Abstract
Plasmablastic lymphoma (PBL) is an aggressive lymphoma characterized by a terminally differentiated B-cell phenotype that usually occurs in immunocompromised or elderly patients. Although the clinical and pathological characteristics of these tumours have been defined the genetic alterations involved in their pathogenesis are not well known. In this study we have investigated the chromosomal alterations of MYC, BCL2, BCL6, MALT1, PAX5, and IGH loci using fluorescence in situ hybridization (FISH) in 42 plasmablastic lymphomas (PBL) and 3 extracavitary primary effusion lymphomas (PEL). MYC rearrangements were identified in 20 of 41 (49%) PBL and the immunoglobulin (IG) genes were the partners in most tumours. MYC rearrangements were more common in EBV-positive (14 of 19, 74%) than EBV-negative (9 of 21, 43%) tumours (p < 0.05). No rearrangements of BCL2, BCL6, MALT1 or PAX5 were detected in any PBL but gains of these loci were observed in 31-41% of the cases examined. Twelve of the 40 PBL in which 3 or more loci could be investigated had multiple simultaneous gains in 3 or more loci. No differences in the survival of the patients according to MYC were observed but the four patients with the longest survival (>50 months) had no or low number of gains (<3). No rearrangements of any of these loci were seen in PEL. In conclusion, PBL are genetically characterized by frequent IG/MYC translocations and gains in multiple chromosomal loci. The oncogenic activation of MYC in these lymphomas may be an important pathogenetic element associated with EBV infection.
Keywords: Plasmablastic lymphoma, MYC, FISH
Introduction
Plasmablastic lymphoma (PBL) is an aggressive lymphoma characterized by a diffuse proliferation of large B-cells usually with immunoblastic morphology and the immunophenotype of a terminally differentiated B-cell with loss of mature B-cell markers and expression of plasma cell related antigens.35 This tumor was initially recognized by Delecluse et al. and proposed as a subtype of diffuse large B-cell lymphoma (DLBCL) presenting in the oral cavity of HIV-infected individuals.16 Subsequent studies have recognized that PBL may present in other locations, mainly extranodal sites, and in patients with other immunodeficiency conditions such as iatrogenic immunosuppression associated with post-transplant therapy or chronic treatments for autoimmune diseases, and aging.14, 33
The clinical and pathological features of PBL have been well defined, and this tumor is now recognized in the current WHO classification. It is distinguished from usual diffuse large B-cell lymphomas, NOS (DLBCL) and other forms of large B-lymphomas expressing a terminally B-cell differentiation phenotype such as ALK positive large B-cell lymphoma, primary effusion lymphoma, and large cell lymphoma arising in multicentric Castelman's disease.35 However, the genetic and molecular mechanisms that may be involved in the pathogenesis of PBL are not well known. One important element in the development of the tumor is the infection of the neoplastic cells by Epstein-Barr virus (EBV), which is present in most the cases with a latency type I.14, 16, 17, 21 Immunohistochemical studies have observed the lack of expression of the cell cycle inhibitors p27 and p16 and strong expression of p53 in a number of cases suggesting that inactivation of these genes may contribute to the high rate of proliferation commonly seen in PBL.40
Conventional and molecular cytogenetic studies have identified a number of primary and secondary chromosomal aberrations associated with many types of lymphoid neoplasms including large B-cell lymphomas 4 but the genetic alterations of PBL have not been fully explored. Some studies have identified complex karyotypes including the presence of MYC rearrangements in occasional cases. However, these reports have included only isolated cases or small series of tumors.7, 13, 15, 36 Thus, the goal of this study was to more fully characterize the cytogenetic alterations that may be involved in the pathogenesis of PBL. We have investigated a large series of these tumors by fluorescence in situ hybridization (FISH) using probes covering genes and chromosomal regions frequently altered in aggressive B-cell lymphomas.
Material and Methods
Case Selection
Forty-two cases of PBL were retrieved from the files of the laboratories of Pathology of the Hospital Clinic of Barcelona, Spain, National Cancer Institute, Bethesda, MD, Howard University Hospital of Washington, DC, and Rikshospitalet-Radiumhospitalet Medical Center of Oslo, Norway. The two cases from Howard University were previously published in part. 7, 13, 15 36 All the tumors were classified as PBL according to the WHO classification35 and further typified as PBL, monomorphic or “oral type” and PBL with plasmacytic differentiation as previously described.14 We also included in the study three extracavitary primary effusion lymphomas (PEL).12 Formalin-fixed and paraffin-embedded tissue was available in all the cases. Clinical information, including previous medical history, clinical presentation, and follow-up of the patients, was obtained to the extent possible from the referring pathologists and clinicians (Table 1).
Table 1. Clinical and pathological features of plasmablastic lymphomas and plasma cell neoplasms with plasmablastic features.
| All PBL cases (n=42) | PBL Monomorphic (n=28) | PBL plasmacytic differentiation (n=14) | PEL (n=3) | |
|---|---|---|---|---|
| Age (range) | 48 (11-86) | 47 (11-86) | 49 (35-82) | 56 (49-62) |
| Sex (M:F) | 33:8 | 22:6 | 11:2 | 3:0 |
| Immunodeficiency | ||||
| HIV+ | 27/37 (73%) | 20/26 (77%) | 8/11 (72%) | 3/3 (100%) |
| Other | 2/37 (5%) | 1/26 (4%)† | 1/11 (9%)‡ | 0/3 |
| Location | ||||
| Nodal | 12/42 (29%) | 6/28 (21%) | 6/14 (43%) | 3/3 (100%)┼ |
| Extranodal | 30/42 (71%) | 22/28 (79%) | 8/14 (57%) | 0 |
| Oral & URT | 11/30 (37%) | 8/22 (36%) | 3/8 (38%) | 0 |
| GI tract | 6/30 (20%) | 5/22 (23%) | 1/8 (13%) | 0 |
| Skin | 3/30 (10%) | 3/22 (14%) | 0 | 0 |
| Soft tissues | 6/30 (20%) | 3/22 (14%) | 3/8 (38%) | 0 |
| Other | 4/30 (13%) | 3/22 (14%) | 1/8 (13%) | 0 |
| IHC | ||||
| CD20+ | 0/39 | 0/25 | 0/14 | 1/3 (33%)** |
| CD79a+ | 14*/32 (66%) | 8*/21 (67%) | 6*/14 (50%) | 1/3 (33%) |
| PAX5+ | 1/38 (3%) | 1/26 (4%) | 0/13 | 0/3 |
| MUM1+ | 20/20 (100%) | 16/16 (100%) | 4/4 | 1/1 |
| CD138+ | 26/32 (81%) | 17/22 (77%) | 9/10 (90%) | 2/3 (67%) |
| CD56+ | 7/37 (19%) | 2/23 (9%) | 5/14 (36%) | 0/3 |
| Viral status | ||||
| EBER+ | 24/41 (59%) | 17/27 (63%) | 7/14 (50%) | 2/3 (67%) |
| LMP1+ | 2/31 (6%) | 2/22 (9%) | 0/9 | 0/3 |
| EBNA2+ | 0/18 | 0/14 | 0/4 | - |
| HHV8+ | 0/27 | 0/16 | 0/11 | 3/3 |
PBL: plasmablastic lymphoma; PEL: primary effusion lymphoma; URT: upper respiratory tract mucosa; G.I.: gastrointestinal tract
Crohn's disease;
kidney transplant;
1 case had simultaneous ascites
Seven additional cases had weak and focal expression
Weak expression
Immunohistochemistry and in situ hybridization
Immunohistochemical studies were performed with a panel of monoclonal and polyclonal antibodies reactive in paraffin-embedded tissue sections using a peroxidase-labeled detection system, standard antigen retrieval protocols, and an automated immunostainer (Ventana Medical System, Tucson, AZ or Dako Autostainer, Dako, Copenhagen, Denmark), as previously described.14 The panel of antibodies used included common B-cell markers such as CD20 (clone L26), CD79a (clone JCB117) and PAX5 (clone 24), MUM1/IRF4 (clone MUM1p), CD138 (clones MI15, 5F-7, B-A38), and CD56 (clones 123C3, NCF-CD56-1B6), from DAKO and Ventana suppliers.
The presence of the Epstein-Barr virus (EBV) genome was examined by in situ hybridization (ISH) to detect EBV-encoded early nuclear RNAs (EBER1 and EBER2) as previously described.3 The EBV antigens LMP-1 and EBNA-2 were examined by immunohistochemistry using the CS1-4 (Dako) and NCL-EBV-Pe2 (Novocastra) antibodies, respectively. The antigen of latency (LANA-1) of HHV-8 was also studied by immunohistochemistry using the clone LN53 (Advanced Biotechnologies).
Conventional cytogenetics
Conventional cytogenetics was available in three cases, two of them (cases 33 and 36) previously published, and the studies were performed as described.36
Fluorescence “in situ” hybridization (FISH)
FISH was performed on 3-4μm thick sections of formalin-fixed paraffin-embedded tissues, using split-signal DNA probes (Dako) specific for the following loci: 8q24 (MYC), 18q21 (BCL2), 18q21 (MALT1), 9p13 (PAX5), 3q27 (BCL6) and 14q32 (IGH). In the cases with MYC rearrangements, we performed additional studies with a dual color-dual fusion MYC/IGH/CEP8 translocation probe (Abbott Molecular, Illinois, USA).
The FISH technique was performed following the guidelines of the Euro-FISH project as previously described (www.euro-fish.org).39 Briefly, a dewaxing step was followed by a pre-treatment process at 95°C, digestion with pepsin, washes with wash buffer, and dehydration with gradual ethanol dilutions. For the split probes, the slides were air dried and 10μl of the probe was applied in every sample. For the fusion probe, a mix of 7 μl of buffer solution, 2 μl of distillated water and 1 μl of probe were used. Denaturation at 82°C for 5 minutes and incubation at 45°C overnight were performed for the split probes. For the fusion probe the denaturation step was 3 minutes at 90°C, and the incubation 37°C overnight. The slides were then transferred to a stringency buffer at 65°C, washed in buffer solution, dehydrated, air dried and mounted with Fluorescent Mounting Medium containing DAPI (Dako). The slides were stored in the dark at 2-8°C until read. An Olympus BX51 microscopy (Olympus, Tokyo, Japan) was used.
The cut-off values for the interphase FISH analyses were established following the criteria of Ventura et al.41 Tonsil sections were used as controls, and for each sample 100 to 200 evaluable nuclei with complete FISH signals were scored. For the break-apart probes, the cut-off values were 3% for the detection of the rearrangements, and 6% to detect gains. For the dual fusion probe we considered a cut-off of 11% for both structural and numerical abnormalities.
Statistical Analysis
Categorical data were compared using Fisher's exact test, two-sided P value, whereas for ordinal data non-parametric tests were used. The actuarial survival analysis was performed according to the method described by Kaplan and Meier. Statistical analysis was done using the SPSS software package version 15 (SPSS, Chicago, IL).
Results
Clinical Characteristics
The main clinical data of the 42 PBL and 3 extracavitary variants of PEL are summarized in Table 1 and Table 2. Patients with PBL were predominantly males (81%) and had a median age of 48 years (range 11-86). HIV was positive in 27 (73%) of 37 patients with available information. Two HIV negative patients developed PBL after iatrogenic immunosuppression for Crohn's disease and renal transplantation, respectively. Five patients were older than 65 years, HIV negative, and did not have other recognized causes of immunodeficiency. Thirty (71%) of the 42 PBL were presented in extranodal sites. The most common single site was the oral mucosa (37%) followed by the gastrointestinal tract (20%), soft tissue (20%) and skin (10%). The clinical presentation in monomorphic PBL and tumors with plasmacytic differentiation was relatively similar with a tendency of the latter to have nodal involvement (43% vs 21%) but the difference was not statistically significant (Table 1). The clinical follow-up was available in 16 patients, 8 monomorphic PBL and 8 showing plasmacytic differentiation (Table 2). No significant differences in the overall survival of the patients with these two variants were observed.
Table 2. Clinical features and FISH results.
| Case | Age/Gender | Immuno defiency. | Sites involved | Follow-up (months) | MYC | IGH | MYC/IGH | BCL2 | BCL6 | MALT1 | PAX5 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Monomorphic PBL | |||||||||||
| 1 | 41/M | HIV+ | Oral mucosa | N | N | Ns | N | N | N | N | |
| 2 ‡ | 48/M | HIV+ | Oral mucosa | R | R | R | N | N | N | N | |
| 3 ‡ | 56/F | HIV+ | Oral mucosa | G | N | Ns | G | G | G | N | |
| 4 ‡ | 55/M | HIV+ | Oral mucosa | Dead (7) | R | R | R | G | G | G | G |
| 5 ‡ | 22/M | HIV + | Oral mucosa | R | G | Ns | G | G | G | Nv | |
| 6 | 83/M | HIV- | Oral mucosa | G | Nv | Ns | Nv | Nv | Nv | Nv | |
| 7 | 42/M | HIV+ | Hypopharynx | N | N | Ns | N | N | N | N | |
| 8 | 51/M | HIV+ | Gall bladder | R | R | R | N | N | N | N | |
| 9 | 46/M | u. | Gastric mucosa | N | N | Ns | N | Nv | N | N | |
| 10 | 42/M | HIV+ | Small bowel | R | R | R | N | N | N | N | |
| 11 ‡ | 86/F | HIV - | Large bowel | Dead (4) | G | G | Ns | G | G | G | Nv |
| 12 ‡ | 47/M | HIV+ | Anal mass | R | R | R | N | N | N | N | |
| 13 ‡ | 11/F | HIV+ | Skin of gastrostomy | R | R | R | N | N | N | N | |
| 14 | 31/M | Crohn | Skin | N | N | Ns | N | N | N | N | |
| 15 | 40/M | HIV+ | Skin | R | R | Ns | N | N | N | N | |
| 16 | 78/M | HIV- | Soft tissue (Thorax) | N | N | Ns | N | N | N | N | |
| 17 | 54/M | HIV- | Soft tissue (Thorax) | Alive (22) | N | N | Ns | N | G | N | N |
| 18 | 39/M | HIV+ | Soft tissue (Abdomen) | Dead (<1) | R | R | R | N | G | N | G |
| 19 | 42/F | HIV+ | Breast | Dead (<1) | R | R | Ns | N | N | N | N |
| 20 | 30/F | HIV- | Vulva | Alive (50) | R | R | R | N | N | N | N |
| 21 | 53/M | HIV - | Thyroid gland | R | R | R | N | Nv | N | N | |
| 22 | 55/M | u. | LN | G | N | Ns | G | G | G | N | |
| 23 | 49/M | HIV+ | LN | N | N | Ns | N | N | Nv | N | |
| 24 | 50/M | HIV+ | LN | R | R | R | N | N | Nv | N | |
| 25 | 41/M | HIV+ | LN | Dead (6) | R | R | R | N | N | N | G |
| 26 | 34/M | HIV+ | LN | R | Nv | R | Nv | G | G | G | |
| 27 | 44/F | HIV+ | Gingiva-jawbone | Alive (83) | R | N | Ns | Nv | Nv | Nv | G |
| 28 ‡ | 47/M | HIV+ | LN | N | N | Ns | N | N | N | Nv | |
| PBL with plasma cell differentiation | |||||||||||
| 29 | 42/M | HIV+ | Oral mucosa | Alive (4) | R | N | Ns | G | N | N | N |
| 30 | 48/M | u. | Nasopharynx | N | N | Ns | N | N | N | N | |
| 31 | 67/M | HIV- | Tonsil | N | N | Ns | N | N | N | G | |
| 32 | 35/M | HIV+ | Rectum | Dead (15) | N | N | Ns | N | N | N | N |
| 33 | 45/M | HIV+ | Soft tissue | Dead (<1) | R | G | N+ | G | G | G | G |
| 34 | u/u | u. | Soft tissue(temporal lesion) | N | R | Ns | N | G | N | N | |
| 35 | 49/F | u. | Soft tissue(forehead) | G | G | Ns | G | G | G | G | |
| 36 | 37/M | HIV+ | Testes, pleural effusion | Dead (24) | R | R | R | G | G | G | G |
| 37 | 53/M | HIV+ | LN | R | R | R | N | G | N | N | |
| 38 | 35/M | HIV+ | LN | Alive (55) | Nv | Nv | Ns | N | N | Nv | N |
| 39 | 82/M | HIV- | LN | Dead (41) | G | G | Ns | G | G | G | G |
| 40 | u/M | Transplant* | LN | Dead (14) | G | G | Ns | G | G | G | G |
| 41 | 37/M | HIV+ | LN | G | G | Ns | G | G | G | G | |
| 42 | 59/F | HIV+ | LN | Alive (105) | N | N | Ns | N | N | Nv | Nv |
| Primary effusion lymphoma | |||||||||||
| 43 | u/M | HIV+ | LN | N | N | Ns | N | N | Nv | N | |
| 44 | 49/M | HIV+ | Soft tissues | Alive (36) | G | N | Ns | N | N | N | N |
| 45 | 62/M | HIV+ | LN | Dead (2) | N | N | Ns | G | N | G | G |
N: no alterations; R: rearrangement; G: gains; Nv: not evaluable; Ns: not studied; u:unknown; LN: lymph node; BM: bone marrow;
kidney transplant;
case with t(2;8); IGH/MYC negative, but IGH gained
are included in Taddesse-Heath et al[12].
All the patients alive at the last follow up were free of disease.
All the patients with PEL were HIV positive. One patient had a solitary paravertebral mass from T12 to L2 (case 44). No other sites of involvement were detected and the patient was alive with no evidence of disease 3 years later. Two patients (cases 43 and 45) presented with a large cervical lymphadenopathy (case 43) and simultaneous axillary and retroperitoneal enlarged lymph nodes (case 45). In case 45, clinical information was available and the patient died rapidly after the diagnosis (2 months).
Pathological features
The 42 PBL were further subclassified as monomorphic PBL (28 cases) and PBL with plasmacytic differentiation (14 cases) as previously defined.14 All cases were negative for CD20 whereas CD79a was expressed in 14 cases (Table 1). PAX-5 was only positive in one case that also expressed focal CD79a. All the other cases were PAX-5 negative or weakly positive. MUM-1 and CD138 were positive in 20 (100%) and 26 (81%) of the tumors examined, respectively, with a similar proportion in monomorphic PBL and tumors with plasmacytic differentiation. CD56 was expressed in 5 (36%) of the 14 PBL with plasmacytic differentiation but only in 2 (9%) of the 23 monomorphic PBL examined (p<0.05). EBV was positive in 24 (59%) of the 41 PBL with a similar distribution in the monomorphic and plasmacytic variants (Table 1). Most of the cases had a latency type 1 with expression of EBER but negative for LMP1 and EBNA2. Only two monomorphic PBL showed occasional cells positive for LMP1.
The extracavitary PEL were cytologically composed of large immunoblasts and plasmablasts admixed with more occasional anaplastic cells. One nodal case (case 43) showed a predominantly sinusoidal involvement whereas the other had a tumoral and destructive pattern of growth. All cases expressed the HHV-8 latency antigen LANA-1 and EBV was present in two of the three cases with a type I latency.
Conventional cytogenetics
Conventional cytogenetics were available in one monomorphic PBL (case 21) and 2 PBL with plasma cell differentiation (cases 33 and 36). Two of these cases were previously reported (cases 33, 36).36 All had complex karyotypes with MYC rearrangements. In case 16 the partner of the 8q24.1 rearrangement was the 2p12 region where the immunoglobulin kappa light chain locus is located, whereas in the other two cases MYC was rearranged with the immunoglobulin heavy chain locus. The complete karyotypes of the cases are shown in supplementary table 1.
FISH analysis in plasmablastic lymphomas
The results of the FISH studies are summarized in table 3. We first studied the MYC locus with a dual-color break-apart probe. MYC rearrangements were observed in 20 (49%) of the 41 PBL evaluable. These rearrangements were more frequent in monomorphic PBL (16 of 28, 57%) than in PBL with plasmacytic differentiation (4 of 13, 31%), but this difference was not statistically significant.
Table 3. Frequency of the genetic alterations detected by FISH.
| Probe | Alteration | All PBL cases | PBL Monomorphic | PBL plasmacytic differentiation | PEL |
|---|---|---|---|---|---|
| MYC | Gain | 8/41 (20%) | 4/28 (15%)† | 4/13 (31%)† | 1/3 (33%) |
| Rearrangement | 20/41 (49%) | 16/28 (57%) | 4/13 (31%) | 0/3 | |
| BCL2 | Gain | 12/39 (31%) | 5/25 (20%) | 7/14 (50%) | 1/3 (33%) |
| Rearrangement | 0/39 | 0/25 | 0/14 | 0/3 | |
| BCL6 | Gain | 16/38 (41%) | 8/24 (33%) | 8/14 (57%) | 0/3 |
| Rearrangement | 0/38 | 0/24 | 0/14 | 0/3 | |
| MALT1 | Gain | 12/36 (33%) | 6/24 (25%) | 6/12 (50%) | 1/2 (50%) |
| Rearrangement | 0/36 | 0/24 | 0/12 | 0/2 | |
| PAX5 | Gain | 12/37 (32%) | 5/24 (21%) | 7/13 (54%) | 1/3 (33%) |
| Rearrangement | 0/36 | 0/23 | 0/13 | 0/3 | |
| IGH | Gain | 7/38 (18%) | 2/25 (8%) | 5/13 (38%) | 0/3 |
| Rearrangement | 16/38 (42%) | 13/25 (52%) | 3/13 (23%) | 0/3 | |
| ≥ 3 gains | 12/40 (30%) | 6/26 (23%) | 6/14 (43%) | 0/3 |
One case with MYC translocated and gained is included in the category of rearrangements
To determine whether the IGH gene was the possible partner of the rearranged MYC, we investigated the MYC/IGH dual color-dual fusion probe in 15 cases (table 2). MYC was translocated with the IGH locus in 14 cases, 12 monomorphic PBL and 2 PBL with plasmacytic differentiation (Figure 1). Two monomorphic PBL in which the fusion probe could not be examined had both MYC and IGH rearrangements with the respective split probes and a normal pattern of the other loci (BCL2, BCL6, MALT1, PAX5) suggesting that IGH could be also the MYC partner in these cases. Four cases had a non-IGH MYC rearrangement and one of them had the t(2;8) detected by conventional cytogenetics. Thus, MYC rearrangements with IG gene loci were confirmed in 15 cases and suggested in two additional tumors. Only one case with a split in the IGH locus did not have MYC rearrangements (case 34). No rearrangements of the other loci studied were observed in this case.
FIGURE 1. HE and FISH images of case 13.
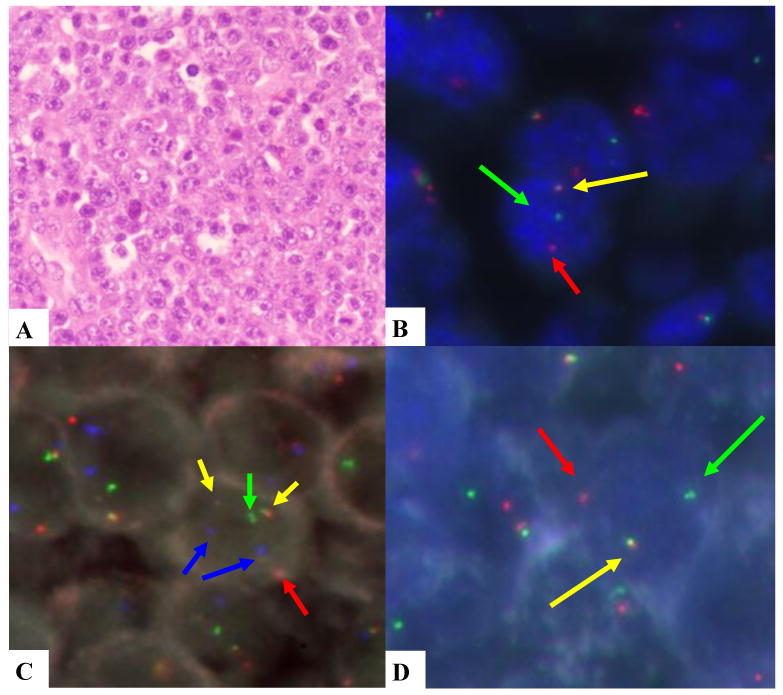
A. monomorphic PBL. B. MYC rearranged (break apart probe). C. IGH/MYC dual fusion probe; yellow signals indicate the IGH/MYC translocation, the green signal is the normal IGH locus, the red signal is the normal MYC locus and the blue signals are the CEP8. D. IGH rearranged (break apart probe).
The MYC rearrangement was the unique genetic alteration observed in 10 of the 39 PBL in which at least 3 of the other loci (BCL2, BCL6, MALT1 and PAX5) could be examined (table 2). All cases with MYC rearrangement as a single alteration were monomorphic PBL. MYC rearrangements were more commonly found in the EBV-positive cases (14 of 19, 74%) than in the EBV-negative cases (9 of 21, 43%) (p < 0.05). The median overall survival of patients with and without MYC rearrangements was 7 and 15 months, respectively. This difference was not statistically significant.
No rearrangements of BCL2, BCL6, MALT1 or PAX5 were detected in any PBL but gains of these loci were observed in 31-41% of the cases examined (Table 3). Gains of PAX5 were significantly more common in PBL with plasmacytic differentiation (7 of 13, 54%) than in monomorphic tumors (5 of 24, 21%) (p=0.041). No other associations were observed. Twelve of the 40 PBL in which 3 or more loci could be investigated had multiple simultaneous gains in 3 or more loci, whereas single gains were detected only in 3 tumors (Fig 2). Multiple gains were more frequent in PBL with plasmacytic differentiation (43%) than in the monomorphic variant (23%), but the difference was not statistically significant (Table 3). BCL2 and MALT1 loci were evaluable simultaneously in 31 cases. Coamplification of both loci was observed in 11 tumors and only one additional case had gains of BCL2 but not of MALT1. The median overall survival of patients with 3 or more gains was 7 months whereas it was not reached in patients with fewer than 3 gains. The difference was not statistically significant, probably due to the low number of patients. Of note, however, the 4 patients with the longest survival (>50 months) had no or fewer than 3 gains.
FIGURE 2. HE and FISH images of case 35.
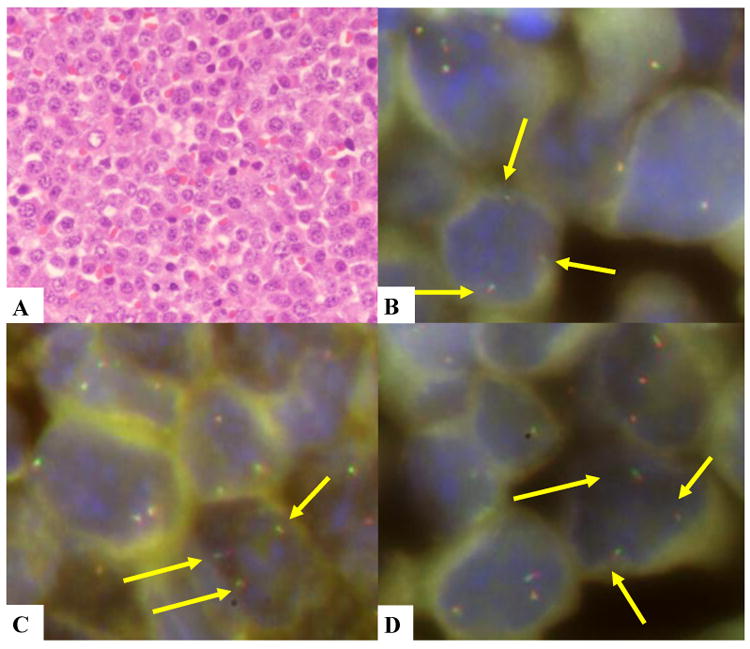
A. PBL with plasmacytic differentiation. B. MYC gained. C. BCL6 gained. D. PAX5 gained. Break apart probes were used in these images.
FISH analysis in primary effusion lymphomas
No rearrangements in any of the loci examined were observed in the three extracavitary PEL. One case had concomitant gains in BCL2, MALT1 and PAX5. Isolated gains of MYC were observed in an additional tumor.
Discussion
Plasmablastic lymphoma is recognized as a distinct category of aggressive lymphoma with specific clinical and pathological features.35 However, little is known about the molecular and cytogenetic abnormalities of this tumor. The incidence of PBL is low and conventional cytogenetics or other techniques are difficult to perform because of the frequent lack of availability of fresh or frozen samples. For these reasons we have conducted a genetic study of these lymphomas using FISH and a panel of probes covering the genes and chromosomal regions most frequently altered in aggressive large B-cell lymphomas.
MYC rearrangements were the most common recurrent structural chromosomal alteration identified in PBL. They occurred in almost 50% of the cases and the immunoglobulin (IG) genes were the MYC partners in most tumors. We identified the t(8;14) using the fusion MYC/IGH probe in the 14 cases that could be examined. In two additional tumors simultaneous split signals of the MYC and IGH loci were observed, with the breakapart probes suggesting also the presence of the MYC/IGH rearrangement. In one of the 4 cases in which the MYC rearrangement was associated with a non-rearranged IGH locus, a t(2;8) translocation was demonstrated by conventional cytogenetics. Thus, these results indicate that IG genes were the partner of MYC rearrangements in at least 17 of the 20 (85%) PBL. These findings are consistent with previous observations of MYC rearrangements in single case reports of PBL13, 15, 42 or small series.7, 36 IG/MYC rearrangements were detected in the 9 cases in which both loci were examined. Conventional cytogenetic studies in PBL are very limited but the previous 7 cases published and our new reported case have been concordant with the FISH observations and show that the MYC translocations occur in the context of very complex karytotypes.7, 13, 36, 42
In addition to MYC translocations, PBL had frequent gains of the other loci examined with 30% of the cases showing gains in three or more of them. These findings are consistent with the complex karyotypes observed in the few conventional cytogenetic studies available. MYC rearrangements were seen more frequently in EBV+ PBL and tumors with a monomorphic morphology, but this latter difference was not statistically significant. Although the number of patients with follow-up information was limited, no differences in the survival related to these genetic alterations were observed. Interestingly, some of our patients exhibited long survival and all of them had no or very few gains in the loci studied. Although the initial descriptions of PBL highlighted the aggressive behavior and poor outcome, some recent reports have observed longer survivals similar to some of our patients.11 This improvement in outcome has been related to new antiretroviral treatments, better immunological status of the patients, and improvements in the supportive care and delivery of chemotherapy.
MYC rearrangement is a genetic feature characteristic of Burkitt's lymphoma (BL) and more recently has been also recognize in subsets of DLBCL with very aggressive clinical behavior.25, 31, 34 Our results indicate that MYC translocations are also a common finding in PBL. Although all three of these lymphomas share the presence of MYC rearrangements, the incidence, MYC partner and genomic context in which this translocation emerges are different. Thus, in BL MYC translocation is present in virtually all cases, the partner is an IG gene, usually IGH and less frequently IGK or IGL, and the translocation is the sole genetic alteration or is associated with very simple karyotypes. By contrast, a MYC translocation has been reported in 4-10% of DLBCL, usually associated with complex karyotypes23, 32, and in around 40% of the cases MYC is rearranged to a non-IG partner.23 MYC rearrangements have been also associated with the t(14;18) or BCL6 rearrangements in a subset of very aggressive lymphomas now recognized in the WHO classification as the provisional category of B-cell lymphoma unclassifiable with features intermediate between DLBCL and BL.13, 35 These translocations are associated with very complex karyotypes.23, 32, 34 Interestingly, the majority of the subtypes of lymphoma with MYC translocations have a germinal centre phenotype. Thus, PBL carrying the MYC translocation show significant differences from other tumors that are MYC-translocation positive. The translocation is relatively common in PBL (50%), it is associated with complex karytotypes but without other translocations considered as “double hits”, the partner is an IG locus, and the translocation occurs in a lymphoma with a terminal B-cell phenotype.
In addition to the MYC translocation we found gains in three or more loci in 30% of the PBL. These gains tended to be more common in tumors with plasmacytic differentiation than in the monomorphic variant. Although 3q27 (BCL6) and 18q21 (BCL2 or MALT1) translocations are common in other types of DLBCL and are associated with the MYC translocation in the “double hit” tumors,38 PBL do not have rearrangements in any of these loci. Gains of chromosome 3, particularly 3q, and 18q are common features of DLBCL of the GCB and ABC types, respectively.5 However, these alterations are mutually exclusive in most of these tumors.5 On the contrary, PBL have frequently simultaneous gains of these loci, reinforcing the idea that these tumors are genetically distinct from conventional DLBCL. Moreover, a recent study shows a different profile of gains in DLBCL arising in HIV-positive patients. This study, based on SNPs array-CGH, obtains a very low frequency of gains of the genes included in our series.10
Plasmablastic lymphoma, particularly the plasmacytic variant, has morphological and immunophenotypic features overlapping with extramedullary plasmablastic transformation of plasma cell neoplasms.14, 36, 40 The clinical context, immunodeficiency status of the patient and EBV infection of the tumor are characteristics that may help to distinguish these two entities. MYC translocations in primary extramedullary plasmacytomas are uncommon but polysomies and IG gene breaks not associated with MYC are frequent.6 In multiple myeloma (MM), MYC rearrangements have been detected in 0-15% of unselected cases2, 29, 37 but they may be seen in 45% of advanced MM and in 65% of MM cell lines19, 20 indicating that this may be an acquired alteration associated with progression of the disease. However, about 40% of these MYC translocations involve a non-IG partner, a finding different from the frequent IG/MYC rearrangements observed in PBL. On the other hand, MM have frequent rearrangements of the IGH locus with partner other than MYC (i.e. CCND1, FGFR3, CCND3, MAF)19, 20 whereas IGH rearrangements in PBL seem to occur almost exclusively with MYC.
Primary effusion lymphomas also share morphological and immunophenotypic characteristics with PBL but the clinical context and the tumor infection by HHV8 are distinctive features. Genetic studies of PEL have shown complex karyotypes but not rearrangements of MYC, BCL2, BCL6 or other recurrent alterations.1, 8, 9MYC translocations have been detected, however, in some cases of DLBCL presenting clinically as primary effusion lymphomas but the mature B-cell phenotype and lack of HHV8 in these tumors rule out the diagnosis of PEL.18, 24
The high frequency of IG/MYC translocations in PBL suggest that MYC activation must play an important role in the pathogenesis of these tumors. Most lymphomas with MYC rearrangements have a germinal center B-cell phenotype whereas in PBL the activation of this gene occurs in the context of a terminal B-cell differentiation stage. One of the master regulators of plasma cell differentiation is the transcription factor BLIMP1 and this transcription factor is highly expressed in PBL.3, 22, 30 BLIMP1 represses genes that maintain mature B-cell identity such as PAX5, and promotes the expression of genes such as XBP1 involved in the plasma cell differentiation program. BLIMP1 also represses genes related to cell proliferation and growth including MYC.28 The frequent incidence of MYC translocation in PBL may be an oncogenic mechanism that overcomes the repressor effect of BLIMP1 on MYC expression.
The IG/MYC translocations in PBL were frequently associated with EBV infection. This finding is similarly to BL in which EBV is also a common finding, particularly in endemic and immunodeficiency associated tumors.13, 35 The cooperative mechanisms between MYC and EBV in lymphomagenesis are not clear.27 Interestingly, experimental studies in tumor cell lines have shown that the full latency III program of EBV infection is incompatible with MYC-driven proliferation.26 Consistent with this in vitro observation, EBV infection in both PBL and BL has a latency I program. Further studies will be required to elucidate the relationship between EBV and MYC activation in PBL.
In conclusion, our study demonstrates that MYC translocations occur in 50% of PBL suggesting that it may play an important role in the pathogenesis of these tumors. MYC is usually rearranged with IG genes and the translocation occurs in the context of complex karyotypes. BCL2, BCL6, MALT1 and PAX5 loci are frequently gained but not rearranged in these tumors. These findings indicate that PBL have a specific profile of genetic alterations that differs from that seen in other aggressive B-cell malignancies.
Supplementary Material
Acknowledgments
We thank Elena Gonzalvo, Concha Muñoz and Laura Gelabert for their excellent technical assistance. We also acknowledge R.Ramos from Hospital Son Dureta, Mallorca, Spain, J.Miró from Clínica Girona, Girona, Spain, M.J.Panadés from Hospital Arnau de Vilanova, Lleida, Spain, C. Martín from Hospital de Sant Camil, Barcelona, Spain, S. Martínez from Hospital Joan XXIII, Tarragona, Spain, and A. Ferrández from Hospital Clínic of Valencia, Spain for their collaboration providing samples of patients with PBL diagnosis.
This study was supported by the Spanish Ministry of Science and Innovation SAF2008/3630, the Instituto de Salud Carlos III “Red Temática de Investigación Cooperativa de Cancer” RD07/0020/2004, Asociación Española Contra el Cáncer AECC_07_011 and Instituto de Salud Carlos III, Fondo de Investigaciones Sanitarias PI080095.
Footnotes
No conflicts of interest were declared.
References
- 1.Ascoli V, Mastroianni CM, Galati V, et al. Primary effusion lymphoma containing human herpesvirus 8 DNA in two AIDS patients with Kaposi's sarcoma. Haematologica. 1998;83:8–12. [PubMed] [Google Scholar]
- 2.Avet-Loiseau H, Gerson F, Magrangeas F, et al. Rearrangements of the c-myc oncogene are present in 15% of primary human multiple myeloma tumors. Blood. 2001;98:3082–6. doi: 10.1182/blood.v98.10.3082. [DOI] [PubMed] [Google Scholar]
- 3.Balague O, Martinez A, Colomo L, et al. Epstein-Barr virus negative clonal plasma cell proliferations and lymphomas in peripheral T-cell lymphomas: a phenomenon with distinctive clinicopathologic features. Am J Surg Pathol. 2007;31:1310–22. doi: 10.1097/PAS.0b013e3180339f18. [DOI] [PubMed] [Google Scholar]
- 4.Bea S, Campo E. Secondary genomic alterations in non-Hodgkin's lymphomas: tumor-specific profiles with impact on clinical behavior. Haematologica. 2008;93:641–5. doi: 10.3324/haematol.13030. [DOI] [PubMed] [Google Scholar]
- 5.Bea S, Zettl A, Wright G, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183–90. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bink K, Haralambieva E, Kremer M, et al. Primary extramedullary plasmacytoma: similarities with and differences from multiple myeloma revealed by interphase cytogenetics. Haematologica. 2008;93:623–6. doi: 10.3324/haematol.12005. [DOI] [PubMed] [Google Scholar]
- 7.Bogusz AM, Seegmiller AC, Garcia R, et al. Plasmablastic lymphomas with MYC/IgH rearrangement: report of three cases and review of the literature. Am J Clin Pathol. 2009;132:597–605. doi: 10.1309/AJCPFUR1BK0UODTS. [DOI] [PubMed] [Google Scholar]
- 8.Boulanger E, Agbalika F, Maarek O, et al. A clinical, molecular and cytogenetic study of 12 cases of human herpesvirus 8 associated primary effusion lymphoma in HIV-infected patients. Hematol J. 2001;2:172–9. doi: 10.1038/sj.thj.6200096. [DOI] [PubMed] [Google Scholar]
- 9.Bubman D, Guasparri I, Cesarman E. Deregulation of c-Myc in primary effusion lymphoma by Kaposi's sarcoma herpesvirus latency-associated nuclear antigen. Oncogene. 2007;26:4979–86. doi: 10.1038/sj.onc.1210299. [DOI] [PubMed] [Google Scholar]
- 10.Capello D, Scandurra M, Poretti G, et al. Genome wide DNA-profiling of HIV-related B-cell lymphomas. Br J Haematol. 2010;148:245–55. doi: 10.1111/j.1365-2141.2009.07943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castillo J, Pantanowitz L, Dezube BJ. HIV-associated plasmablastic lymphoma: lessons learned from 112 published cases. Am J Hematol. 2008;83:804–9. doi: 10.1002/ajh.21250. [DOI] [PubMed] [Google Scholar]
- 12.Chadburn A, Hyjek E, Mathew S, et al. KSHV-positive solid lymphomas represent an extra-cavitary variant of primary effusion lymphoma. Am J Surg Pathol. 2004;28:1401–16. doi: 10.1097/01.pas.0000138177.10829.5c. [DOI] [PubMed] [Google Scholar]
- 13.Chuah KL, Ng SB, Poon L, et al. Plasmablastic lymphoma affecting the lung and bone marrow with CD10 expression and t(8;14)(q24;q32) translocation. Int J Surg Pathol. 2009;17:163–6. doi: 10.1177/1066896908316785. [DOI] [PubMed] [Google Scholar]
- 14.Colomo L, Loong F, Rives S, et al. Diffuse large B-cell lymphomas with plasmablastic differentiation represent a heterogeneous group of disease entities. Am J Surg Pathol. 2004;28:736–47. doi: 10.1097/01.pas.0000126781.87158.e3. [DOI] [PubMed] [Google Scholar]
- 15.Dawson MA, Schwarer AP, McLean C, et al. AIDS-related plasmablastic lymphoma of the oral cavity associated with an IGH/MYC translocation--treatment with autologous stem-cell transplantation in a patient with severe haemophilia-A. Haematologica. 2007;92:e11–e12. doi: 10.3324/haematol.10933. [DOI] [PubMed] [Google Scholar]
- 16.Delecluse HJ, Anagnostopoulos I, Dallenbach F, et al. Plasmablastic lymphomas of the oral cavity: a new entity associated with the human immunodeficiency virus infection. Blood. 1997;89:1413–20. [PubMed] [Google Scholar]
- 17.Dong HY, Scadden DT, de Leval L, et al. Plasmablastic lymphoma in HIV-positive patients: an aggressive Epstein-Barr virus-associated extramedullary plasmacytic neoplasm. Am J Surg Pathol. 2005;29:1633–41. doi: 10.1097/01.pas.0000173023.02724.1f. [DOI] [PubMed] [Google Scholar]
- 18.Fujisawa S, Tanioka F, Matsuoka T, et al. CD5+ diffuse large B-cell lymphoma with c-myc/IgH rearrangement presenting as primary effusion lymphoma. Int J Hematol. 2005;81:315–8. doi: 10.1532/IJH97.04155. [DOI] [PubMed] [Google Scholar]
- 19.Gabrea A, Leif BP, Michael KW. Distinguishing primary and secondary translocations in multiple myeloma. DNA Repair (Amst) 2006;5:1225–33. doi: 10.1016/j.dnarep.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Gabrea A, Martelli ML, Qi Y, et al. Secondary genomic rearrangements involving immunoglobulin or MYC loci show similar prevalences in hyperdiploid and nonhyperdiploid myeloma tumors. Genes Chromosomes Cancer. 2008;47:573–90. doi: 10.1002/gcc.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaidano G, Cerri M, Capello D, et al. Molecular histogenesis of plasmablastic lymphoma of the oral cavity. Br J Haematol. 2002;119:622–8. doi: 10.1046/j.1365-2141.2002.03872.x. [DOI] [PubMed] [Google Scholar]
- 22.Garcia JF, Roncador G, Garcia JF, et al. PRDM1/BLIMP-1 expression in multiple B and T-cell lymphoma. Haematologica. 2006;91:467–74. [PubMed] [Google Scholar]
- 23.Hummel M, Bentink S, Berger H, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–30. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 24.Ichinohasama R, Miura I, Kobayashi N, et al. Herpes virus type 8-negative primary effusion lymphoma associated with PAX-5 gene rearrangement and hepatitis C virus: a case report and review of the literature. Am J Surg Pathol. 1998;22:1528–37. doi: 10.1097/00000478-199812000-00010. [DOI] [PubMed] [Google Scholar]
- 25.Johnson NA, Savage KJ, Ludkovski O, et al. Lymphomas with concurrent BCL2 and MYC translocations: the critical factors associated with survival. Blood. 2009;114:2273–9. doi: 10.1182/blood-2009-03-212191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly GL, Milner AE, Baldwin GS, et al. Three restricted forms of Epstein-Barr virus latency counteracting apoptosis in c-myc-expressing Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 2006;103:14935–40. doi: 10.1073/pnas.0509988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klapproth K, Wirth T. Advances in the understanding of MYC-induced lymphomagenesis. Br J Haematol. 2010;149:484–97. doi: 10.1111/j.1365-2141.2010.08159.x. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–9. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- 29.Mohamed AN, Bentley G, Bonnett ML, et al. Chromosome aberrations in a series of 120 multiple myeloma cases with abnormal karyotypes. Am J Hematol. 2007;82:1080–7. doi: 10.1002/ajh.20998. [DOI] [PubMed] [Google Scholar]
- 30.Montes-Moreno S, Gonzalez-Medina AR, Rodriguez Pinilla SM, et al. Aggressive large B cell lymphoma with plasma cell differentiation: immunohistochemical characterization of plasmablastic lymphoma and diffuse large B cell lymphoma with partial plasmablastic phenotype. Haematologica. 2010 doi: 10.3324/haematol.2009.016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niitsu N, Okamoto M, Miura I, et al. Clinical features and prognosis of de novo diffuse large B-cell lymphoma with t(14;18) and 8q24/c-MYC translocations. Leukemia. 2009;23:777–83. doi: 10.1038/leu.2008.344. [DOI] [PubMed] [Google Scholar]
- 32.Salaverria I, Zettl A, Bea S, et al. Chromosomal alterations detected by comparative genomic hybridization in subgroups of gene expression-defined Burkitt's lymphoma. Haematologica. 2008;93:1327–34. doi: 10.3324/haematol.13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawyer JR, Lukacs JL, Munshi N, et al. Identification of new nonrandom translocations in multiple myeloma with multicolor spectral karyotyping. Blood. 1998;92:4269–78. [PubMed] [Google Scholar]
- 34.Snuderl M, Kolman OK, Chen YB, et al. B-cell lymphomas with concurrent IGH-BCL2 and MYC rearrangements are aggressive neoplasms with clinical and pathologic features distinct from Burkitt lymphoma and diffuse large B-cell lymphoma. Am J Surg Pathol. 2010;34:327–40. doi: 10.1097/PAS.0b013e3181cd3aeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC; Lyon: 2008. [Google Scholar]
- 36.Taddesse-Heath L, Meloni-Ehrig A, Scheerle J, et al. Plasmablastic lymphoma with MYC translocation: evidence for a common pathway in the generation of plasmablastic features. Mod Pathol. 2010 doi: 10.1038/modpathol.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tchinda J, Volpert S, Kropff M, et al. Frequent gains of the short arm of chromosome 9 in multiple myeloma with normal G-banded karyotype detected by comparative genomic hybridization. Am J Clin Pathol. 2004;122:875–82. doi: 10.1309/5KWK-P6UK-GNXX-HMYH. [DOI] [PubMed] [Google Scholar]
- 38.Tomita N, Tokunaka M, Nakamura N, et al. Clinicopathological features of lymphoma/leukemia patients carrying both BCL2 and MYC translocations. Haematologica. 2009;94:935–43. doi: 10.3324/haematol.2008.005355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van RA, Mason D, Jones M, et al. Translocation detection in lymphoma diagnosis by split-signal FISH: a standardised approach. J Hematop. 2008;1:119–26. doi: 10.1007/s12308-008-0017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vega F, Chang CC, Medeiros LJ, et al. Plasmablastic lymphomas and plasmablastic plasma cell myelomas have nearly identical immunophenotypic profiles. Mod Pathol. 2005;18:806–15. doi: 10.1038/modpathol.3800355. [DOI] [PubMed] [Google Scholar]
- 41.Ventura RA, Martin-Subero JI, Jones M, et al. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn. 2006;8:141–51. doi: 10.2353/jmoldx.2006.050083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yotsumoto M, Ichikawa N, Ueno M, et al. CD20-negative CD138-positive leukemic large cell lymphoma with plasmablastic differentiation with an IgH/MYC translocation in an HIV-positive patient. Intern Med. 2009;48:559–62. doi: 10.2169/internalmedicine.48.1359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


