Abstract
PALI (release 1.2) contains three-dimensional (3-D) structure-dependent sequence alignments as well as structure-based phylogenetic trees of homologous protein domains in various families. The data set of homologous protein structures has been derived by consulting the SCOP database (release 1.50) and the data set comprises 604 families of homologous proteins involving 2739 protein domain structures with each family made up of at least two members. Each member in a family has been structurally aligned with every other member in the same family (pairwise alignment) and all the members in the family are also aligned using simultaneous superposition (multiple alignment). The structural alignments are performed largely automatically, with manual interventions especially in the cases of distantly related proteins, using the program STAMP (version 4.2). Every family is also associated with two dendrograms, calculated using PHYLIP (version 3.5), one based on a structural dissimilarity metric defined for every pairwise alignment and the other based on similarity of topologically equivalent residues. These dendrograms enable easy comparison of sequence and structure-based relationships among the members in a family. Structure-based alignments with the details of structural and sequence similarities, superposed coordinate sets and dendrograms can be accessed conveniently using a web interface. The database can be queried for protein pairs with sequence or structural similarities falling within a specified range. Thus PALI forms a useful resource to help in analysing the relationship between sequence and structure variation at a given level of sequence similarity. PALI also contains over 653 ‘orphans’ (single member families). Using the web interface involving PSI_BLAST and PHYLIP it is possible to associate the sequence of a new protein with one of the families in PALI and generate a phylogenetic tree combining the query sequence and proteins of known 3-D structure. The database with the web interfaced search and dendrogram generation tools can be accessed at http://pa uling.mbu.iisc.ernet.in/~pali.
INTRODUCTION
The three-dimensional (3-D) structure of a protein could suggest a molecular basis for the function and biological role of the protein. Similarity in gross 3-D structures could exist for proteins with insignificant sequence similarity (1–3). Proteins with no similarity in their amino acid sequences, but with a common fold may or may not have similar function. However, 3-D structures of homologous proteins with clear sequence similarity have highly similar structures and often have similar biological roles in the living systems (for examples see 4–6).
Variation in the amino acid sequences of homologous proteins in a family is constrained by the high similarity in their structures and functions. This feature is exploited in the comparative modelling wherein a 3-D model of a protein is generated on the basis of homologues of known structure (7–9). Indeed comparative modelling is performed to model a large number of proteins coded in genomes (10,11).
The accuracy of the model, in terms of fine features of the 3-D structure, generated using comparative modelling could be low if the sequence similarity between the modelled protein and the basis structures is low (under ∼30%) (12). Hence utility of such a model is limited. This is an important problem, as the necessity to generate a 3-D model of a sequence of a new protein based on known structures of distantly related proteins is a common situation. There is a need for special attention to improve the comparative modelling for proteins with sequence identities, with known related structures, falling under the ‘twilight zone’ defined by Doolittle (13). With improvements in the recognition of protein folds in the absence of significant similarity in the amino acid sequences (14), there is a clear necessity to improve the accuracy of models generated using comparative modelling.
Some of the reasons for the low accuracy of the models generated on the basis of template structures with low sequence similarity with the modelled protein are (8):
(i) Inaccurate alignment between modelled protein and basis structures.
(ii) Difficulties in modelling insertion and deletion regions in the alignment
(iii) Variations in the lengths and geometry of helices and β-strands in distantly related proteins.
(iv) Major variations in the relative orientation of helices and β-sheets in the distantly related proteins
(v) High variations in the conformation of ‘equivalent’ sidechains within distantly related homologues.
Further, after association of the sequence of a new protein to more than one protein of known structure, it is not trivial to choose proper template structure(s) for comparative modelling. On the one hand, use of several template structures can maximise the chances of modelling various regions on the homologous structures and hence contribute towards the accuracy of the model. But, use of several basis structures can reduce the number and lengths of structurally conserved regions and can also reduce the number of conserved structural features that can be responsible for adding errors in the model (12). A proper balance between these factors can be achieved by identifying optimal set of basis structures that are usually most closely related to the protein to be modelled.
The PALI (Phylogeny and ALIgnment of homologous protein structures) database is a step towards learning rules relating variations in amino acid sequences and homologous structures. Incorporation of the rules in suites of programs for comparative modelling might improve the accuracy of the models.
PALI comprises a compendium of homologous protein structures with pairwise and multiple structural alignments and phylogenetic dendrograms. Thus, PALI forms a source of value-added information on protein structures to understand sequence determinants of the fold and evolution. PALI is also equipped with a user-friendly web interface for associating a query sequence with one of the PALI families and can also automatically generate a dendrogram combining the query sequence and homologous structures. This provides a quick overview of the relationships of the query sequence with homologues of known structure and also aids in choosing most closely related proteins to use as template structures in the comparative modelling of the query sequence. It is also possible to query PALI for pairs of proteins with a specific range of sequence and structural similarity. Thus, PALI can aid in investigations on the relationship between sequence variation and structural variation at various levels of sequence or structural similarity.
GENERAL FEATURES OF PALI
(i) Structural comparison of proteins is made at the level of domains using the domain boundaries suggested in SCOP. The fold and superfamily assignment by SCOP is documented for every family in PALI. The families are classified as α, β, α/β, α+β, small proteins and multi-domain systems.
(ii) in various families are available. Structure-dependent sequence alignments, quantification of sequence and structural similarity, and superposed coordinate sets are readily available for every superposition.
(iii) Structural similarity-based and structure-dependent, sequence similarity-based dendrograms for all the PALI families with at least three members are available.
(iv) A user-friendly tool is integrated with the database to identify pairs of proteins with a given range of sequence identity calculated using either all the residue–residue aligned positions or topologically equivalent residues.
(v) A search tool is available to recognise protein pairs with a specific range of structural similarity using either root mean square deviation (RMSD) of topologically equivalent Cα atoms or a structural distance metric which combines RMSD and number of equivalences.
(vi) Amino acid sequences of proteins in the single member families (‘orphans’) are available.
(vii) A PSI_BLAST (15) interface enables a sequence search to be made on all the proteins in PALI including ‘orphans’.
(viii) A dendrogram generation tool enables a query sequence to be associated with a family in PALI. If the associated family has at least two members then a dendrogram is automatically generated combining the query sequence and homologous structures.
Several of the features such as simultaneous availability of pairwise and multiple alignments, structural similarity and sequence similarity-based dendrograms and web-based tools to generate a dendrogram can be viewed as complementary to other databases of aligned homologous protein structures (6,16–20).
DATABASE STATISTICS
The list of homologous protein families and the members in each family have been extracted from the SCOP database (release 1.50) (2, 21). The number of families, with at least two proteins in a family, in the present release of PALI is 604. The number of protein domains in these families is 2739 and hence the average number of members per family is between 4 and 5. There are 230 families with only two members. The largest family is the globins with 37 members.
The families in PALI extracted from SCOP are characterised by six classes of protein folds: predominantly α, predominantly β, α/β, α+β, multi-domain and small proteins (other classes such as membrane and cell surface proteins, low resolution protein structures and peptides are not included in the PALI database). The number of families in these six classes of folds are 128, 134, 135, 138, 16 and 53, respectively. Hence the number of families in α, β, α/β and α+β classes are similar. PALI also contains single member families (‘orphans’) for the purposes of including them in the PSI_BLAST searches of query sequences. The number of ‘orphans’ in α, β, α/β, α+β, multidomain and small proteins classes of folds are: 146, 109, 157, 186, 13 and 42, respectively, making of total of 653 ‘orphans’.
There are 9510 pairwise structure-based alignments (taking two homologous proteins in a family) in the present version of PALI. Obviously the pairwise and multiple structural alignments are identical in families with two members. There are 374 families, with at least three members in each family, with multiple structural alignments performed by simultaneous superposition of all the structures in a family. The 9510 pairwise structure-based alignments in PALI correspond to 1 278 404 residue–residue positional alignments.
Pre-calculated dendrograms are available for all the families with at least three members in a family. The two types of dendrograms are available for each one of these families resulting in 748 dendrograms in the present release of PALI. The structure-based dendrogram has been generated from pairwise alignments using a structural distance metric (see below). The other dendrogram has been generated using sequence similarity obtained from pairwise structure-based alignments.
STRUCTURAL ALIGNMENTS
Pairwise and multiple structural alignments have been performed using the latest version (4.2) of the STAMP suite of programs developed by Russell and Barton (22). STAMP uses the procedure of Rossmann and Argos (23). STAMP aligns structures and produces a corresponding sequence alignment with confidence values associated with each aligned position. For the overwhelming majority of the alignments SCAN option of STAMP has been used to obtain a set of initial equivalences followed by preliminary alignment which is optimised by STAMP to obtain the final rigid body superposition of topologically equivalent Cα atoms in the structures. Although the procedure is automated to suit the large-scale application as in setting-up PALI, the result files of the superposition program have been manually inspected to ensure that there are no erroneous results. The results of STAMP include superposed coordinate sets of proteins and also several other parameters that characterise the similarity between the protein structures considered. These include (i) the number of residues in the proteins compared; (ii) number of fitted Cα atoms; (iii) number of topologically equivalent Cα atoms defined by a distance cut-off and structural similarity in the flanking residues; (iv) root mean square deviation (RMSD); (v) STAMP score; (vi) percentage sequence identity for the structure-based sequence alignment; (vii) percentage secondary structure identity; and (viii) confidence values at various residue positions. Most of these parameters are stored and available in PALI.
One of the common measures of structural divergence between two homologous protein structures is RMSD of topologically equivalent Cα atoms. Length dependence can be seen in structural comparisons using RMSD (for examples see 24). Further, identical RMSD in two superpositions does not guarantee the same extent of structural divergences since the number of topologically equivalent Cα atoms in the two pairs could be highly different. Hence, a combination of RMSD and number of equivalent Cα atoms could give a better picture about structural divergence between two proteins. In addition to calculating various similarity measures in STAMP we have also calculated Structural Distance Metric (SDM) (25,26) for every pairwise alignment in PALI. SDM combines the RMSD and the number of equivalences and it was defined by Johnson et al. (25,26) as:
SDM = –100 × log [(w1 × SRMS) + (w2 × PFTE)]
Where SRMS = 1–RMSD (in Å)/3.0 and PFTE = (Number of equivalent Cα atoms)/(Number of residues in the smallest protein)
w1 = (1–SRMS + 1–PFTE)/2 and w2 = (SRMS + PFTE)/2
The definition of the weights w1 and w2 are such that SDM is more effective representation than RMSD particularly in the case of distantly related protein structures.
Multiple structural alignments for representative families in PALI have been compared with the alignments obtained from COMPARER (27,28) which uses structural features such as solvent accessibility and secondary structures and relationships such as hydrogen bonding. The alignments from PALI (performed using a rigid-body superposition program, STAMP) are virtually identical to those from COMPARER except for families with <25% sequence identity. In the low sequence similarity cases the differences in alignments occurred only in and around loops that are known to be variable among homologous proteins. Every pairwise alignment has been compared with the alignment extracted from the multiple structural superposition involving all the members in the family. For the overwhelming majority of pairs the two kinds of alignments are almost identical (S.Balaji and N.Srinivasan, manuscript submitted).
Sequence identities for every pair of homologous proteins has been calculated using three methods: (i) using all the aligned positions; (ii) using the topologically equivalent residues defined by 3 Å cut-off distance between Cα atoms; and (iii) using topologically equivalent residues as defined by STAMP. STAMP considers the extent of structural similarity of the flanking residues as well in defining the topological equivalence of the aligned residues.
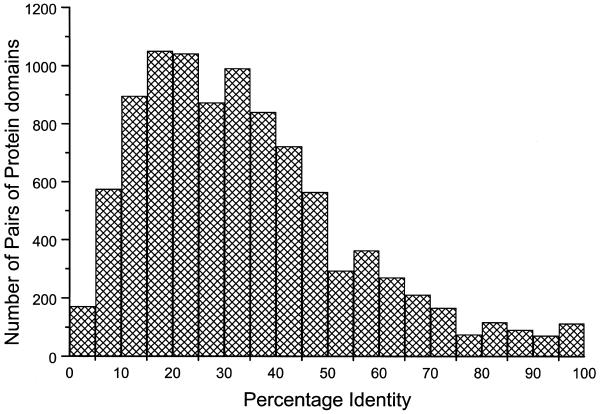
Figure 1 shows a histogram of the number of pairwise alignments represented at every 5% of the sequence identity calculated using the topologically equivalent Cα atoms defined by the 3 Å cut-off distance after optimal superposition. Although the PALI consists of homologous proteins that are expected to often have high sequence identity (over ∼35%), the entire range of 0–100% is represented. Indeed peaks with a comparable number of examples occur in the ranges 15–20% and 20–25%, which are in the twilight zone defined by Doolittle (13). The number of pairwise alignments between 0 and 35% is 5601 representing ∼59% of all the pairwise alignments. Hence most of the pairwise alignments have structure-based sequence identities under the common threshold of 35% for homology detection. Analyses of variation in various structural features in pairwise aligned protein structures at difference ranges of sequence identity could provide rules useful in improved comparative model building.
Figure 1.
Histogram showing the distribution of number of pairwise alignments in PALI at every 5% interval of sequence identity.
DENDROGRAMS FOR PROTEIN FAMILIES
Structure-based and structure-dependent, sequence-based phylogenetic tree diagrams have been generated for every family in PALI, with at least three members. The PHYLIP package of programs (version 3.5) has been used for the purpose (29). The KITSCH program employing the Fitch–Margoliash and least squares methods was used to generate phylogenies. The input to structure-based phylogeny of a family is a symmetric matrix of SDM between various proteins in the family. The percentage sequence non-identity matrix has been generated considering residues corresponding to topologically equivalent Cα atoms in the two proteins. Such a matrix has been used to generate structure-dependent, sequence-based phylogeny. The dendrograms were generated using the program DRAWGRAM within the suite of PHYLIP.
ACCESS TO THE PALI DATABASE AND INTERFACED TOOLS
PALI can be accessed at http://pauling.mbu.iisc.ernet.in/~pali. A specific family may be reached by browsing or by a search using key words. Pairwise structure-based sequence alignments are available for all the two-member families. Pairwise and multiple structural alignments are separately available for all the families with at least three members. Structural meaning at the level of residue–residue alignment is indicated for every aligned position, using the output from STAMP. Superimposed coordinate sets along with various details of overall structural and sequence similarities are provided for every alignment in PALI.
Searches can be made for pairs of proteins falling within a specific range of sequence identity or structural similarity. The user has the option of choosing the kind of sequence identity (calculated using all the aligned positions or by considering topologically equivalences only) or structural similarity (SDM or RMSD). Links are available to the appropriate protein families in the output of searches.
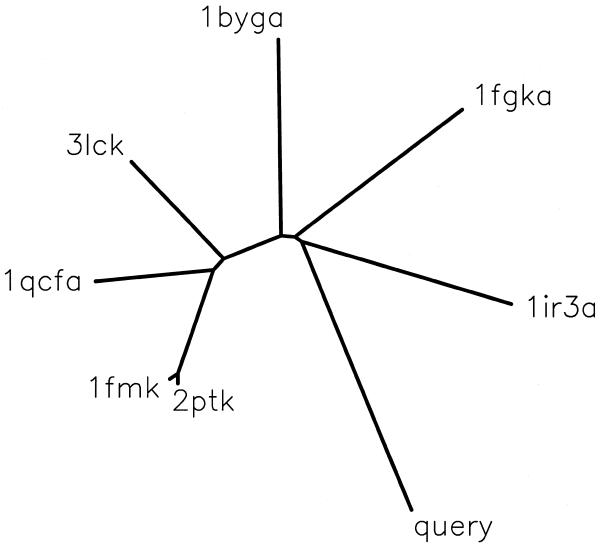
A PSI_BLAST (15) interface allows query sequences to be searched against all the proteins (including ‘orphans’) in PALI. The user may also generate a dendrogram combining a query sequence with members of a family in PALI. For this purpose the PSI_BLAST is employed with a stringent cut-off of 0.0005 for the E-value in order to add to the reliability of the family assignment of the new sequence. Once a family can be assigned the query sequence is aligned with the members in the family and the sequence dissimilarity of the query sequence with each one of the homologues of known structure is calculated. The pre-calculated sequence dissimilarity between homologues of known structures defined using structural alignments is maintained while generating a dendrogram, using PHYLIP package (29). Figure 2 shows the sample result for the protein kinase-like domain of guanylyl cyclase C, which has no experimental structure available yet. The protein domain could be associated with the family of tyrosine kinases and the codes given in the figure correspond to the tyrosine kinases of known 3-D structure. It can be seen that insulin receptor kinase is closest to the query sequence and is likely to be the best template structure to model the 3-D structure of the query sequence. The dendrogram also provides an overview of the query sequence in relation to the homologues of known structure.
Figure 2.
Dendrogram combining a query sequence and homologues of known 3-D structure. The query sequence corresponds to that of protein kinase-like domain of guanylyl cyclase receptor. The protein data bank (32) codes correspond to various tyrosine kinases of known structure: 1ir3, insulin receptor kinase; 1fgk, fibroblast growth factor receptor kinase 1; 1byg, human csk src kinase; 3lck, Lck kinase; 1qcf, Hck kinase; 1fmk, Human c-src kinase; 2ptk, chicken src kinase. The fifth character in the code, where present, corresponds to the chain identifier in the protein data bank file.
OUTLOOK
In the future PALI will be periodically updated with the new releases of SCOP. We will also make a number of improvements in the database such as inclusion of protein domain superfamilies. Different measures of structural divergence such as the one suggested by Levitt and Gerstein (30) will be provided. Recently RasMol (31) has been interfaced with PALI to facilitate interactive view and analysis of structural overlays. While PALI can be accessed at http://pauling.mbu.iisc.ernet.in/~pali, the database containing machine-parseable files may be obtained from the authors upon request.
The ongoing work aims at associating every family and orphans in the database with new gene products being suggested from genome sequencing projects. The objective is to align the amino acid sequences of putative protein domains coded in genome sequences with homologues of known structures using a variety of homology detection and alignment tools. Phylogenetic tree diagrams combining protein domains, coded in genome sequences and known 3-D structural families in PALI will also be generated.
The analysis of variation in several structural features of homologous proteins in PALI is also in progress with a view to devising rules in improved comparative modelling of protein structures.
Acknowledgments
ACKNOWLEDGEMENTS
S.B. and S.S. are supported by Council of Scientific and Industrial Research, India and The Wellcome Trust, UK, respectively. This research is supported by the award of International Senior Fellowship in Biomedical Sciences to N.S. from the Wellcome Trust, UK.
References
- 1.Holm L. and Sander,C. (1994) The FSSP database of structurally aligned protein fold families. Nucleic Acids Res., 22, 3600–3609. [PMC free article] [PubMed] [Google Scholar]
- 2.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 3.Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M (1997) CATH–a hierarchic classification of protein domain structures. Structure, 5, 1093–1108. [DOI] [PubMed] [Google Scholar]
- 4.Lesk A.M. and Chothia,C. (1980) How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J. Mol. Biol., 136, 225–270. [DOI] [PubMed] [Google Scholar]
- 5.Chothia C. and Lesk,A.M. (1986) The relation between the divergence of sequence and structure in proteins. EMBO J., 5, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overington,J.P., Johnson,M.S., ali,A. and Blundell,T.L. (1990) Tertiary structural constraints on protein evolutionary diversity: templates, key residues and structure prediction. Proc R. Soc. Lond., 241, 132–145. [DOI] [PubMed] [Google Scholar]
- 7.Johnson M.S., Srinivasan,N., Sowdhamini,R. and Blundell,T.L. (1994) Knowledge-based protein modeling. Crit. Rev. Biochem. Mol. Biol., 29, 1–68. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan N., Guruprasad,K. and Blundell,T.L. (1996) In Sternberg,M.J.E. (ed.) Protein Structure Prediction–A Practical Approach. Oxford University Press, Oxford, UK, pp. 111–140.
- 9.Sanchez R. and ali,A. (1997) Advances in comparative protein-structure modelling. Curr. Opin. Struct. Biol., 7, 206–214. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez R. and ali,A. (1998) Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl Acad. Sci. USA, 95, 13597–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ali A. (1998) 100,000 protein structures for the biologist. Nat. Struct. Biol., 5, 1029–1032. [DOI] [PubMed] [Google Scholar]
- 12.Srinivasan N. and Blundell,T.L. (1993) An evaluation of the performance of an automated procedure for comparative modelling of protein tertiary structure. Protein Eng., 6, 501–512. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle R.F. (1981) Similar amino acid sequences: chance or common ancestry? Science, 214, 149–159. [DOI] [PubMed] [Google Scholar]
- 14.Jones D.T. (2000) Protein structure prediction in the postgenomic era. Curr. Opin. Struct. Biol., 10, 371–379. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhanng,Z., Miller,W. and Lipman,D.J.(1997) N Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sander C. and Schneider,R. (1991) Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 17.ali A. and Overington,J.P. (1994) Derivation of rules for comparative protein modeling from a database of protein structure alignments. Protein Sci., 3, 1582–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pascarella S., Milpetz,F. and Argos,P. (1996) A databank (3D-ali) collecting related protein sequences and structures. Protein Eng., 9, 249–251. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt R., Gerstein,M. and Altman,R. (1997). LPFC: an Internet library of protein family core structures. Protein Sci., 6, 246–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuguchi K., Deane,C.M., Blundell,T.L. and Overington,J.P. (1998) HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci., 7, 2469–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo Conte L., Ailey,B., Hubbard,T.J.P., Brenner,S.E., Murzin,A.G. and Chothia,C. (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res., 28, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell R.B. and Barton,G.J. (1992) Multiple protein sequence alignment from tertiary structure comparison: assignment of global and residue confidence levels. Proteins, 2, 309–323. [DOI] [PubMed] [Google Scholar]
- 23.Rossmann M.G. and Argos,P. (1976) Exploring structural homology of proteins. J. Mol. Biol., 105, 75–95. [DOI] [PubMed] [Google Scholar]
- 24.Swindells M.B. (1996) Detecting structural similarities: a user’s guide. Methods Enzymol., 266, 643–653. [DOI] [PubMed] [Google Scholar]
- 25.Johnson M.S., Sutcliffe,M.J. and Blundell,T.L. (1990) Molecular anatomy: phyletic relationships derived from three-dimensional structures of proteins. J. Mol. Evol., 1, 43–59. [DOI] [PubMed] [Google Scholar]
- 26.Johnson M.S., ali,A. and Blundell,T.L. (1990) Phylogenetic relationships from three-dimensional protein structures. Methods Enzymol., 183, 670–690. [DOI] [PubMed] [Google Scholar]
- 27.ali A. and Blundell,T.L. (1990) Definition of general topological equivalence in protein structures. A procedure involving comparison of properties and relationships through simulated annealing and dynamic programming. J. Mol. Biol., 212, 403–428. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Z.-Y., ali,A. and Blundell,T.L. (1992) A variable gap penalty function and feature weights for protein 3-D structure comparisons. Protein Eng., 5, 43–51. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. (1995) PHYLIP (Phylogeny Inference Package) Version 3.57c. Department of Genetics, University of Washington, Seattle, WA.
- 30.Levitt M. and Gerstein,M. (1998) A unified structural framework for sequence comparison and structure comparison. Proc. Natl Acad. Sci. USA, 95, 5913–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayle R.A. and Milner-White,E.J. (1995) RasMol: Biomolecular graphics for all. Trends Biochem. Sci., 20, 374–376. [DOI] [PubMed] [Google Scholar]
- 32.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. Updated article in this issue: Nucleic Acids Res. (2001), 29, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]