Abstract
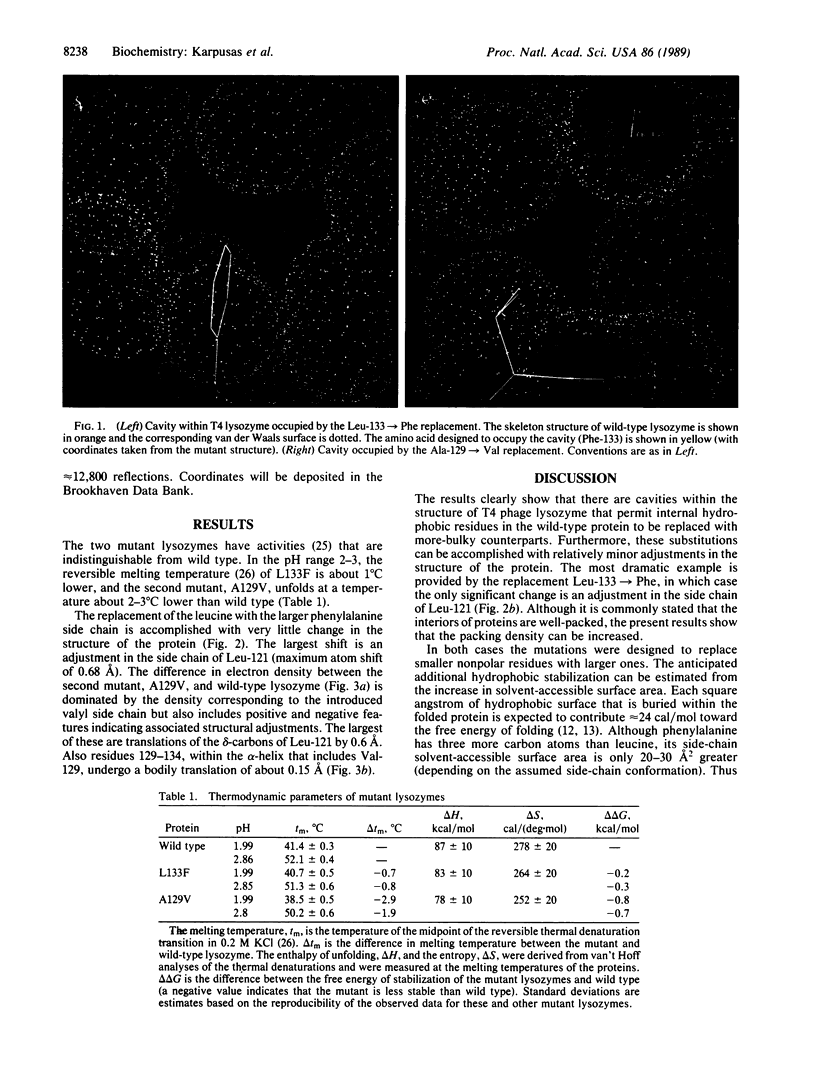
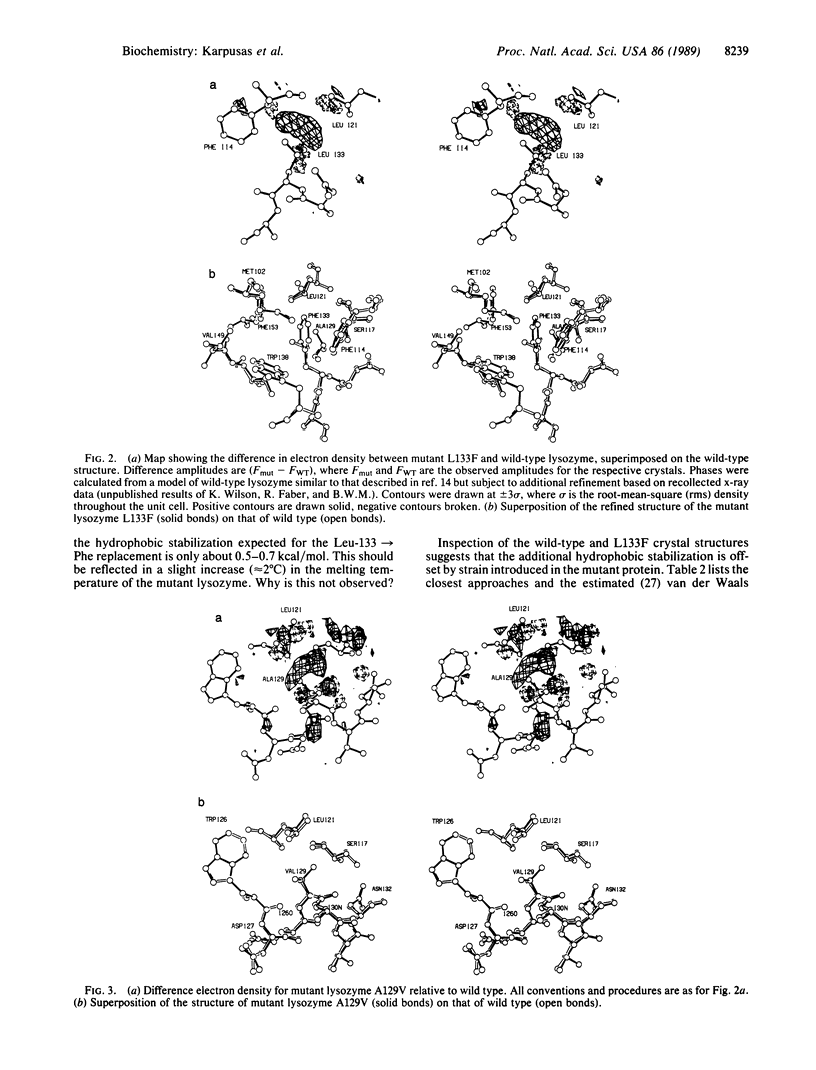
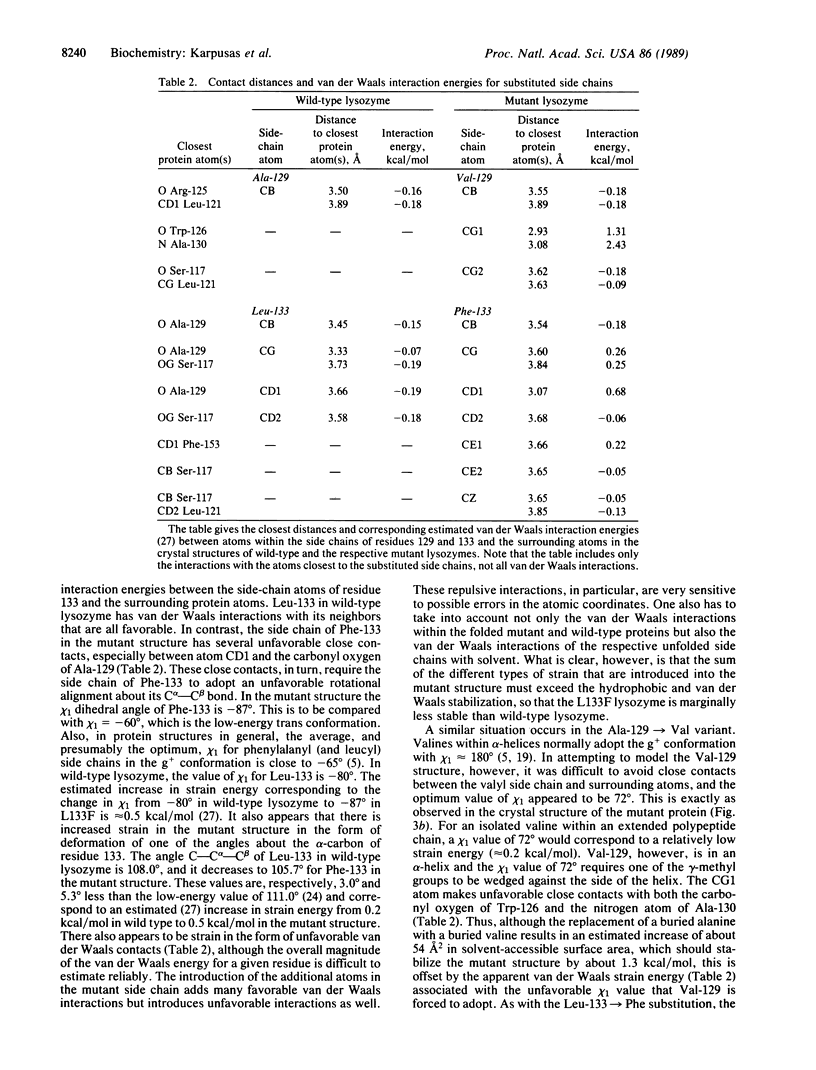
To probe the nature of the hydrophobic cores of proteins and to test potential ways of increasing protein thermostability, an attempt was made to improve the packing within T4 bacteriophage lysozyme by engineered amino acid replacements. Two mutations, Leu-133----Phe and Ala-129----Val, which were designed to fill the largest cavities that exist in the folded structure of the native protein, were constructed. The mutant proteins have normal activities and their thermal stabilities are marginally lower than that of wild-type lysozyme. Crystal structure analysis of the mutant proteins shows that the introduced amino acids are accommodated with very little perturbation of the three-dimensional structure. Incorporation of the more bulky hydrophobic residues within the core of the protein is expected to provide an increase in hydrophobic stabilization, but this is seen to be offset by the introduction of strain. Inspection of the mutant structures shows that in each case the introduced amino acid side chain is forced to adopt a non-optimal dihedral angle X1. Strain is also observed in the form of bond angle distortion and in unfavorable van der Waals contacts. The results illustrate how the observed core structures of proteins represent a compromise between the hydrophobic effect, which will tend to maximize the core packing density, and the strain energy that would be incurred in eliminating all packing defects. The results also suggest that mutations designed to increase protein stability by filling existing cavities may be effective in some cases but are unlikely to provide a general method for increasing protein stability.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Matthews B. W. Structure and thermal stability of phage T4 lysozyme. Methods Enzymol. 1987;154:511–533. doi: 10.1016/0076-6879(87)54093-9. [DOI] [PubMed] [Google Scholar]
- Alber T., Sun D. P., Nye J. A., Muchmore D. C., Matthews B. W. Temperature-sensitive mutations of bacteriophage T4 lysozyme occur at sites with low mobility and low solvent accessibility in the folded protein. Biochemistry. 1987 Jun 30;26(13):3754–3758. doi: 10.1021/bi00387a002. [DOI] [PubMed] [Google Scholar]
- Becktel W. J., Baase W. A. A lysoplate assay for Escherichia coli cell wall-active enzymes. Anal Biochem. 1985 Nov 1;150(2):258–263. doi: 10.1016/0003-2697(85)90508-1. [DOI] [PubMed] [Google Scholar]
- Becktel W. J., Baase W. A. Thermal denaturation of bacteriophage T4 lysozyme at neutral pH. Biopolymers. 1987 May;26(5):619–623. doi: 10.1002/bip.360260505. [DOI] [PubMed] [Google Scholar]
- Chothia C. Hydrophobic bonding and accessible surface area in proteins. Nature. 1974 Mar 22;248(446):338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- Chothia C. Structural invariants in protein folding. Nature. 1975 Mar 27;254(5498):304–308. doi: 10.1038/254304a0. [DOI] [PubMed] [Google Scholar]
- Connolly M. L. Atomic size packing defects in proteins. Int J Pept Protein Res. 1986 Oct;28(4):360–363. doi: 10.1111/j.1399-3011.1986.tb03266.x. [DOI] [PubMed] [Google Scholar]
- Connolly M. L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983 Aug 19;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Nelson H. C., Sauer R. T. Mutations in lambda repressor's amino-terminal domain: implications for protein stability and DNA binding. Proc Natl Acad Sci U S A. 1983 May;80(9):2676–2680. doi: 10.1073/pnas.80.9.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis J. T., Jr, Nyberg K., Sali D., Fersht A. R. Contribution of hydrophobic interactions to protein stability. Nature. 1988 Jun 23;333(6175):784–786. doi: 10.1038/333784a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Levitt M. Energy refinement of hen egg-white lysozyme. J Mol Biol. 1974 Jan 25;82(3):393–420. doi: 10.1016/0022-2836(74)90599-3. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature. 1989 May 4;339(6219):31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Becktel W. J., Matthews B. W. Hydrophobic stabilization in T4 lysozyme determined directly by multiple substitutions of Ile 3. Nature. 1988 Aug 4;334(6181):406–410. doi: 10.1038/334406a0. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Genetic and structural analysis of the protein stability problem. Biochemistry. 1987 Nov 3;26(22):6885–6888. doi: 10.1021/bi00396a001. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Wolynes P. G., Karplus M. Picosecond dynamics of tyrosine side chains in proteins. Biochemistry. 1979 Mar 20;18(6):927–942. doi: 10.1021/bi00573a001. [DOI] [PubMed] [Google Scholar]
- McGregor M. J., Islam S. A., Sternberg M. J. Analysis of the relationship between side-chain conformation and secondary structure in globular proteins. J Mol Biol. 1987 Nov 20;198(2):295–310. doi: 10.1016/0022-2836(87)90314-7. [DOI] [PubMed] [Google Scholar]
- Muchmore D. C., McIntosh L. P., Russell C. B., Anderson D. E., Dahlquist F. W. Expression and nitrogen-15 labeling of proteins for proton and nitrogen-15 nuclear magnetic resonance. Methods Enzymol. 1989;177:44–73. doi: 10.1016/0076-6879(89)77005-1. [DOI] [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Reidhaar-Olson J. F., Sauer R. T. Combinatorial cassette mutagenesis as a probe of the informational content of protein sequences. Science. 1988 Jul 1;241(4861):53–57. doi: 10.1126/science.3388019. [DOI] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Richards F. M. The interpretation of protein structures: total volume, group volume distributions and packing density. J Mol Biol. 1974 Jan 5;82(1):1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- Weaver L. H., Matthews B. W. Structure of bacteriophage T4 lysozyme refined at 1.7 A resolution. J Mol Biol. 1987 Jan 5;193(1):189–199. doi: 10.1016/0022-2836(87)90636-x. [DOI] [PubMed] [Google Scholar]




