Editor: The nonnucleoside reverse transcriptase inhibitor (NNRTI) class of drugs is a widely used component of highly active antiretroviral therapy, but a low barrier to resistance is a major limitation of this drug class.1 Etravirine, a new NNRTI, has potent antiviral efficacy against both wild-type and NNRTI-resistant subtype B HIV-1 strains.2 Rather than one or two mutations conferring resistance to a specific drug, such as nevirapine or efavirenz, resistance to etravirine develops through the accumulation of multiple NNRTI resistance-associated mutations (RAMs), i.e., higher genetic barrier to resistance.3,4 Specifically, 17 RAMs have been associated with virologic responses to etravirine use and a relative weighted score has been proposed; 74%, 52%, and 38% patients receiving etravirine achieved viral suppression (<50 copies/ml) in the presence of 0–2, 2.5–3.5, and >3.5 of scores, respectively.5
Unlike the subtype B epidemic found in western countries where etravirine has been evaluated, the HIV epidemic in the Southeast Asian countries is mainly caused by subtype CRF_01 AE, and etravirine resistance has not been evaluated in relation to this or other non-B subtypes.6–8 This is important, as the NNRTIs nevirapine and efavirenz are extensively used in this region where viral load monitoring is limited, thereby allowing for prolonged periods of undetected virologic failure during exposure to these drugs, which would increase the accumulation of NNRTI-RAMs.9 In addition, the genetic background of the infecting HIV-1 subtype may also influence the types and cross-resistance of the NNRTI-RAMs that emerge.10,11 To investigate these issues, we examined the associated factors and frequency of etravirine cross-resistance in a clinical practice in which viral loads are available and among patients infected with CRF_01 AE failing first-line efavirenz- and nevirapine-based regimens in Thailand.
All patients followed at Bamrasnaradura Infectious Diseases Institute, Ministry of Public Health, Thailand, between January 2005 and June 2008 were evaluated for antiretroviral therapy failure based on guidelines for antiretroviral therapy of the Thai AIDS Society, which define failure as viral load >1000 copies/ml after 6 months of receiving treatment or a rebound of viral load to >1000 copies/ml in any duration after undetectable viral load.12 For cohort patients identified with failing first-line antiretroviral therapy regimens, the HIV-1 RNA pol gene was genotyped (TRUGENE HIV-1). All sequences were aligned and analyzed using Geneious Pro 4.5.4. Genotypic NNRTI susceptibility was determined using the Stanford Resistance Database (http://hivdb.stanford.edu/ accessed February 2009), which included the following list of 17 etravirine-RAMs: V90I, A98G, L100I, K101E/H/P, V106I, E138A, V179D/F/T, Y181C/I/V, G190A/S, and M230L.9 A weighted etravirine-RAM score of 0–2, 2.5–3.5, and >3.5 was computed for each sequence,5 and RAM codons were compared across subtype B and CRF01_AE consensus sequences (http://www.hiv.lanl.gov accessed February 2009). Frequencies (%) and median (interquartile range, IQR) were used to describe demographic characteristics, and odds ratios were presented. Fisher's exact test was used to compare frequencies of etravirine-RAMs between weighted score 0–2 versus >2, 0–3.5 versus >3.5, and among those patients receiving nevirapine versus efavirenz. Pearson's correlation was used to analyze the correlation of weighted scores and plasma HIV-1 RNA at the time of virologic failure. All analyses were performed using SPSS software version 11.5 (SPSS Inc., Chicago, IL). A two-tailed p value less than 0.05 was considered statistically significant. All sequences are available at http://id.ucsd.edu/Faculty/DaveySmithMD/DATA/tabid/338/Default.aspx.
A total of 147 pol sequences were obtained from patients with first virologic failure who received nevirapine- and efavirenz-based regimens. Thirteen non-CRF01_AE sequences were excluded. A total of 134 sequences were included into the final analysis. In all, 110 (82%) patients were receiving nevirapine-based regimens and 24 (18%) were receiving efavirenz. At time of failure, median (IQR) viral load was 4.1 (3.5–4.8) log10 copies/ml, median (IQR) CD4 cell count was 142 (72–206) cells/μl, and median treatment duration was 2.1 years. The most recent viral load measurement before recognition of virologic failure was obtained after a median of 6.1 months. The median nadir CD4 cell count was 33 (5–86) cells/μl. Backbone nucleoside reverse transcriptase (NRTI) regimens included 76% stavudine + lamivudine, 15% zidovudine + lamivudine, 6% tenofovir + lamivudine, 2% zidovudine + didanosine, and 2% stavudine + didanosine.
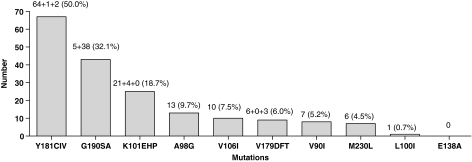
All etravirine-RAMs except K101P, E138A, and V179F were found, and the frequency of each etravirine-RAM is shown in Fig. 1. By the weighted scoring, 59 (44%), 58 (43%), and 17 (13%) of all sequences had scores of 0–2, 2.5–3.5, and >3.5, respectively. An etravirine-weighted score >2.0 was associated with nevirapine-based regimens (p < 0.001, OR = 6.7), viral load ≥4.5 log10 copies/ml (p = 0.025, OR = 2.5), and Y181C mutations (p < 0.001, OR = 333.3). An etravirine-weighted score >3.5 was associated with mutations: Y181C (p < 0.001, OR = 1.4), V106I (p < 0.001, OR = 26.3), and G190A (p = 0.020, OR = 3.4). In addition, a weighted score of >2.0 was also associated with failing nevirapine (p = 0.001, OR = 5.3). By correlation analysis, higher weighted scores trended toward an association (p = 0.063, r = 0.163) with higher viral loads at time of virologic failure. The patients who received a nevirapine-based regimen at time of failure had a higher proportion of Y181C (56% vs. 8%, p < 0.001 and OR = 14.2). Conversely, those who received an efavirenz-based regimen had a higher proportion of A98G (21% vs. 6%, p = 0.040 and OR = 3.9). All thymidine analogue mutations were not found to be associated with etravirine-RAMs (p > 0.05). Comparing the wild-type consensus sequences of subtype B to that of CRF01_AE, the percent similarity of nucleotide codons influencing etravirine-RAMs was 100% concordant; however, the percent similarity for other subtypes was 98%, 94%, 90%, 90%, 88%, and 88% for subtypes D, H, C, G, A, and F, respectively. In our sample, the maximum likelihood estimate of the mean pairwise genetic similarity between CRF01_AE pol sequences, based on the fit of the codon model to a neighbor joining tree, was 94.2%.
FIG. 1.
Frequency of etravirine-RAMs among 134 CRF01_AE sequences.
This study shows that the prevalence of high-level etravirine resistance in clinical practice is substantial after virologic failure of first-line NNRTI-based regimens, particularly nevirapine, among patients infected with the CRF01_AE subtype in a tertiary care HIV referral center in Thailand. Of note, mutation Y181C was the most frequent etravirine-RAM and was detected in almost half of all sequences, which is probably because this RAM is frequently isolated from patients treated with nevirapine13 and approximately 80% of our study patients were experiencing nevirapine-based regimen failure. The K103N mutation, commonly isolated from patients experiencing efavirenz-based regimen failure,14 is not a recognized etravirine RAM.5 Moreover, a study from Italy among individuals most likely infected with subtype B virus demonstrated that a delayed diagnosis of virologic failure contributes to a 5% increase in risk of etravirine-RAMs for each month after the first detectable viral load.15 In our clinical setting, we were fortunate to have access to virologic monitoring, approximately every 6 months; some regions in Southeast Asia are not as fortunate and use WHO immunologic criteria to define ART failure,16 which most likely exacerbates the observed situation of increased development of etravirine RAMs during first-line NNRTI-based ART.
Although evaluating etravirine response by weighted score is not widely accepted, this study can provide some beneficial non-subtype B data; therefore we also investigated how the infecting subtype could influence the development of etravirine RAMs. We found that the consensus nucleotide codons influencing etravirine-RAMs were not different between subtype CRF01_AE and subtype B, which may explain the similarity between our results and the Italian study.15 In our survey of other subtypes, we also found considerable similarity in viral genetic backgrounds for the development of etravirine RAMs; however, subtypes A and F might be the most likely to demonstrate varying patterns of NNRTI drug resistance and should be further evaluated.11
Given these results, etravirine should most likely be avoided in salvage regimens in the setting of first-line nevirapine failure and where drug resistance testing is not performed. Because etravirine-RAM codons are relatively conserved across subtypes, the prevalence of resistance is likely to be high in other resource-limited settings with limited virologic monitoring and high use of nevirapine-based regimens. Similarly in these settings, transmitted drug resistance is most likely to be highly prevalent or on its way to be highly prevalent,17,18 and therefore the usefulness of etravirine as even a first-line ART drug is suspect.
Acknowledgment
The authors would like to thank Samruay Nilkamhang for her coordination of the study.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hammer SM. Eron JJ., Jr Reiss P, et al. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society-USA panel. JAMA. 2008;300(5):555–570. doi: 10.1001/jama.300.5.555. [DOI] [PubMed] [Google Scholar]
- 2.Vingerhoets J. Azijn H. Fransen E, et al. TMC125 displays a high genetic barrier to the development of resistance: Evidence from in vitro selection experiments. J Virol. 2005;79(20):12773–12782. doi: 10.1128/JVI.79.20.12773-12782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazzarin A. Campbell T. Clotet B, et al. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-2: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet. 2007;370(9581):39–48. doi: 10.1016/S0140-6736(07)61048-4. [DOI] [PubMed] [Google Scholar]
- 4.Madruga JV. Cahn P. Grinsztejn B, et al. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet. 2007;370(9581):29–38. doi: 10.1016/S0140-6736(07)61047-2. [DOI] [PubMed] [Google Scholar]
- 5.Vingerhoets J. Peeters M. Azijn H, et al. An update on the list of NNRTI mutations associated with decreased virological response to etravirine: Multivariate analysis on the pooled DUET-1 and DUET-2 clinical trial data. Antivir Ther. 2008;13(Suppl 3):A26. [Google Scholar]
- 6.Hemelaar J. Gouws E. Ghys PD. Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 7.Llibre JM. Santos JR. Puig T, et al. Prevalence of etravirine-associated mutations in clinical samples with resistance to nevirapine and efavirenz. J Antimicrob Chemother. 2008;62(5):909–913. doi: 10.1093/jac/dkn297. [DOI] [PubMed] [Google Scholar]
- 8.Sungkanuparph S. Manosuthi W. Kiertiburanakul S. Piyavong B. Chantratita W. Evaluating the role of etravirine in the second-line antiretroviral therapy after failing an initial non-nucleoside reverse transcriptase inhibitor-based regimen in a resource-limited setting. Curr HIV Res. 2008;6(5):474–476. doi: 10.2174/157016208785861230. [DOI] [PubMed] [Google Scholar]
- 9.Hirsch MS. Gunthard HF. Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47(2):266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Cajas JL. Pant-Pai N. Klein MB. Wainberg MA. Role of genetic diversity amongst HIV-1 non-B subtypes in drug resistance: A systematic review of virologic and biochemical evidence. AIDS Rev. 2008;10(4):212–223. [PubMed] [Google Scholar]
- 11.Kosakovsky Pond SL. Smith DM. Are all subtypes created equal? The effectiveness of antiretroviral therapy against non-subtype B HIV-1. Clin Infect Dis. 2009;48(9):1306–1309. doi: 10.1086/598503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sungkanuparph S. Anekthananon T. Hiransuthikul N, et al. Guidelines for antiretroviral therapy in HIV-1 infected adults and adolescents: The recommendations of the Thai AIDS Society (TAS) 2008. J Med Assoc Thai. 2008;91(12):1925–1935. [PubMed] [Google Scholar]
- 13.Richman DD. Havlir D. Corbeil J, et al. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68(3):1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bacheler LT. Anton ED. Kudish P, et al. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob Agents Chemother. 2000;44(9):2475–2484. doi: 10.1128/aac.44.9.2475-2484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapadula G. Calabresi A. Castelnuovo F, et al. Prevalence and risk factors for etravirine resistance among patients failing on non-nucleoside reverse transcriptase inhibitors. Antivir Ther. 2008;13(4):601–605. [PubMed] [Google Scholar]
- 16.Scaling up Antiretroviral Therapy in Resource-Limited Settings. Treatment Guidelines for a Public Health Approach. Geneva: Joint United Nations Programme on HIV/AIDS (UNAIDS) and World Health Organization (WHO); 2006. revision. [Google Scholar]
- 17.Booth CL. Geretti AM. Prevalence and determinants of transmitted antiretroviral drug resistance in HIV-1 infection. J Antimicrob Chemother. 2007;59(6):1047–1056. doi: 10.1093/jac/dkm082. [DOI] [PubMed] [Google Scholar]
- 18.Smith DM. Wong JK. Shao H, et al. Long-term persistence of transmitted HIV drug resistance in male genital tract secretions: Implications for secondary transmission. J Infect Dis. 2007;196(3):356–360. doi: 10.1086/519164. [DOI] [PubMed] [Google Scholar]