016/0169-5347(96)10039-2) [DOI] [PubMed] [Google Scholar]
- Shoemaker K. L., Parsons N. M., Adamo S. A.2006Mating enhances parasite resistance in the cricket Gryllus texensis. Anim. Behav. 71, 371–380 (doi:10.1016/j.anbehav.2005.05.007) [Google Scholar]
- Siva-Jothy M. T., Tsubaki Y., Hooper R. E.1998Decreased immune response as a proximate cost of copulation and oviposition in a damselfly. Physiol. Entomol. 23, 274–277 (doi:10.1046/j.1365-3032.1998.233090.x) [Google Scholar]
- Sokal R. R., Rohlf F. J.1995Biometry, 3rd edn, pp. 413–415 New York, NY: W. H. Freeman and Company [Google Scholar]
- Swanson W. J., Clark A. G., Waldrip-Dail H. M., Wolfner M. F., Aquadro C. F.2001Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl Acad. Sci. USA 98, 7375–7379 (doi:10.1073pnas.1315681983) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley M. C., Blanford S., Jiggins F. M.2006Genetic variation in Drosophila melanogaster pathogen susceptibility. Parasitology 132, 767–773 (doi:10.1017/S0031182006009929) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J.199116S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigby S., Domanitskaya E. V., Choffat Y., Kubli E., Chapman T.2008The effect of mating on immunity can be masked by experimental piercing in female Drosophila melanogaster. J. Insect Physiol. 54, 414–420 (doi:10.1016/j.jinsphys.2007.10.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaher T. L., Parsek M. R.2006Pseudomonas aeruginosa In The prokaryotes, vol. 6 (ed. Dworkin M.), pp. 704–713 New York, NY: Springer [Google Scholar]
- Ye Y. H., Chenoweth S. F., McGraw E. A.2009Effective but costly, evolved mechanisms of defense against a virulent opportunistic pathogen in Drosophila melanogaster. PLoS Pathog. 5, e1000 385 (doi:10.1371/journal.ppat.1000385) [DOI] [PMC free article] [PubMed] [Google Scholar]