Abstract
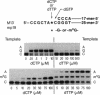
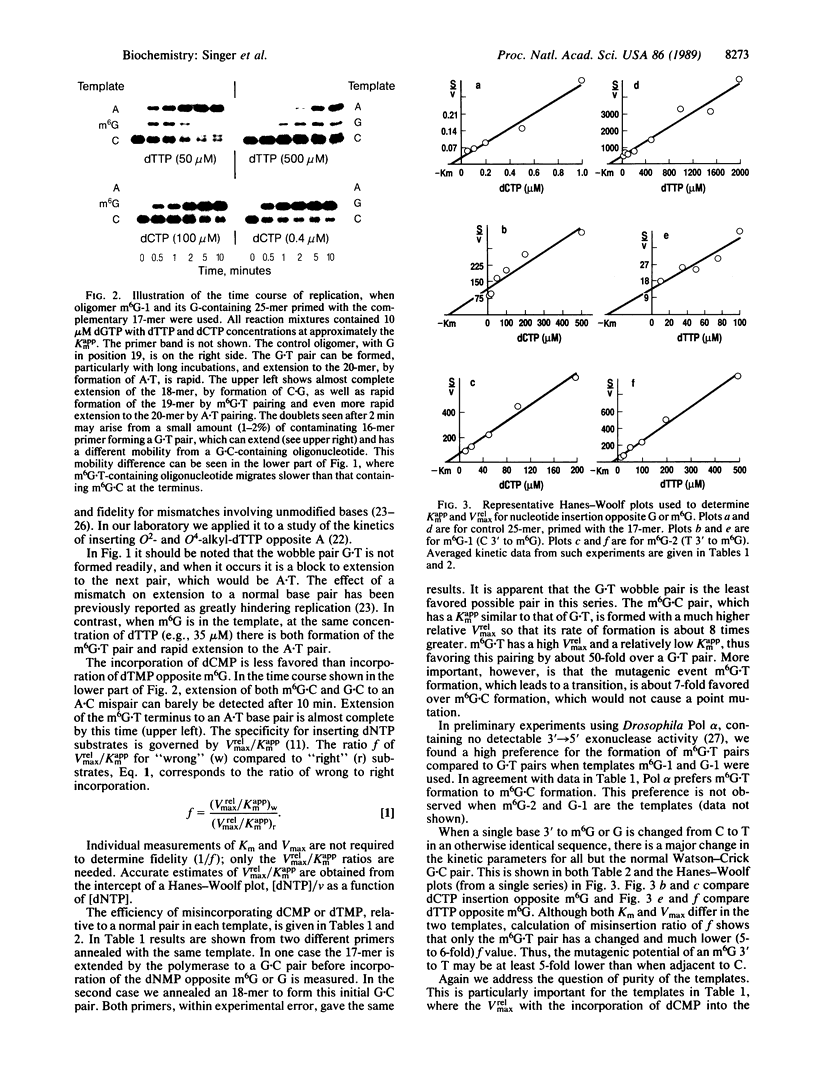
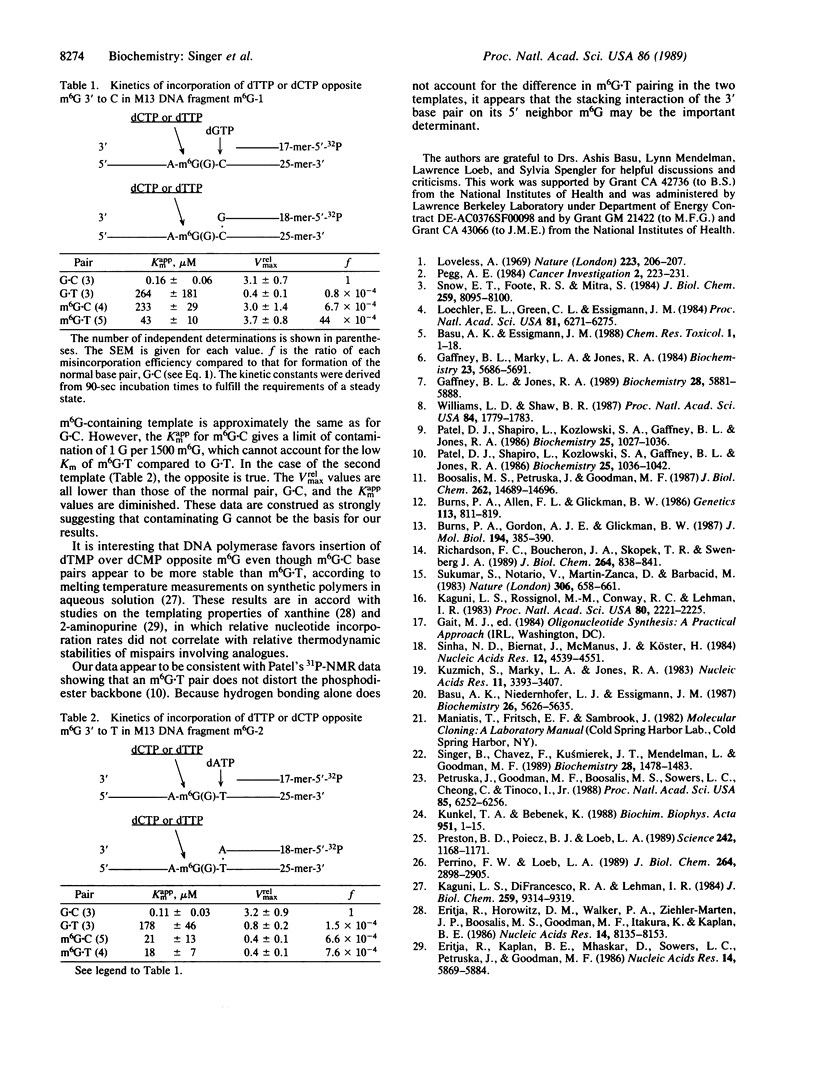
O6-Methylguanine (m6G) was incorporated site-specifically into two 25-base oligonucleotides differing only in the nucleotide on the 3' side of the modified base. Templates were primed with oligonucleotides terminating one or two bases prior to the site at which incorporation kinetics were to be investigated. Escherichia coli DNA polymerase I (Klenow fragment) was used to determine the apparent Km and relative Vmax of incorporation of either dCTP or dTTP opposite m6G or G. These data were used to calculate the relative frequency of incorporation opposite the m6G or the unmodified G. When the sequence was 3'-Cm6G-5', there was a 6- to 7-fold preference for formation of a m6G.T pair compared with m6G.C. The m6G.T frequency, based on Vmax/Km, was at least 50-fold greater than that of a G.T pair at the same site. Changing the sequence to 3'-Tm6G-5' had a marked effect on both Km and Vmax of pairs containing m6G and on the incorporation frequency of T opposite m6G, which was then only slightly favored over m6G.C. When replication was started directly opposite m6G, the kinetics appeared unaffected. These data indicate that the frequency of incorporation of C or T opposite m6G in a DNA template is dependent on the flanking neighbors and that a change of even a single base at the 3' position can have a major effect on mutagenic efficiency. Replication using Drosophila Pol alpha gave the same values for relative frequencies. Pairing of either C or T with m6G on the primer terminus did not significantly inhibit extension of the next normal base pair, in contrast to terminal mismatches of unmodified bases. It is concluded that, in the absence of repair, m6G can exhibit widely differing mutation frequencies which, in these experiments, can be as high as 85% of the replicated base. This variation in frequency of changed pairing could contribute to the occurrence of mutational "'hot spots" after replication of damaged DNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basu A. K., Essigmann J. M. Site-specifically modified oligodeoxynucleotides as probes for the structural and biological effects of DNA-damaging agents. Chem Res Toxicol. 1988 Jan-Feb;1(1):1–18. doi: 10.1021/tx00001a001. [DOI] [PubMed] [Google Scholar]
- Basu A. K., Niedernhofer L. J., Essigmann J. M. Deoxyhexanucleotide containing a vinyl chloride induced DNA lesion, 1,N6-ethenoadenine: synthesis, physical characterization, and incorporation into a duplex bacteriophage M13 genome as part of an amber codon. Biochemistry. 1987 Sep 8;26(18):5626–5635. doi: 10.1021/bi00392a007. [DOI] [PubMed] [Google Scholar]
- Boosalis M. S., Petruska J., Goodman M. F. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J Biol Chem. 1987 Oct 25;262(30):14689–14696. [PubMed] [Google Scholar]
- Burns P. A., Allen F. L., Glickman B. W. DNA sequence analysis of mutagenicity and site specificity of ethyl methanesulfonate in Uvr+ and UvrB- strains of Escherichia coli. Genetics. 1986 Aug;113(4):811–819. doi: 10.1093/genetics/113.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns P. A., Gordon A. J., Glickman B. W. Influence of neighbouring base sequence on N-methyl-N'-nitro-N-nitrosoguanidine mutagenesis in the lacI gene of Escherichia coli. J Mol Biol. 1987 Apr 5;194(3):385–390. doi: 10.1016/0022-2836(87)90668-1. [DOI] [PubMed] [Google Scholar]
- Eritja R., Horowitz D. M., Walker P. A., Ziehler-Martin J. P., Boosalis M. S., Goodman M. F., Itakura K., Kaplan B. E. Synthesis and properties of oligonucleotides containing 2'-deoxynebularine and 2'-deoxyxanthosine. Nucleic Acids Res. 1986 Oct 24;14(20):8135–8153. doi: 10.1093/nar/14.20.8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eritja R., Kaplan B. E., Mhaskar D., Sowers L. C., Petruska J., Goodman M. F. Synthesis and properties of defined DNA oligomers containing base mispairs involving 2-aminopurine. Nucleic Acids Res. 1986 Jul 25;14(14):5869–5884. doi: 10.1093/nar/14.14.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney B. L., Jones R. A. Thermodynamic comparison of the base pairs formed by the carcinogenic lesion O6-methylguanine with reference both to Watson-Crick pairs and to mismatched pairs. Biochemistry. 1989 Jul 11;28(14):5881–5889. doi: 10.1021/bi00440a026. [DOI] [PubMed] [Google Scholar]
- Gaffney B. L., Marky L. A., Jones R. A. Synthesis and characterization of a set of four dodecadeoxyribonucleoside undecaphosphates containing O6-methylguanine opposite adenine, cytosine, guanine, and thymine. Biochemistry. 1984 Nov 20;23(24):5686–5691. doi: 10.1021/bi00319a004. [DOI] [PubMed] [Google Scholar]
- Kaguni L. S., DiFrancesco R. A., Lehman I. R. The DNA polymerase-primase from drosophila melanogaster embryos. Rate and fidelity of polymerization on single-stranded DNA templates. J Biol Chem. 1984 Jul 25;259(14):9314–9319. [PubMed] [Google Scholar]
- Kaguni L. S., Rossignol J. M., Conaway R. C., Lehman I. R. Isolation of an intact DNA polymerase-primase from embryos of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2221–2225. doi: 10.1073/pnas.80.8.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim Biophys Acta. 1988 Nov 10;951(1):1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- Kuzmich S., Marky L. A., Jones R. A. Specifically alkylated DNA fragments. Synthesis and physical characterization of d[CGC(O6Me)GCG] and d[CGT(O6Me)GCG]. Nucleic Acids Res. 1983 May 25;11(10):3393–3403. doi: 10.1093/nar/11.10.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loechler E. L., Green C. L., Essigmann J. M. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveless A. Possible relevance of O-6 alkylation of deoxyguanosine to the mutagenicity and carcinogenicity of nitrosamines and nitrosamides. Nature. 1969 Jul 12;223(5202):206–207. doi: 10.1038/223206a0. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Kozlowski S. A., Gaffney B. L., Jones R. A. Structural studies of the O6meG.C interaction in the d(C-G-C-G-A-A-T-T-C-O6meG-C-G) duplex. Biochemistry. 1986 Mar 11;25(5):1027–1036. doi: 10.1021/bi00353a012. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Shapiro L., Kozlowski S. A., Gaffney B. L., Jones R. A. Structural studies of the O6meG.T interaction in the d(C-G-T-G-A-A-T-T-C-O6meG-C-G) duplex. Biochemistry. 1986 Mar 11;25(5):1036–1042. doi: 10.1021/bi00353a013. [DOI] [PubMed] [Google Scholar]
- Pegg A. E. Methylation of the O6 position of guanine in DNA is the most likely initiating event in carcinogenesis by methylating agents. Cancer Invest. 1984;2(3):223–231. doi: 10.3109/07357908409104376. [DOI] [PubMed] [Google Scholar]
- Perrino F. W., Loeb L. A. Differential extension of 3' mispairs is a major contribution to the high fidelity of calf thymus DNA polymerase-alpha. J Biol Chem. 1989 Feb 15;264(5):2898–2905. [PubMed] [Google Scholar]
- Petruska J., Goodman M. F., Boosalis M. S., Sowers L. C., Cheong C., Tinoco I., Jr Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston B. D., Poiesz B. J., Loeb L. A. Fidelity of HIV-1 reverse transcriptase. Science. 1988 Nov 25;242(4882):1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- Richardson F. C., Boucheron J. A., Skopek T. R., Swenberg J. A. Formation of O6-methyldeoxyguanosine at specific sites in a synthetic oligonucleotide designed to resemble a known mutagenic hotspot. J Biol Chem. 1989 Jan 15;264(2):838–841. [PubMed] [Google Scholar]
- Singer B., Chavez F., Spengler S. J., Kuśmierek J. T., Mendelman L., Goodman M. F. Comparison of polymerase insertion and extension kinetics of a series of O2-alkyldeoxythymidine triphosphates and O4-methyldeoxythymidine triphosphate. Biochemistry. 1989 Feb 21;28(4):1478–1483. doi: 10.1021/bi00430a008. [DOI] [PubMed] [Google Scholar]
- Sinha N. D., Biernat J., McManus J., Köster H. Polymer support oligonucleotide synthesis XVIII: use of beta-cyanoethyl-N,N-dialkylamino-/N-morpholino phosphoramidite of deoxynucleosides for the synthesis of DNA fragments simplifying deprotection and isolation of the final product. Nucleic Acids Res. 1984 Jun 11;12(11):4539–4557. doi: 10.1093/nar/12.11.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow E. T., Foote R. S., Mitra S. Base-pairing properties of O6-methylguanine in template DNA during in vitro DNA replication. J Biol Chem. 1984 Jul 10;259(13):8095–8100. [PubMed] [Google Scholar]
- Sukumar S., Notario V., Martin-Zanca D., Barbacid M. Induction of mammary carcinomas in rats by nitroso-methylurea involves malignant activation of H-ras-1 locus by single point mutations. Nature. 1983 Dec 15;306(5944):658–661. doi: 10.1038/306658a0. [DOI] [PubMed] [Google Scholar]
- Williams L. D., Shaw B. R. Protonated base pairs explain the ambiguous pairing properties of O6-methylguanine. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1779–1783. doi: 10.1073/pnas.84.7.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]