Summary
Although ADAMTS13, the von Willebrand factor-cleaving protease, is expressed in a range of tissues, the physiological significance of tissue-specific ADAMTS13 alternative splicing isoforms have yet to be clarified. Screening a panel of human tissues and cell lines revealed a spliced ADAMTS13 transcript in hepatic stellate cells and a hepatoma cell line that retains the 25th intron. A nonsense codon within the intron truncates the protease, which gains 64 novel amino acids in lieu of both CUB domains. This isoform, while retaining VWF-cleaving capability, accumulates intracellularly and its biological inaccessibility may prevent its participation in regulating hemostasis and other physiologic functions.
Keywords: ADAMTS13, alternative splicing, intron retention, TTP, CUB domain
Introduction
The ADAMTS13 gene, located at human chromosomal position 9q34, encodes for A Disintegrin-like and Metalloprotease with Thrombospondin type 1 motif, member 13 (ADAMTS13) (1). ADAMTS13 plays an integral role in vascular homeostasis of ultra-large von Willebrand factor multimers. A severe deficiency of ADAMTS13 results in increased VWF thrombogenic potential and thrombotic thrombocytopenic purpura (TTP) (2). The open reading frame for ADAMTS13 contains 29 exons (28 introns), spanning 37 kb in all, encoding a protein product of 1427 amino acids with a calculated molecular weight of ~150 kDa. The metalloprotease domain and the proximal carboxyl-terminus regions (Tsp1-1, cys-rich, spacer) have all been established as indispensable for ADAMTS13 activity (3). The relevance of the distal thrombospondin repeats and CUB (complement C1r/C1s, Uegf, Bmp1) domains to overall function have been more heavily disputed, even though TTP-associated mutations have been found in these regions. Several studies have shown that ADAMTS13 mutants lacking these domains are still active (4). Recent reports, however, have demonstrated that the CUB domains have importance in both apical sorting of ADAMTS13 and docking of the protease to the VWF substrate (5). Alternative pre-mRNA splicing, a mechanism that expands transcriptional repertoire, is known to vary among tissues, during different developmental stages, or under different physiological conditions (6,7). It is estimated that a large proportion of disease-associated mutations involve aberrant splicing. Tissue-based screening for ADAMTS13 alternative splicing has suggested splice isoforms specific to the liver, prostate and brain (8), although the functional qualities of these variants were not investigated. Here, we analyze alternative splicing of ADAMTS13 in several tissues in order to assess whether this might play a role during its expression. We also investigate the biological characteristics of such alternative splicing forms and suggest possible consequence to cancer.
Materials and methods
MaxEntScan analysis
MaxEntScan was used to score the splice site signals of each exon-intron junction. MaxEntScan is based on the approach for modeling the sequences of short sequence motifs such as those involved in RNA splicing which simultaneously accounts for non-adjacent as well as adjacent dependencies between positions (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html).
Cell lines and cell culture
HEK293 cells (human embryonic kidney cells), Hep3b (ATCC), human lung cancer cells, human normal lung cell line (kind gift from Dr. Samelson) and LX2 cells (9,10) were cultured as specified earlier (11,12). Chinese Hamster Ovary (CHO) cells were cultured under standard temperature, CO2 and humidity conditions in cellgro FREE Serum-Free/Protein-Free Medium, 1X (Mediatech) supplemented with 10% v/v fetal bovine serum, 1% L-glutamine and 1% penicillin-streptomycin prior to transfection. Following transfection, cellgro FREE-only was utilized to aide in protein concentration and analysis.
Analysis of splicing isoforms and cloning of cDNA fragments
RNA was isolated using the Qiagen RNeasy kit with a 15-minute on-column DNase incubation step. Total human RNAs were purchased from Ambion Inc. and Clontech, Mountain View, CA, or as a gift from Dr. Jean-Pierre Gillet (NCI, NIH) (cancer tissues). After total RNA (0.2–0.4 μg) was reverse transcribed using the Advantage RT-for-PCR (Clontech), specific regions of ADAMTS13 cDNA were amplified by PCR. For PCR analyses focused on exon 25–26 boundary, forward and reverse primers for exon 24–27 were 5′-CCAGTGTCCCCTGTCTCATT-3′ and 5′-AAGGTTTCAGGAGCAAGCTG-3′ (Fig. 1).
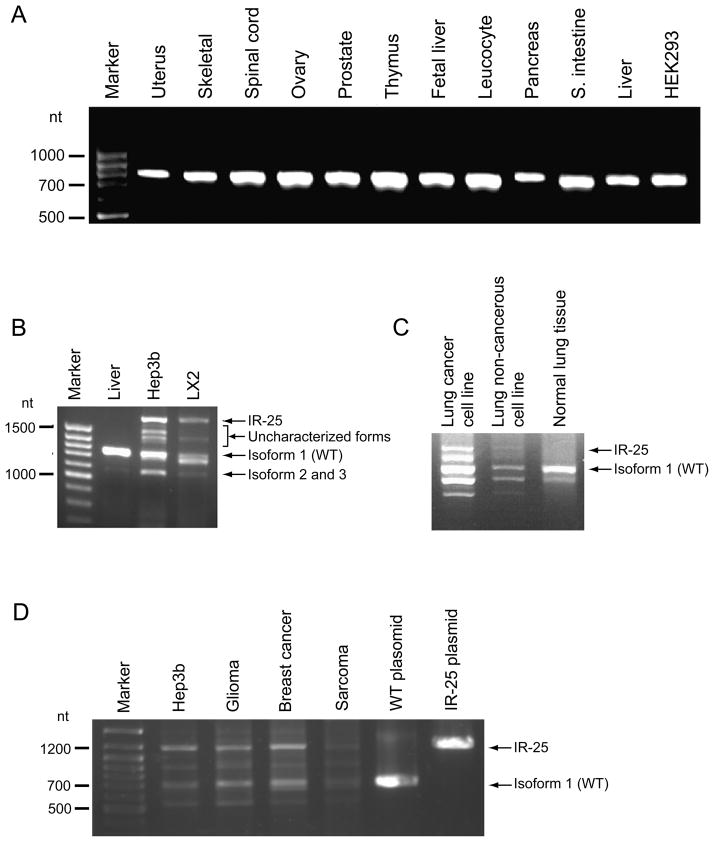
Fig. 1.
Characterization of known and novel isoforms of human ADAMTS13 focused on exon 25–26 boundary. Primers for exons 24–27 were used in panels A, C, D; Primers 23–24F and 29R+ were used in panel B. (A) RT-PCR products of reverse-transcribed RNA isolated from various human tissues and an embryonic kidney cell line. No potential splicing variants are evident among these whole tissue samples at the exon 25–26 boundary. (B) RT-PCR products of reverse-transcribed RNA isolated from whole liver, hepatoma cells (Hep3b) and hepatic stellate cells (LX2). Both conventional (1.3 kb) and ectopic splicing (1.8 kb) are evident in the two liver subpopulations. At least two retention forms are discernable (between 1.4 and 1.6 kb) in addition to at least one truncated form (~1.1 kb). (C) RT-PCR products of reverse-transcribed RNA isolated from lung cancer cell line, lung non-cancerous cell line and normal lung tissue. (D) RT-PCR products of reverse-transcribed RNA isolated from breast, glioma and sarcoma tissue from cancers.
The cDNA fragments of novel splice variants were cloned by RT-PCR from the Hep3b cell line using forward and reverse primers, 23–24F: 5′-CCCTGCCCACCTAGGTGGAAA-3′ (AvrII restriction site underlined) and 29+R: 5′-TGG AAA AAG TTC GAA TTG GAA AGC-3′ (BstBI restriction site underlined). After a fragment of 1.8 kb (~ 0.5 kb longer than expected size for isoform 1) was isolated, it was served for additional identical PCR reaction, cloned into pCR4-TOPO (Clontech) and sequenced. This fragment was cloned, at AvrII/BstBI sites, into pcDNA-ADAMTS13 (isoform 1) in which one of two AvrII sites downstream from the SV40 promoter was eliminated by partial AvrII digestion and Klenow DNA polymerase treatment. The new construct was sequenced using Bigdye-3.1 reaction system and ABI-3130xl sequencer and the results were aligned with Genbank ADAMTS13 sequence by EMBOSS-Lite software.
Quantification of ADAMTS13 using Western blotting
The cell lysates were prepared and quantified as described earlier (11,12). Immunostaining was performed using the primary monoclonal antibody Wh2-11-1 which detects the TSP-1-4 domain in ADAMTS13 (12) followed by secondary anti-mouse IgG HRP antibody (Invitrogen).
Measurement of Protease Activity using FRETS-VWF73 Assay
Sample preparation and protease activity assay were carried out according to the manufacturer of FRETS-VWF73 (Peptide Institute) as described earlier (11,12).
Results and discussion
At least three human ADAMTS13 splice variants involving combinations of exons 8 and 25 are currently known and sequenced (isoform 1 (NM_139025), isoform 2 (NM_139027) and isoform 3 (NM_139026)). In order to better understand ADAMTS13 splicing patterns and guide our investigation of unidentified splicing patterns, we first computationally examined the underlying mechanisms and characterized another previously unidentified splicing pattern. We specifically analyzed: (i) splice site strengths (using MAXENT (13)), (ii) evolutionary conservation, (14), (iii) splicing regulators, (iv) published splicing-related mutations (1) and (v) SNP databases.
We first calculated, using MaxEntScan, the splice site strengths by scoring ADAMTS13 exon-intron boundaries. Notably, the 5′ ss (splice site) in the 25th intron of wild type (isoform 1) showed a large score reduction (Supplement Fig. 1), suggesting that alternative splicing is likely at exon 25 and could contribute to transcript heterogeneity in this area due to a weak 5′ intronic ss. In fact, an upstream alternative 5′ ss was identified (Supplement Fig. 2, 5′ ss (i)) which gives rise to a shorter 25th exon (longer corresponding intron) seen in isoforms 2 and 3. This second splice site has a higher MAXENT score relative to its counterpart (Supplement Fig. 1, compare 25_L to 25_S). We next analyzed the conservation of intron 25 in ADAMTS13, which is very weak (Supplement Fig. 2), suggesting that either the intron retention event is not regulated through intronic sequences or that this event is unique to humans. Because splicing regulatory sequences participate in defining exon-intron boundaries, we then asked whether intron 25 or its flanking exons contain splicing signals that impact the use of intron 25 splice sites. We analyzed known regulatory sequences from high-throughput studies (15). Within the human mRNA transcript, splicing regulatory sequences in the retained intron were not relatively more frequent than in any other intron of the transcript. We did find several insertions in the human intron that might lead to an intron inclusion event. In particular, in Supplement Fig. 3, the regions marked as 2, 3 and 5 exhibit 6, 6, and 4 potential exonic enhancers, respectively, in the human intron, compared to none in the mouse intron.
In order to validate our computational analysis, we systematically scanned 11 tissues and 3 cell lines for the existence of intron 25 splicing isoforms. Interestingly, splice variants 2 and 3 were not detected in whole tissue screens (Fig. 1A). However, an ensuing study of hepatic stellate cells (LX2), a cell line that represents an important non-parenchymal population of liver cells (10) which significantly contributes to plasma ADAMTS13 (9), revealed a new splice isoform in which the entire 25th ADAMTS13 intron is retained (termed here IR-25) (Fig. 1B). Because hepatic stellate cells represent a minor portion (5–8%) of the cells that comprise the liver, the apparent lack of IR-25 in total liver tissue likely represents dilution of this isoform amidst a large amount of RNA derived from hepatocytes and other resident populations. A hepatoma cell line, Hep3B, also displayed this novel transcript (Fig. 1B). At least two other shorter, less abundant transcripts were observed in Fig. 1B.. One of these transcripts was identified as an additional new variant that not only retains intron 25, but also lacks exon 28 (data not shown). The resulting protein is biologically indistinct from IR-25 at the protein level due to a premature stop codon.
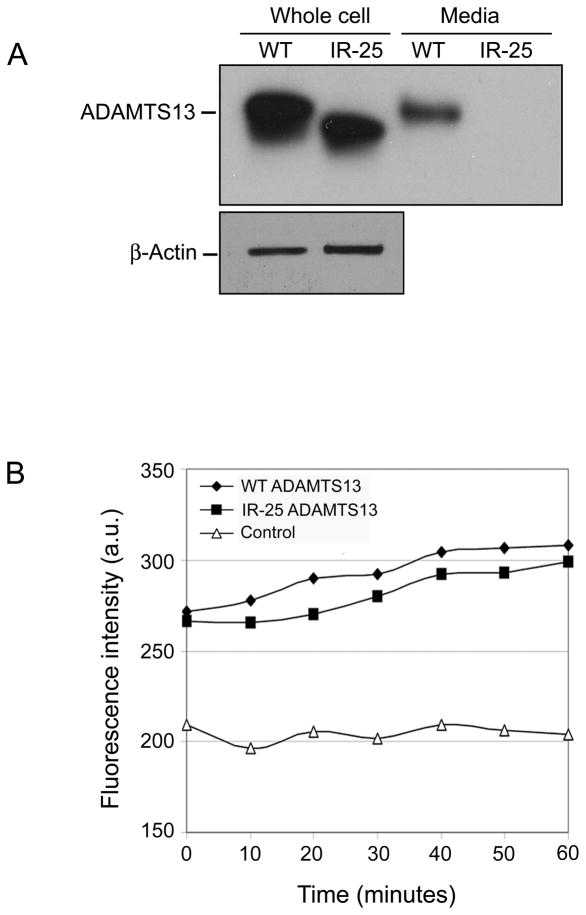
Sequencing the amplified exonic boundary confirmed that the entire 25th intron (465 bps) is included in IR-25. The intron retention is unique in that the retained intron includes a premature stop codon in the open reading frame following exon 25. As a result, IR-25 is truncated, lacking both C-terminal CUB domains and instead carries 64 novel amino acids, starting from amino acid #1190: SYVLSSFLSG SCCRRGGQRH LPLGRTGTST WSLGCVPGRP GSGLALFLPG KAKPPFYYYQ GEVT. There are two possible scenarios following this intron retention event: (i) mRNA degradation through Nonsense Mediated mRNA Decay; (ii) synthesis of a truncated protein. If the IR-25 mRNA is translated, a shortened protein product would be generated which lacks both CUB domains and instead carries 64 intron-encoded amino acids (the first intronic amino acid matches the first amino acid in exon 26) and bears 134 kDa. To test these possibilities, isoform 1 (WT) or IR-25 were transiently transfected into HEK293 cells, and harvested 24 hours later. We have demonstrated earlier that Western blotting can not detect endogenous expression of ADAMTS13 in Hek293 [9] and therefore we have use a specific monoclonal antibody to detect the recombinant forms in these cells. Western blotting revealed that cells transduced with IR-25 maintain most of ADAMTS13 intracellularly (Fig. 2A, Whole cell). In fact, IR-25 intracellular expression is only slightly lower (~15%) than its WT counterpart according to band densitometry. The aberrant splicing form, however, is not secreted at appreciable levels into culture media (Fig. 2A, Media). Similar expression patterns were obtained when using Chinese hamster ovary cells for transfection, indicating that this phenomenon is generally applicable. The intracellularly accumulated protein does retain its ability to recognize and specifically cleave VWF substrate (Fig. 2B). These findings are consistent with previous studies of a frameshift mutation within the first CUB domain of ADAMTS13 as well as another mutation in the second CUB domain, both of which produce truncated forms of ADAMTS13 that are not sorted nor secreted properly yet display specific activity against a VWF substrate (1,5,16). In accordance with the literature and the results presented here, an absence of both CUB domains in IR-25 may result in impaired structural integrity and render it non-processive within the cellular secretory system. The additional 64 amino acids found on the C-terminus of IR-25 are unlikely to adversely affect production or impart abnormal cellular localization. Indeed, an application that predicts subcellular localization signals, PSORT (www.psort.org), indicates that the 64 novel amino acids are unlikely to include a new sorting motif (data not shown).
Fig. 2.
Expression and functional characterization of the human ADAMTS13 wild type (WT) and intron retention isoforms (IR-25). (A) Western blot analysis of WT and IR-25 protein expression 24 hours following transient transfection of HEK293 cells. Thirty μg total protein of whole cell lysates or concentrated media were loaded on the gel. β-Actin levels in lysate samples are shown to be comparable (lower panel). (B) Fluorescogenic activity assay of intracellular WT and IR-25 over the course of a 60-minute incubation.
IR-25 ADAMTS13’s extracellular exclusion underscores its potential clinical relevance. A decreased level of plasma ADAMTS13 stemming from changes to the relative abundance of WT ADAMTS13 and IR-25 could induce thrombotic conditions such as TTP. An imbalance towards increasing amounts of IR-25 could arise in two ways. It could develop under the influence of mutations or SNPs in ADAMTS13 that promote intron retention at the exon 25–26 junction, but those have yet to be identified. The imbalance could also occur via trans-factors affecting splicing characteristics, namely pre-mRNA splicing machinery. We were able to detect IR-25 within human lung cancer cell line, but not in normal lung cells (Fig. 1C) as well as in three other cancer tissues (glioma, breast and weak signal in sarcoma) (Fig. 1D). It is therefore probable that a characteristic inherent to many types of cancer could contribute to the manifestation of IR-25. Indeed, cancerous cells are specifically known to produce a higher number of aberrant intron retention events (17). This feature is conjectured to result from changes in the abundance of splicing factors or their abnormal phosphorylation. The appearance of IR-25 in hepatoma cells can thus be reasonably inferred, but its presence in hepatic stellate cells remains mechanistically unclear.
Twenty percent of cancer patients experience thrombosis at some time during the course of their disease (18). The risk of venous thromboembolic events is particularly higher in cancers of the liver, pancreas, ovary and lung. The exact mechanism underlying the cancer-associated hypercoagulable state is not fully understood. In this report we describe in vitro experiments which show a decreased secretion of IR-25 ADAMTS13 form. It is possible to speculate that this altered form of ADAMTS13 may contribute to the hypercoagulable state in cancer patients in different mechanisms, such as competitive inhibition of the normal form at various levels. A physiologically dominate defected ADAMTS13 which appears in various cancers might explain the broad reports regarding high chances of cancer patients developing blood clots. Further clinical studies will be needed to assess the correlation between the presence of IR-25 ASAMTS13 form and venous thromboembolic events.
Supplementary Material
Supplement Fig. 1 Probability of RNA splicing for known splice sites in the human ADAMTS13 gene based on the ‘Maximum Entropy Principle’. MaxEntScan was used to score the splice site signals of each exon-intron junction (see Design and methods). L (in isoforms 2 and 3) and S (in isoform 1) refer to the long or short version of intron 7 and 25 (see Fig. 1A).
Supplement Fig. 2 A schematic representation intron 25 retention. Alternative 5′ ss are marked (i defines exon 25 in WT and ii defines exon 25 in isoform 2 and 3). Conservation along this region is depicted below in the mountain-like graph reproduced from the UCSC Genome Browser (MultiZ alignment). Peak height corresponds to conservation level among 17 animal species.
Supplement Fig. 3 Alignment of human (isoform 1), mouse intron 25 and flanking exons. Unique human sequences are indicated with black bars and numbered 1–6. Matched nucleotides between human and mouse are indicated with asterisks.
Table 1.
Summary of the work in the perspective of other data
| What is known on this topic | What this paper adds |
|---|---|
|
|
Acknowledgments
We would like to thank Mr. George Leiman and Mr. Javaid, Atiqur for helping with preparation of the manuscript and Dr. Reed, CBER, FDA, for insightful commentary. We are grateful to Dr. Sadler, Washington University, St. Louis, for proving us with pcDNA-ADAMTS13 and to Dr. Samelson, NCI, NIH, for providing us with the lung cells.
Footnotes
Disclaimer
The findings and conclusions in this article have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any Agency determination or policy.
References
- 1.Levy GG, Nichols WC, Lian EC, Foroud T, McClintick JN, Mcgee BM, Yang AY, Siemieniak DR, Stark KR, Gruppo R, Sarode R, Shurin SB, Chandrasekaran V, Stabler SP, Sabio H, Bouhassira EE, Upshaw JD, Ginsburg D, Tsai HM. Nature. 2001;413:488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 2.Tsai HM. Journal of Thrombosis and Haemostasis. 2003;1:2038–2040. doi: 10.1046/j.1538-7836.2003.t01-1-00377.x. [DOI] [PubMed] [Google Scholar]
- 3.Majerus EM, Anderson PJ, Sadler JE. Journal of Biological Chemistry. 2005;280:21773–21778. doi: 10.1074/jbc.M502529200. [DOI] [PubMed] [Google Scholar]
- 4.Banno F, Kaminaka K, Soejima K, Kokame K, Miyata T. Journal of Biological Chemistry. 2004;279:30896–30903. doi: 10.1074/jbc.M314184200. [DOI] [PubMed] [Google Scholar]
- 5.Shang DZ, Zheng XW, Niiya M, Zheng XL. Blood. 2006;108:2207–2215. doi: 10.1182/blood-2006-02-002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan SO, Homer VM, Davis RL, Meyer M, George PM. Thrombosis and Haemostasis. 2006;96:535–537. [PubMed] [Google Scholar]
- 7.Censarek P, Steger G, Paolini C, Hohlfeld T, Grosser T, Zimmermann N, Fleckenstein D, Schror K, Weber AA. Thrombosis and Haemostasis. 2007;98:1309–1315. [PubMed] [Google Scholar]
- 8.Zheng XL, Chung D, Takayama TK, Majerus EM, Sadler JE, Fujikawa K. Journal of Biological Chemistry. 2001;276:41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 9.Uemura M, Tatsumi K, Matsumoto M, Fujimoto M, Matsuyama T, Ishikawa M, Iwamoto TA, Mori T, Wanaka A, Fukui H, Fujimura Y. Blood. 2005;106:922–924. doi: 10.1182/blood-2005-01-0152. [DOI] [PubMed] [Google Scholar]
- 10.Xu L, Hui AY, Albanis E, Arthur MJ, O’Byrne SM, Blaner WS, Mukherjee P, Friedman SL, Eng FJ. Gut. 2005;54:142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geetha S, Allen CE, Hunt RC, Plum E, Garfield S, Friedman SL, Soejima K, Sauna ZE, Kimchi-Sarfaty C. Cytometry Part A. 2009;75A:675–681. doi: 10.1002/cyto.a.20748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauna ZE, Okunji C, Hunt RC, Gupta T, Allen C, Plum E, Blaisdell A, Grigoryan V, Geetha S, Fathke R, Soejima K, Kimchi-Sarfaty C. Plos One. 2009;4:e6506. doi: 10.1371/journal.pone.0006506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeo G, Burge CB. Journal of Computational Biology. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 14.Ishikawa Y, Wirz J, Vranka JA, Nagata K, Bachinger HP. J Biol Chem. 2009;284:17641–17647. doi: 10.1074/jbc.M109.007070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Molecular Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 16.Pimanda JE, Maekawa A, Wind T, Paxton J, Chesterman CN, Hogg PJ. Blood. 2004;103:627–629. doi: 10.1182/blood-2003-04-1346. [DOI] [PubMed] [Google Scholar]
- 17.Kim E, Goren A, Ast G. Trends in Genetics. 2008;24:7–10. doi: 10.1016/j.tig.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Donati MB. Haemostasis. 1994;24:128–131. doi: 10.1159/000217092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Fig. 1 Probability of RNA splicing for known splice sites in the human ADAMTS13 gene based on the ‘Maximum Entropy Principle’. MaxEntScan was used to score the splice site signals of each exon-intron junction (see Design and methods). L (in isoforms 2 and 3) and S (in isoform 1) refer to the long or short version of intron 7 and 25 (see Fig. 1A).
Supplement Fig. 2 A schematic representation intron 25 retention. Alternative 5′ ss are marked (i defines exon 25 in WT and ii defines exon 25 in isoform 2 and 3). Conservation along this region is depicted below in the mountain-like graph reproduced from the UCSC Genome Browser (MultiZ alignment). Peak height corresponds to conservation level among 17 animal species.
Supplement Fig. 3 Alignment of human (isoform 1), mouse intron 25 and flanking exons. Unique human sequences are indicated with black bars and numbered 1–6. Matched nucleotides between human and mouse are indicated with asterisks.