Abstract
The Ribosomal RNA Operon Copy Number Database (rrndb) is an Internet-accessible database containing annotated information on rRNA operon copy number among prokaryotes. Gene redundancy is uncommon in prokaryotic genomes, yet the rRNA genes can vary from one to as many as 15 copies. Despite the widespread use of 16S rRNA gene sequences for identification of prokaryotes, information on the number and sequence of individual rRNA genes in a genome is not readily accessible. In an attempt to understand the evolutionary implications of rRNA operon redundancy, we have created a phylogenetically arranged report on rRNA gene copy number for a diverse collection of prokaryotic microorganisms. Each entry (organism) in the rrndb contains detailed information linked directly to external websites including the Ribosomal Database Project, GenBank, PubMed and several culture collections. Data contained in the rrndb will be valuable to researchers investigating microbial ecology and evolution using 16S rRNA gene sequences. The rrndb web site is directly accessible on the WWW at http://rrndb.cme.msu.edu.
INTRODUCTION
Microbes are the most abundant and most diverse forms of life on earth (1). Despite their ubiquity, it is clear that only a small percentage of microbes (0.1–0.5%) has been cultivated in the laboratory (2). Identification and classification of microbes is further confounded by a general absence of morphologically distinct features—thousands of bacterial species can be categorized by a few different (∼17) morphologies. For the past 10–15 years, microbiologists have relied upon DNA sequence information for microbial identification, based primarily on the genes encoding the small subunit RNA molecule of the ribosome (16S rRNA or SSU rRNA). Functional constraints on the translational apparatus limit variability in the 16S rRNA molecule, resulting in a high degree of sequence conservation. The conservation of the rRNA gene sequence permits bacterial characterization based on sequence information obtained from pure cultures or cloned genes from mixed communities. A priori knowledge of rRNA sequence data can be used to design phylogenetically conserved probes that target both individual and closely related groups of microorganisms without cultivation. A principle repository of 16S rRNA sequences, the Ribosomal Database Project, currently maintains over 17 000 aligned entries (12 425 sequences ≥900 bp) representing 850 of 940 formally recognized prokaryotic genera, which are placed into 1149 phylogenetic groups (3).
The ribosomal RNA genes (encoding 16S, 23S and 5S rRNAs) are typically linked together with tRNA molecules into operons that are coordinately transcribed to produce equimolar quantities of each gene product. During rapid exponential growth (µ = 2.5 h–1), the effective number of rRNA operons in Escherichia coli can be as high as 36 copies (4). Sequence heterogeneity exists among multiple rRNA genes encoded on a single genome, yet little evidence exists suggesting functional independence (5,6). While reports of intra-genomic variability of 16S rRNA range as high as 6.5% (7), an analysis of complete genome sequences stored in the rrndb indicates a maximum of 1.23% (E.coli) among the 14 species examined (Table 1). Both rRNA operon redundancy and intra-genomic sequence heterogeneity have important practical implications for researchers attempting to identify and quantify bacteria using rRNA sequence data (8,9).
Table 1. Intra-genomic 16S rRNA variability for Bacteria and Archaea with full-genome sequence availability.
| Organism |
No. rRNAa operons |
Diff. (nt)b |
% differencec |
| Aquifex aeolicus VF5 | 2 | – | – |
| Bacillus subtilis ATCC 23857 | 10 | 1–15 | 0.97 |
| Campylobacter jejuni ATCC 700819 | 3 | – | – |
| Deinococcus radiodurans ATCC 13939 | 3 | 0–2 | 0.13 |
| Escherichia coli ATCC 10798 | 7 | 0–19 | 1.23 |
| Haemophilus influenzae ATCC 51907 | 6 | – | – |
| Helicobacter pylori 26695 | 2 | – | – |
| Methanococcus jannaschii DSMZ 2661 | 2 | 3 | 0.20 |
| Methanococcus themoautotrophicum ATCC 29096 | 2 | 2 | 0.14 |
| Neisseria menigitidis MC 58 | 4 | – | – |
| Treponema pallidum ATCC 25870 | 2 | – | – |
| Ureaplasma urealyticum serovar 3 | 2 | 1 | 0.07 |
| Vibrio cholerae ATCC 39315 | 8 | 0–14 | 0.91 |
| Xyella fastidosa 9a5c | 2 | – | – |
aNumber of rRNA operons per genome.
bPairwise difference range between 16S rRNA genes per genome.
cPairwise difference range between 16S rRNA genes per genome calculated as a percentage. –, no nucleotide differences.
Molecular methods for microbial diversity assessment rely primarily on PCR-amplification of 16S rRNA genes from complex samples followed by (i) cloning and sequencing of unique amplicons, (ii) separation of amplicons based on chemical composition via denaturing- or temperature-gradient gel electrophoresis (10,11), or (iii) separation of amplicons after restriction digestion based on size via terminal restriction fragment length polymorphism analysis (12). The number of unique sequences or bands detected by these methods is often considered a proxy for organismal diversity. Rather, due to intra-genomic 16S rRNA heterogeneity, these methods are more accurately a measure of 16S rRNA sequence diversity. Similarly, intra-genomic sequence heterogeneity limits the phylogenetic resolution of the 16S rRNA gene (13,14). The majority of 16S rRNA entries in public databases, such as GenBank and the Ribosomal Database Project, are ‘composite’ sequences obtained from sequencing PCR amplicons generated through simultaneous amplification of all 16S rRNA gene copies on a genome (15).
In an attempt to understand the evolutionary implications of rRNA operon gene redundancy, our laboratory has maintained an internal database of rRNA operon copy number values for both Bacteria and Archaea. Mapping of this information onto a phylogenetic tree indicates that phylogenetic relatedness is not the sole determinant of rRNA operon copy number (16). Rather, bacteria with the same number of rRNA operons appear to have arisen convergently in several phylogenetic lineages. While our primary interest resides in elucidating the underlying physiological and evolutionary consequences of rRNA operon multiplicity, rRNA operon copy number information has become increasingly valuable to researchers performing emerging technologies such as quantitative real-time PCR (17). Working closely with the Ribosomal Database Project at Michigan State University, we have created an Internet-based interactive database of rRNA operon copy number values for a diverse collection prokaryotic microorganisms: The Ribosomal RNA Operon Copy Number Database (rrndb).
DATABASE DESCRIPTION
The rrndb provides information pertaining to the number of rRNA operons contained on the genomes of prokaryotic microorganisms in a phylogenetic context. The rrndb is co-located with the RDP server at the Center for Microbial Ecology at Michigan State University and is accessible via the WWW at http://rrndb.cme.msu.edu. The initial release of our database (December, 2000) contains over 250 annotated entries, including information from all full-genome sequencing projects completed at the time of release. An internal database management system (described below) permits entry of data from any WWW browser, facilitating public release of information shortly after entry and verification. The rrndb WWW site also contains answers to frequently asked questions, an opportunity to provide feedback and a form for direct submission of new data.
WEB INTERFACE
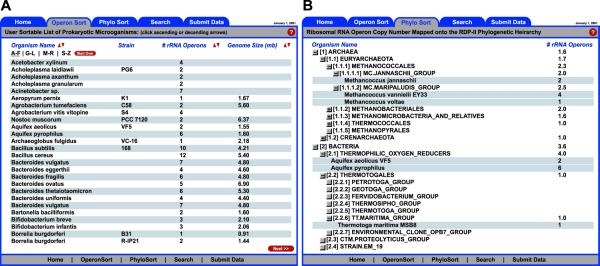
Information contained within the rrndb is accessible via three main interfaces: (i) ‘Operon Sort’, a complete list of organisms in the database presented in alphabetical order; (ii) ‘Phylo Sort’, rRNA operon copy number mapped onto the RDP organismal hierarchy; and (iii) a ‘Search’ page. The ‘Operon Sort’ list can be sorted in ascending and descending order by organism name, rRNA operon copy number or genome size (Fig. 1A). rRNA operon copy number is mapped onto the RDP organismal hierarchy presented on the ‘Phylo Sort’ page. The hierarchy is expandable and collapsible and mean rRNA operon copy number is displayed for each phylogenetic group (Fig. 1B). User queries can be entered on the ‘Search’ page, which also offers advanced searches limited to rRNA operon copy number and genome size. Each entry in the rrndb is linked to an individual page containing detailed information about the selected organism, including: genus, species, sub-species, strain, culture deposit, 16S, 23S and 5S rRNA gene copy number, phylogenetic position, genome size, genome sequence availability, 16S rRNA sequence records and literature reference(s). Sequence deposits are linked directly to GenBank and the RDP, culture deposits to the American Type Culture Collection (ATCC) and Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) and references to the National Library of Medicine’s PubMed database. 16S rRNA genes from individual rRNA operons are denoted when available and include gene start and stop locations within GenBank entries from full-genome sequence deposits.
Figure 1.
HTML interfaces of the rrndb. (A) ‘Operon Sort’ page offering complete list of database entries sortable by organism name, rRNA operon copy number and genome size fields. (B) ‘Phylo Sort’ page with rRNA operon copy number mapped onto the RDP phylogenetic hierarchy. Mean rRNA operon copy number displayed for each phylogenetic group.
DATA CURATION
All entries in the rrndb possess at least a genus name, strain designation or culture deposit number and a literature or electronic reference describing rRNA operon copy number determination. The entries for each organism are obtained from computerized searches of reference databases (PubMed, ISI Current Contents, etc.), literature articles, full genome sequencing projects listed at The Institute for Genomic Research (http://www.tigr.org) and the National Center for Biotechnology Information’s web sites (http://www.ncbi.nlm.nih.gov) and from direct web site submission. Effort is made to include all pertinent references to the determination of rRNA operon copy number for an organism. Literature references for a particular organism may not be reported due to an article predating electronic database records or to the absence of relevant search terms in a database entry. If the complete genome sequence of an organism is available, those data are considered to be the most accurate determination of rRNA operon copy number and is the only reported reference. In certain instances the 16S, 23S and 5S rRNA genes are not present in equal numbers per genome (18). Laboratory methods to determine rRNA operon copy number typically rely upon Southern hybridization of a 16S rRNA-based probe to restriction-digested genomic DNA; in these instances, the number of 16S rRNA genes serves as an estimate for rRNA operon copy number.
DATABASE MANAGEMENT SYSTEM
The rrndb data are stored using the MySQL relational database management system (RDBMS), which supports the structured query language (SQL) standard (http://www.mysql.com). The WWW interface to the rrndb is generated by Java Server Pages and Java Servlets that retrieve information to be displayed by employing custom designed JavaBean objects (http://java.sun.com). These objects access the database using MM MySQL JDBC drivers (http://www.worldserver.com/mm.mysql). The rrndb website is hosted on a Sun Ultra 60 server running the Solaris 2.6 operating system and Apache Software Foundation’s Apache HTTP and Tomcat servers (http://jakarta.apache.org).
FUTURE CHANGES AND ADDITIONS
Planned additions to the rrndb include interface tools to select and download individual organism entries from both the ‘Operon Sort’ list and the ‘Phylo Sort’ pages. Information on intra-genomic rRNA sequence variability, such as presented in Table 1, will be added for organisms with full genome sequences. Further changes will be dictated by feedback obtained from users of the rrndb website. It is anticipated that the rrndb will be updated on a quarterly basis as new information becomes available through electronic databases and full-genome sequencing projects.
Acknowledgments
ACKNOWLEDGEMENTS
We thank C. T. Parker and J. M. Stredwick for early prototype development and hardware support for the rrndb. Funds provided by the National Science Foundation (IBN-9875254), the Center for Microbial Ecology at Michigan State University (BIR91-20006) and the US Department of Energy (to the RDP) supported the development of the rrndb.
References
- 1.Whitman W.B., Coleman,D.C. and Wiebe,W.J. (1998) Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA, 95, 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torsvik V., Goksoyr,J. and Daae,F.L. (1990) High diversity in DNA of soil bacteria. Appl. Environ. Microbiol., 56, 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maidak B.L., Cole,J.R., Lilburn,T.G., Parker,C.T.,Jr, Saxman,P.R., Stredwick,J.M., Garrity,G.M., Li,B., Olsen,G.J., Pramanik,S., et al. (2000) The RDP (Ribosomal Database Project) continues. Nucleic Acids Res., 28, 173–174. Updated article in this issue: Nucleic Acids Res. (2001), 29, 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bremer H. and Dennis,P.P. (1996) In Neidhardt,F.C., Curtiss,R.,III, Ingraham,J.L., Lin,E.C.C., Low,K.B. et al. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 1553–1569.
- 5.Dryden S.C. and Kaplan,S. (1990) Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res., 18, 7267–7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condon C., Philips,J., Fu,Z.Y., Squires,C. and Squires,C.L. (1992) Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J., 11, 4175–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Zhang,Z. and Ramanan,N. (1997) The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J. Bacteriol ., 179, 3270–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wintzingerode F.v., Gobel,U.B. and Stackebrandt,E. (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev., 21, 213–229. [DOI] [PubMed] [Google Scholar]
- 9.Fogel G.B., Collins,C.R., Li,J. and Brunk,C.F. (1999) Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb. Ecol., 38, 93–113. [DOI] [PubMed] [Google Scholar]
- 10.Muyzer G., de Waal,E.C. and Uitterlinden,A.G. (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol., 59, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muyzer G. and Smalla,K. (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek, 73, 127–141. [DOI] [PubMed] [Google Scholar]
- 12.Liu W.T., Marsh,T.L., Cheng,H. and Forney,L.J. (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol., 63, 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cilia V., Lafay,B. and Christen,R. (1996) Sequence heterogeneities among 16S ribosomal RNA sequences and their effect on phylogenetic analyses at the species level. Mol. Biol. Evol., 13, 451–461. [DOI] [PubMed] [Google Scholar]
- 14.Dahllöf I., Baillie,H. and Kjelleberg,S. (2000) rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol., 66, 3376–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton R.A., Sutton,G., Hinkle,P.S.,Jr, Bult,C. and Fields,C. (1995) Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int. J. Syst. Bacteriol., 45, 595–599. [DOI] [PubMed] [Google Scholar]
- 16.Klappenbach J.A., Dunbar,J.M. and Schmidt,T.M. (2000) rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol., 66, 1328–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyons S.R., Griffen,A.L. and Leys,E.J. (2000) Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol., 38, 2362–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser C.M., Casjens,S., Huang,W.M., Sutton,G.G., Clayton,R., Lathigra,R., White,O., Ketchum,K.A., Dodson,R., Hickey,E.K. et al. (1997) Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature, 390, 580–586. [DOI] [PubMed] [Google Scholar]