Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding
Internal initiation of translation on picornavirus IRESs requires canonical initiation factors as well as specific IRES trans-acting factors, such as the pyrimidine tract-binding protein PTB, whose mode of action has remained largely unclear. Now, Richard Jackson et al elucidate the mechanistic function of PTB in poliovirus type 1 IRES-mediated translation initiation.
Keywords: eIF4G, IRES, poliovirus, polypyrmidine tract-binding protein (PTB), tethered hydroxyl-radical probing
Abstract
Tethered hydroxyl-radical probing has been used to determine the orientation of binding of polypyrimidine tract-binding protein (PTB) to the poliovirus type 1 (Mahoney) (PV-1(M)) internal ribosome entry site/segment (IRES)—the question of which RNA-binding domain (RBD) binds to which sites on the IRES. The results show that under conditions in which PTB strongly stimulates IRES activity, a single PTB is binding to the IRES, a finding which was confirmed by mass spectrometry of PTB/IRES complexes. RBDs1 and 2 interact with the basal part of the Domain V irregular stem loop, very close to the binding site of eIF4G, and RBDs3 and 4 interact with the single-stranded regions flanking Domain V. The binding of PTB is subtly altered in the presence of the central domain (p50) of eIF4G, and p50 binding is likewise modified if PTB is present. This suggests that PTB stimulates PV-1(M) IRES activity by inducing eIF4G to bind in the optimal position and orientation to promote internal ribosome entry, which, in PV-1(M), is at an AUG triplet 30 nt downstream of the base of Domain V.
Introduction
Polypyrimidine tract-binding protein (PTB), which is also known as hnRNP-I, is a predominantly nuclear protein and an important regulator of alternative splicing (reviewed in Valcarcel and Gebauer, 1997; Sawicka et al, 2008). It is known to be a monomeric protein with a somewhat extended conformation (Simpson et al, 2004; Monie et al, 2005) and has four RNA-binding domains (RBDs) of the RNP1/RNP2 class, although the amino-acid sequences of these motifs in PTB are somewhat non-canonical. Alternative splicing results in two variants of the prototypic PTB1: PTB2 and PTB4, which differ from PTB1 by the insertion of 19 or 26 amino acids, respectively, in the linker between RBDs2 and 3. In addition, there are two paralogues encoded by different genes: nPTB, which is enriched in cells of neuronal origin and ROD1, which is especially abundant in haematopoietic cells (Spellman et al, 2005).
There is considerably more PTB in the cytoplasm than is typical of hnRNPs (Ghetti et al, 1992), which suggests the possibility that it may also have a function in cytoplasmic events. Stimulation of translation initiation dependent on picornaviral internal ribosome entry sites/segments (IRESs) was the first such cytoplasmic function to be discovered (reviewed in Jackson and Kaminski, 1995). Subsequently, PTB has been reported to stimulate the activity of many cellular mRNA IRESs, and it has been suggested that PTB may be a general trans-acting factor for IRES-dependent initiation (Sawicka et al, 2008).
The binding of PTB to its pre-mRNA splicing targets and to various IRESs has been extensively studied using UV-crosslinking, gel-shift and foot-printing assays. However, as there are four RBDs, there is the possibility of multi-site interactions with its target RNAs in a variety of orientations. To explore the orientation of binding, we have recently developed a series of PTB derivatives suitable for tethered hydroxyl-radical probing (Kafasla et al, 2009). In this approach, PTB mutants, each with a single cysteine at a strategically chosen site, are conjugated with Fe(II)-BABE, followed by verification that the conjugated derivatives retain functional biological activity towards the particular RNA target under investigation. After forming the RNA–protein complex with the conjugated PTB derivative, hydroxyl radicals are generated at the Fe(II) by the Fenton reaction. These radicals cause RNA backbone cleavage, irrespective of structure or sequence, but because they are very short lived, cleavage is limited to sites in close proximity to the Fe(II), facilitating identification of which RNA segment is nearest that particular conjugated cysteine residue (Culver and Noller, 2000). By placing the cysteine residues in different RBDs in PTB, this approach allows the sites of binding of each RBD to be mapped (Kafasla et al, 2009).
We have recently reported the use of this method, in conjunction with mass spectrometry of the PTB/RNA complexes, to analyse the interactions of PTB1 with the encephalomyocarditis virus (EMCV) IRES (Kafasla et al, 2009). The complete 5′-UTR downstream of the poly(C) tract (i.e. nt 280–842) bound two PTBs: one to the nt 280–400 segment, which is not an essential part of the IRES and the other binding with slightly lower affinity to the core IRES (nt 400–842). In the latter case, RBDs1 and 2 bound near the 3′-end of the core IRES, and RBDs3 and 4 near the 5′-end. This suggests that PTB binding to the core IRES places constraints on the flexibility of the three-dimensional structure of the IRES, thus stabilising this structure in a form, which is favourable for internal initiation of translation.
In the work reported here, we have used the same approaches to examine the interaction of PTB1 with the poliovirus type 1 (PV-1) IRES, which belongs to a different class of picornavirus IRESs than EMCV. Picornavirus IRESs have traditionally been classified into two main groups (Alexander et al, 1994; Jackson and Kaminski, 1995): type I, exemplified by poliovirus and human rhinoviruses, and type II, exemplified by EMCV and foot-and-mouth disease virus. Within each group, there is fairly strong conservation of IRES primary sequence, and even stronger conservation of secondary structure, but there is little in common between the two groups apart from a conserved ∼25 nt pyrimidine-rich tract ending with an AUG triplet at the extreme 3′-end of the IRES as defined by deletion mapping (Jackson and Kaminski, 1995). In the case of the EMCV IRES, this AUG is the authentic initiation site for viral polyprotein synthesis, and there is good evidence that 43S complexes (40S ribosomal subunits with associated initiation factors) bind directly to this AUG and all initiate there (Kaminski et al, 1990, 1994). In contrast, the corresponding AUG triplet in type I IRESs (at nt 586 in PV-1) is not the initiation site for viral polyprotein synthesis, which is the next AUG further downstream, at a distance of ∼35 nt in human rhinoviruses, and ∼150 nt in the polioviruses. Nevertheless, AUG-586 in PV-1, and its counterpart in other type I IRESs, is very important for internal initiation as its mutation severely reduces initiation at the correct site (Pelletier et al, 1988; Meerovitch et al, 1991; Nicholson et al, 1991; Pilipenko et al, 1992). This has led to the idea that all (or most) ribosomes enter or land at AUG-586, but very few, if any of them initiate at this site, and instead they are transferred, most likely by linear scanning, to the next AUG further downstream (Jackson and Kaminski, 1995).
Apart from this difference in the detailed mechanism of internal initiation on the two types of picornavirus IRES, there are also differences in the stringency of the PTB requirement. It is absolutely required for all the type I (entero- and rhinovirus) IRESs that have been tested (Hunt and Jackson, 1999), but it is stimulatory rather than essential for internal initiation on the EMCV IRES, with the degree of stimulation depending on the particular reporter and IRES variant used (Kaminski and Jackson, 1998).
We show here that, in contrast to the binding of PTB at widely dispersed sites in the generally accepted two-dimensional structure of the EMCV IRES, a single PTB binds to the poliovirus IRES in a highly localised manner, mainly to the basal half of Domain V (an irregularly base-paired stem loop just upstream of the pyrmidine-rich tract), and principally through RBDs1 and 2, with RBDs3 and 4 binding to the short single-stranded regions flanking this domain. Domain V is where the attenuating mutations in the Sabin vaccine strains are located, as well as being where eIF4G binds (de Breyne et al, 2009), and we further show that the binding sites of PTB and the central domain of eIF4G on Domain V partially overlap, with the consequence that each protein reciprocally modifies the binding of the other.
Results
Design, assay and derivatisation of PTB mutants
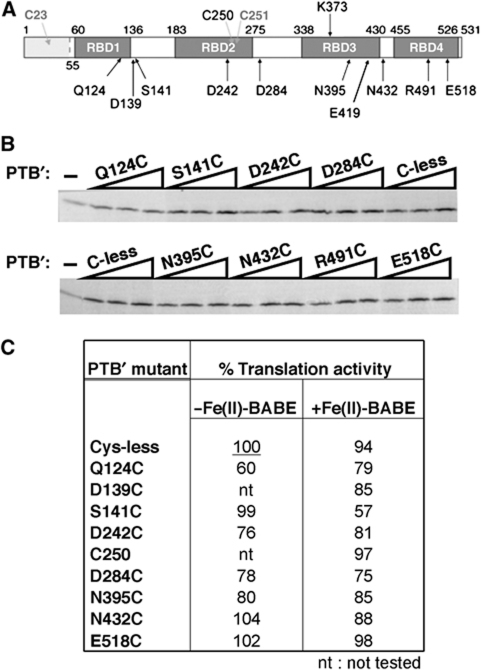
To map the interaction of PTB with the PV IRES, we used the single Cys-replacement PTB mutants described in detail previously (Kafasla et al, 2009). All the derivatives were made in the background of PTB1 with a deletion of the N-terminal domain (amino acids 1–54), which has nuclear import and export signals, but is not involved in the interaction of PTB with RNA. This parent ΔNTD-PTB construct (hereafter designated PTB′) also has an N-terminal His-tag, and the two remaining cysteine residues (C250 and C251 in RBD2) have been changed to serines by point mutagenesis (Kafasla et al, 2009). Seventeen single cysteine substitution mutants of PTB′ were constructed, and two further derivatives were generated by reinstating either C250 or C251. The choice of positions for introducing the cysteines was based on the NMR structures of each RBD complexed with an oligonucleotide ligand (Oberstrass et al, 2005), with the aim that the cysteines should be sufficiently exposed to be reactive towards Fe(II)-BABE, and near enough to the RNA-binding surface that the Fe(II) would likely be close to the RNA backbone, yet far enough away from this surface that the bulkiness of the Fe(II)-BABE moiety would not interfere with RNA binding. In practice, only three of the mutants (including the C251 derivative) failed to be derivatised efficiently (Kafasla et al, 2009). For mapping the interactions of PTB′ with the PV-1 IRES, we mainly used the following subset of mutants (Figure 1A), on the grounds that these gave the strongest cleavage signals in tethered hydroxyl-radical probing assays: Q124C and S141C in RBD1; D242C, C250 and D284C in RBD2; N395C and N432C in RBD3; and E518C in RBD4. Interestingly, these were not precisely the same as the mutants, which gave the strongest signals with the EMCV IRES, which were Q124C and D139C in RBD1; N219C and D284C in RBD2; N395C, E419C and N432C in RBD3; and R491C and E518C in RBD4. It should be noted that although we have assigned N432C to RBD3, this is somewhat arbitrary as it is located approximately in the middle of the short linker joining RBDs 3 and 4 (Oberstrass et al, 2005).
Figure 1.
Activity of single Cys-replacement mutants of PTB1 in PV IRES-driven translation. (A) Schematic representation of PTB1 showing the positions of the four RBDs and of the amino acids that were replaced by Cys to generate the single Cys-replacement mutants used in this study. Residues 1–54, which are deleted in ΔNTD-PTB (PTB′), are shown by light shading. The native cysteine residues (C250, C251) that were replaced by Ser in PTB′ are also shown: a mutant with C250 reinstated as the sole Cys residue was used in probing assays. (B) Representative assay of the designated single Cys mutant derivatives of PTB′ for their ability to enhance PV IRES-driven translation. Uncapped monocistronic mRNA (12 nM) with the PV IRES linked to CAT-coding sequence was translated in standard reticulocyte lysate (−) and in lysate supplemented with purified recombinant hexahistidine-tagged PTB′ mutants at final concentrations of 47, 94 and 188 nM. Translation products were analysed by SDS–PAGE, and the resulting autoradiograph is shown. (C) Summary of translation assay results for the designated mutants before and after their reaction with Fe(II)-BABE. After subtracting the background yield obtained in the absence of any added PTB, the maximum value of the stimulation observed for each mutant was expressed as a percentage of the maximum seen with the Cys-less PTB′ control derivative (set at 100%) that had not been taken through the mock-conjugation procedure. The results shown for each mutant before and after conjugation are the mean of the values from three independent experiments.
We assessed the ability of each of the single Cys PTB′ mutants, and their Fe(II)-BABE derivatives, to enhance the activity of the PV IRES in rabbit reticulocyte lysate, using an RNA transcript (12 nM) with the CAT-coding sequence downstream of the PV-1 (Mahoney) (PV-1(M)) 5′-UTR (nucleotides 1–743; Figure 2A). Although we used a PTB-depleted reticulocyte lysate for assaying the effect of the mutants on EMCV IRES activity, this was not feasible for the PV IRES, presumably because of co-depletion of other essential proteins. However, the standard nuclease-treated lysate was considered appropriate, because the PTB requirement is more stringent and its apparent affinity lower for the poliovirus IRES than the EMCV IRES (Kaminski and Jackson, 1998), so that even though this batch of lysate was estimated to contribute 40 nM endogenous PTB to the assay (Supplementary Figure S1A), added PTB nevertheless elicited a significant stimulation. A minimum of three concentrations of each PTB′ mutant (47, 94 and 188 nM as in the representative assays shown in Figure 1B) and up to five different concentrations (24–376 nM) were used. With full-length wild-type PTB, the maximum stimulation (generally ∼three-fold) was achieved at 94 or 188 nM added protein, and, as expected, the Cys-less PTB′ control gave almost exactly the same outcome (Supplementary Figure S1B). Unlike the case with the EMCV IRES (Kafasla et al, 2009), higher concentrations (above 188 nM) of wild-type PTB or PTB′ mutants generally resulted in quite a sharp downturn in translation yield (Supplementary Figure S1B), for unknown reasons. For each PTB′ mutant, the maximum stimulation was expressed as a percentage of the maximum stimulation observed with the Cys-less PTB′ derivative, and the results for all the mutants that provided the most informative tethered hydroxyl-radical probing data are summarised in Figure 1C. It is evident that all of them retained significant activity (>∼75% of C-less PTB′), apart from Q124C, and that conjugation with Fe(II)-BABE had little negative effect on the stimulatory activity, with the exception of S141C (Figure 1C), which may be due to some steric hindrance resulting from the bulkiness of the Fe(II)-BABE moiety. Nevertheless, the activity of the Fe(II)-BABE derivatives of Q124C and S141C was considered high enough to justify their use in probing assays.
Figure 2.
Representative primer extension analysis of directed hydroxyl-radical probing of PV IRES with Fe(II)-BABE PTB′ mutants. (A) Schematic representation of the PV-1(M) 5′-UTR, showing the cloverleaf domain (I), the putative ribosome entry site AUG (at nt 586), the initiation codon for viral polyprotein synthesis (at nt 743) and the individual domains (I–VI) (adapted from Balvay et al, 2009). The annealing positions of the oligonucleotide primers (PVpr1–PVpr8) used in the primer extension analysis of directed hydroxyl-radical probing of the PV IRES are also shown. (B, C) PV IRES RNA (100 nM) was incubated with the specified derivatised PTB′ mutants (700 nM), or without any protein added (−). The RNA cleavage products were analysed by primer extension using (B) primer PVpr6 to analyse nucleotides 470–570 of the PV IRES, and (C) primer PVpr5 to analyse nucleotides 390–490. Sites of hydroxyl-radical cleavage are indicated by the vertical lines to the left of each lane. The brackets on the left of the two gels (B, C) indicate the same set of cleavages detected by both primers. Lanes T, C, G, A depict sequencing ladders generated from the same primers. The background bands used as ‘reference standards' (see text) for normalisation in the determination of cleavage signal strength are highlighted by arrows.
Directed hydroxyl-radical probing of the PV IRES
The derivatised mutants described above were used in directed hydroxyl-radical probing assays to map the interaction of PTB with the PV IRES. The Fe(II)-BABE PTB′ derivatives, as well as the mock-conjugated Cys-less control, were pre-incubated at 37°C with the PV IRES RNA for 30 min, then ascorbic acid and hydrogen peroxide were added, and incubation continued for 15 min on ice, before the reaction was quenched by phenol extraction of the RNA fragments. The cleavage sites were identified by primer extension analysis, using oligonucleotide primers PVpr1–PVpr8 designed to hybridise to the RNA at the positions shown in Figure 2A. Use of the complete set of these primers enables detection of cleavages produced along the whole length of the 5′-UTR of PV-1(M). Representative examples of the primer extension gels with two different primers, PVpr6 and PVpr5, which detected the strongest cleavages, are shown in Figure 2B and C, respectively. As noted previously (Kafasla et al, 2009), a number of cleavages were detected after addition of hydrogen peroxide and ascorbic acid, either in the absence of any protein, or in the presence of the mock-derivatised Cys-less PTB′ (Figure 2B and C). The bands in the Cys-less lane were treated as the background, and bands in lanes with Fe(II)-BABE PTB′ mutants were taken to be genuine products of hydroxyl-radical cleavage only if they were entirely absent from the Cys-less control lane (or were present in this control lane, but at decidedly lower intensity).
The RNA probe used in the first experiments encompassed nt 94–630 of the PV-1(M) 5′-UTR, which consists of five secondary structure domains (Domains II–VI) as shown in Figure 2A. This is considered to include the complete core IRES, because deletions, which removed most or all of the PV-1(M) 5′-UTR downstream of AUG-586, had very little effect on IRES activity, and 5′-deletions that removed the first ∼100 nt (including all of the Domain I cloverleaf) reduced IRES activity by <40%, whereas 5′-deletions extending into Domain II or 3′-deletions extending into the oligopyrimidine tract resulted in extremely defective IRESs (Kuge and Nomoto, 1987; Iizuka et al, 1989; Nicholson et al, 1991). With our standard probe concentration of 100 nM, appreciable backbone cleavage was generally seen at a molar ratio of Fe(II)-PTB′/IRES of 3.5. Increasing the Fe(II)-PTB′/IRES molar ratio to 7, which was generally used as the standard, resulted in slightly stronger cleavage product bands on the gels. Raising the Fe(II)-PTB′/IRES molar ratio further resulted in an increase of the non-specific background cleavages and a reduction in the ratio of specific cleavage signal over non-specific noise, as we observed with the EMCV IRES at high PTB/RNA ratios (Kafasla et al, 2009). In an attempt to detect the possibility of low-affinity binding of PTB to other specific sites, we raised the PTB′/RNA molar ratio as high as 21 (at 100 nM IRES RNA), but no additional cleavage product bands were clearly detected over and above those seen at a ratio of 7 (data not shown). The primers used for primer extension analysis of the cleavage products were designed so that the cleavage bands, which were seen in the upper part of the gel with one particular primer, could be verified by using the next primer annealing to an upstream site in the IRES, which revealed the same cleavage sites in the lower part of the gel (as shown for PVpr6 and PVpr5 in Figure 2B and C).
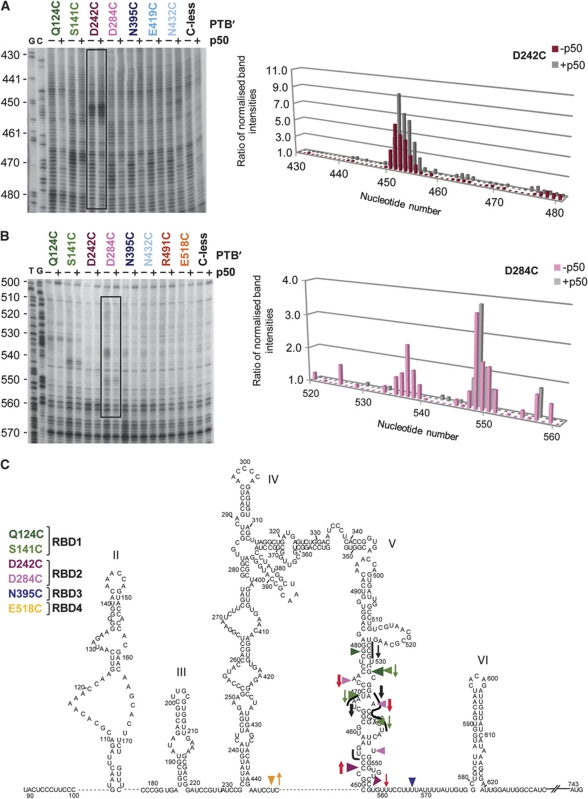
To estimate the relative efficiency of cleavage at different sites, we used the Semi-Automated Foot-printing Analysis (SAFA) software (Das et al, 2005; Laederach et al, 2008), which facilitated a high throughput quantification of the intensity of all bands on the primer extension gels. To compensate for possible variations in loading, these band intensities were normalised with respect to background bands (arrowed in Figures 2B, C and 3A), whose intensity was largely invariant in all lanes (including the Cys-less control lane), but sometimes fluctuated in a few lanes in parallel with each other (and with the full-length primer extension product when it was visible). This normalised intensity for each individual band in a given lane was compared with the intensity of the corresponding normalised band in the Cys-less control lane. The derived ratios were plotted against the nucleotide position in the PV IRES, as illustrated in the representative example of Figure 3B, which shows the ratios derived from the primer extension gel of Figure 3A.
Figure 3.
Quantitation of the directed hydroxyl-radical probing results using SAFA software. (A) PV IRES RNA (100 nM) was incubated with the specified derivatised PTB′ mutants (700 nM), or without any protein added (−). The RNA cleavage products were analysed by primer extension using primer PVpr5. Sites of hydroxyl-radical cleavage are indicated by vertical lines to the left of each lane. Lanes T, C, G, A depict sequencing ladders generated by the same primer. The invariant background bands used as ‘reference standards' for normalisation in the determination of cleavage signal strength are highlighted by arrows. (B) Graph derived from quantification of the gel shown in (A) using the SAFA software, as described in the text. After normalisation to adjust for small loading variations (see text), the intensity of each band in lanes with an Fe(II)-PTB′ mutant was compared, as a ratio, with the intensity of the same band in the lane loaded with the mock-conjugated Cys-less control. (C) Schematic representation of the location of hydroxyl-radical cleavages in the PV-1(M) IRES produced by Fe(II)-PTB′ derivatives. The predicted secondary structure shown is as presented in Figure 2A. Cleavage strength assessed as described in the text is indicated as strong (large, filled arrowhead), medium (medium-sized, filled arrowhead) or weak (smallest open arrowhead). For clarity, clusters of cleavages at contiguous phosphodiester bonds are denoted by a single arrowhead at the site of strongest cleavage. The strength of cleavage at nt 566 by N395C-PTB′ was rather borderline and in different experiments varied between weak and medium.
The calculated ratios for all cleavage product bands were ranked, and the ranking list was divided into three groups (representing strong, medium and weak cleavages), following common practice and our previous analysis of PTB interactions with the EMCV IRES (Culver and Noller, 2000; Lancaster et al, 2002; Kafasla et al, 2009). If the ratio was <2.5 (but >1.0) that particular band was classed as a weak cleavage, whereas if the ratio was 4.5 or greater it was classed as a strong cleavage, with medium cleavage assigned to ratios in the range 2.5–4.4.
Mapping the cleavage sites produced by Fe(II)-PTB′ mutants on the PV IRES
Figure 3C summarises cleavage site data obtained with the full set of primers from all our probing experiments with the eight Fe(II)-BABE PTB′ derivatives that consistently gave the strongest signals. Where a given mutant produced cleavages in a number of adjacent phosphodiester bonds (as is usually the case, except for some very weak cleavages), for clarity, a single arrowhead (sized according to cleavage efficiency) is used to indicate either the strongest signal, or the central cleavage site in cases in which there was little difference in signal strength.
In mapping the interaction of PTB with the EMCV IRES, it was noted that the strong and medium cleavages generated by all mutants in a particular RBD are closely clustered in the conventional two-dimensional secondary structure map (Kafasla et al, 2009). In the case of the PV IRES, the same is clearly true of the RBD1 cleavages due to Q124C and S141C as well as the weaker cleavages seen with D139C (Figures 2B, C and 3C), and it is also true of the RBD2 cleavages due to D242C and D284C as well as the weaker cleavages seen with C250 (Figures 2B, C and 3C). However, RBD4 mutants gave medium strength cleavages at just one region (in the linker between Domains IV and V), with additional weak cleavages at the base of Domain III and in Domain IV (Figure 3C). N395C in RBD3 gave only one somewhat variable medium–weak cleavage in the first part of the oligopyrimidine tract, and no strong or medium strength cleavages were seen with two other RBD3 mutants tested: K373C and E419C (data not shown). In addition, the only cleavages seen with N432C-PTB′, which has the Cys in the short linker between RBDs 3 and 4 (Kafasla et al, 2009), were weak cuts at the base of Domains V and IV, quite close to the slightly stronger cleavages produced by RBD3 N395C and, more especially, RBD4 E518C. This pattern of rather weak cleavages generated by PTB′ mutants in RBD3 and the linker between RBDs3 and 4 suggests that RBD3 may not be in close proximity to the IRES RNA.
The finding of numerous, but closely clustered cleavages generated by the mutants in RBDs1 and 2, coupled with no more than one medium strength cleavage from mutants in each of RBDs3 and 4, suggests that under our probing conditions, the PV IRES is binding a single PTB in a unique orientation. For the purposes of deducing this orientation (depicted later in Figure 6), we focused on the strong and medium cleavages, as these will arise from high occupancy PTB binding with the RNA reasonably close to the relevant RBD residue, which will not be the case for most, if not all, of the weak cleavages.
The same directed hydroxyl-radical probing experiments were performed with RNA probes encompassing nt 1–630 (the PV core IRES plus the Domain I cloverleaf structure; Figure 2A), or nt 1–743 plus an addition of 114 nt of the CAT-coding sequence. The pattern of the cleavages presented in Figure 3C did not change when the cloverleaf domain was included in the RNA probe, not even when the complete 5′-UTR sequence up to and including the authentic initiation codon was included, indicating that Domains II–VI are sufficient for strong binding of PTB to the PV IRES and that the orientation of PTB binding does not change in the presence of these additional sequences. When the probe included the cloverleaf structure, some additional very weak cleavages produced by S141C in the cloverleaf were observed, at nt 53, 80 and 90, possibly because of folding of the RNA such that the cloverleaf came fairly close to Domain V. Similarly, the weak cleavages at the top of Domain IV caused by RBD1 mutants (Figure 3C) are possibly because of the top of Domain IV folding towards Domain V.
Mass spectrometric analysis of the complex formed between PTB and PV IRES RNA
In our previous work on mapping the interaction of PTB with the EMCV IRES (Kafasla et al, 2009), mass spectrometric analysis proved to be a very precise and efficient way of determining the stoichiometry of PTB binding to the IRES, showing that the core EMCV IRES (nt 400–848) binds a single PTB, whereas the longer RNA probe (nt 280–848) binds two PTBs. We, therefore, applied this approach to analyse PTB interactions with the PV-1(M) IRES, using wild-type ΔNTD-PTB and the complete core IRES (nt 94-630) at nominal input PTB/RNA molar ratios of 1:1 and 2:1. The PTB/IRES complexes were assembled in 88 mM ammonium acetate (pH 7.4), 2 mM Mg2+, with RNA at 7.4 μM, followed by buffer exchange into 200 mM ammonium acetate with dilution to an RNA concentration of 3.7 μM. In view of the fact that the analysis was carried out at an RNA concentration ∼300-fold greater than in the in vitro translation assays (or 37-fold greater than for directed hydroxyl-radical probing assays), and that the apparent Kd of the functionally relevant PTB/IRES interaction is ∼70 nM as judged from the translation assay data (Supplementary Figure S1), we consider that the 1:1 PTB/RNA molar input ratio will approximate most closely to the degree of saturation of the RNA with bound PTB pertaining in the translation and probing assays.
As shown in Figure 4, at this 1:1 input molar ratio, a slight majority of the RNA was complexed with PTB, and only complexes formed by one molecule of RNA with one molecule of PTB could be detected, as expected from our hydroxyl-radical probing results (summarised in Figure 3C). This is very similar to the result obtained with the core EMCV IRES at the same PTB/RNA input ratio (Kafasla et al, 2009), except that 90% of the input EMCV IRES was assimilated into a 1:1 complex with PTB, as against only 60% of the PV IRES (Figure 4). This difference may be in part due to the slightly lower RNA concentration used for the PV IRES, but a more important parameter is likely to be the lower affinity of the functionally relevant PTB interaction with the PV IRES than with the EMCV IRES (Supplementary Figure S1B; Kafasla et al, 2009).
Figure 4.
Mass spectra of PV-1(M) IRES RNA, and complexes of the IRES with wild-type ΔNTD-PTB. The spectra were taken with RNA only (A), a molar PTB/RNA input ratio of 1:1 (B) and a molar input ratio of 2:1 (C). The charge states are shown in italics for RNA alone, in bold for the 1:1 PTB/RNA complex and in smaller font for the 2:1 PTB/RNA complex. The spectra are shown after background subtraction and smoothing, to improve clarity. The table shows the relative proportions of free RNA, 1:1 PTB/RNA complex and 2:1 PTB/RNA complex observed under the two conditions. These values were obtained using the method described in Natan et al (2009).
When the input molar ratio was increased to 2:1, all the PV IRES was complexed with PTB, with ∼50% of the RNA binding one PTB molecule and the rest participating in a complex with two PTB molecules per RNA. However, as mentioned previously, hydroxyl-radical probing at a molar ratio of PTB/IRES of 21 failed to reveal cleavages at additional sites, which were not cleaved at a PTB/IRES ratio of 7. This suggests that the second PTB, which binds to the IRES at the higher PTB/IRES input ratio under the conditions of the mass spectrophotometric analysis, is probably not binding at a specific site, or in a specific orientation.
PTB and the central domain of eIF4G reciprocally cause slight modification of each other's binding to the PV IRES Domain V
We went on to test whether other IRES-binding proteins modify the interaction of PTB with the IRES, starting with poly rC-binding protein 2 (PCBP2), and ‘upstream of N-ras' (unr), both of which have been shown to stimulate type I IRES activity. PCBP2 binds to the cloverleaf Domain I and to Domain IV, and is required for all type I IRESs tested so far (Blyn et al, 1997; Walter et al, 1999). Unr has been shown to stimulate poliovirus replication in vivo (Boussadia et al, 2003), but in cell-free systems the stimulation of PV-1(M) IRES activity is minimal, especially in comparison with the human rhinovirus IRES, which is strongly dependent on unr as well as PTB (Hunt et al, 1999). The binding sites of unr on the rhinovirus IRES have been shown to be principally Domain II and the upper (apical) half of Domain V (Anderson et al, 2007). When added to the probing assays neither PCBP2 on its own nor PCBP2 plus unr caused any changes in the intensity or position of the cleavages generated by all the Fe(II)-PTB′ mutants tested (data not shown), showing that PTB binding to the PV IRES is independent of these other ITAFs (IRES trans-acting factors).
However, the sites in which PTB RBDs1 and 2 interact with Domain V (Figure 3C) seem very close to the sites in which the central domain (p50) of eIF4G binds, according to hydroxyl-radical probing results of de Breyne et al (2009). We, therefore, examined whether the cleavage product patterns were different if eIF4G p50 was also present, using PV IRES RNA (100 nM), with or without the cloverleaf Domain I, and a variety of Fe(II)-PTB′ derivatives (700 nM), in the presence or absence of 700 nM or 1 μM eIF4G p50. The only changes observed were in and around Domain V, and they are evident in the representative gels presented in Figure 5A and B, together with the graphs generated using the SAFA software. It is immediately obvious that in the presence of p50, the cleavage bands produced by the derivatised PTB′ mutants in Domain V downstream of nt 460 were still present, but most of them were at reduced intensity (Figure 5A and B). In the extreme case, the cleavages generated by D284C at nt 537 usually disappeared almost completely (Figure 5B), which is likely due to the fact that nt 536–543 was found to be a site in which eIF4G p50 interacts with the IRES (de Breyne et al, 2009). On the other hand, the intensity of the cleavage bands produced around nt 453 (D242C) was enhanced by the presence of p50 (Figure 5A), as was also observed for the cleavage by E518C at nt 446 (data not shown), and the weak N432C cleavage at nt 451 (Figure 5A). These probing experiments were also carried out in the presence of 700 nM eIF4G p50 plus 700 nM eIF4A and 1 mM ATP, but the position and intensity of the cleavages were the same as with p50 alone (data not shown).
Figure 5.
Representative primer extension analysis of directed hydroxyl-radical probing of PV IRES nucleotides 430–480 (A) and 500–570 (B) using the designated Fe(II)-BABE PTB′ mutants in the presence or absence of eIF4G p50 fragment (the central one-third domain of eIF4G). Hydroxyl-radical probing was performed as in Figure 3A, using the specified derivatised PTB′ mutants (700 nM) in the presence (+) or absence (−) of eIF4G p50 (700 nM). The gels were quantified by SAFA as described in the text and the legend to Figure 3. Representative graphs corresponding to the areas of the gel enclosed in the boxes are shown next to each gel. The height of the histograms represents the ratio of the normalised band intensity in the experimental lane, with an Fe(II)-PTB′ mutant, to the normalised intensity of the same band in the mock-conjugated Cys-less control lane. (C) Simultaneous binding of PTB and central domain of eIF4G on PV IRES. The strong and medium cleavages caused by the designated PTB′ mutants are shown by coloured arrowheads as in Figure 3C. Green and red arrows (for RBD1 and RBD2 mutants, respectively) pointing down denote cleavages that were weaker when eIF4G p50 was present, and arrows pointing up signify cleavages that were stronger in the presence of p50. Thick arrows denote major changes, and thin arrows relatively small changes, as determined by the SAFA software output. The cleavage sites observed in tethered hydroxyl-radical probing assays using Fe(II)-BABE derivatives of eIF4G p50 (de Breyne et al, 2009) are shown as short black lines, and the black arrows pointing down show the sites in which a reduction in band intensity was seen when ΔNTD-PTB was present (see Figure 6), with the thickness of the arrow denoting the magnitude of the reduction. The cleavages seen at nt 467–470 in the absence of PTB were slightly displaced to nt 467–474 when PTB was present. No significant change in cleavage intensity was observed at nt 455–456.
These changes in cleavage band intensity are summarised in Figure 5C, in which upward pointing coloured arrows indicate enhanced cleavage and downward arrows reduced band intensity, with the thickness of the arrow signifying the magnitude of the changes. The fact that there were both increases and decreases in band intensity, with no change at some sites, such as nt 548 (Figure 5B and C), implies that the presence of p50 has not significantly changed the amount of PTB bound to Domain V. It should also be noted that there were no changes in the actual sites in which cleavage occurred, indicating that there was no major repositioning or reorientation of the PTB. Rather, the results suggest that PTB (strictly speaking the conjugated Fe moiety and the relevant Cys residue) moved closer to the RNA strand at the 5′-side of the base of Domain V and the Domain IV–V linker, and further away from the RNA at sites higher up Domain V, especially nt 535–540. An alternative explanation for the decreases in band intensity is that they could be due to shielding of the RNA backbone by p50 or quenching of hydroxyl radicals (Lancaster et al, 2002), as might occur if part of the bound p50 lies between the PTB′-associated Fe(II) moiety and the RNA backbone.
These results with eIF4G p50 prompted us to examine whether similar changes occurred in the presence of the complete (native) eIF4F complex, in view of the fact that it is eIF4F that drives poliovirus RNA translation in the early stages of infection until sufficient viral 2A protease has accumulated to promote cleavage of all the eIF4G component into an N-terminal one-third fragment (with the eIF4E interaction site) and a C-terminal two-thirds (p100) fragment; p100 is thought to have similar IRES-binding properties as the p50 central domain and similar activity in promoting picornavirus IRES functionality, at least with the EMCV IRES, although it appears slightly more potent than p50 (Ali and Jackson, 2001). The presence of native eIF4F resulted in qualitatively the same changes in the pattern of cleavages generated by the Fe(II) PTB′ derivatives as were seen with the eIF4G p50 domain (compare Figure 5 and Supplementary Figure S2): a strong decrease in cleavage intensity at nt 537 (D284C), with a lesser decrease at nt 473 (D284C), and increased cleavage band intensity at nt 453 (D242C). As with p50, there was no shift in the position of the cleavage bands, and no new cleavage products were observed that were not seen when eIF4G p50 was added to the assays. This implies that when the native eIF4F complex binds to the PV IRES, it is only its eIF4G central (p50) domain that interacts closely with the basal part of Domain V. This conclusion is consistent with the results showing that the eIF4F complex and eIF4G p50 (or p100) all give precisely the same toe prints on the EMCV IRES and protect the same residues against chemical reagents (Pestova et al, 1996; Kolupaeva et al, 1998).
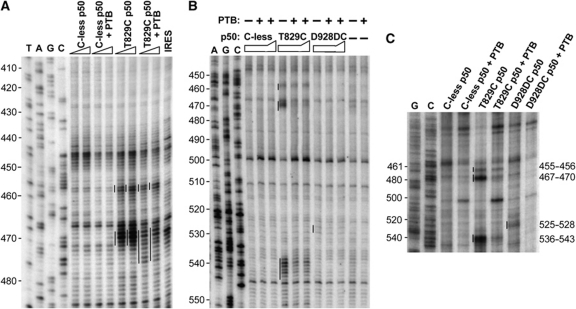
In view of the finding that eIF4G p50 and the eIF4F complex modified PTB′ binding to Domain V, we used tethered hydroxyl-radical probing to investigate whether PTB reciprocally modified eIF4G p50 binding, using Fe(II) derivatives of two single cysteine p50 mutants that were found to cleave in Domain V (de Breyne et al, 2009), designated T830C and D929DC in that publication, but now T829C and D928DC because of slight revision of the eIF4GI sequence. We found that in the absence of PTB these cleaved in exactly the same positions and with similar relative intensities (Figure 6) as described by de Breyne et al (2009). However, in the presence of wild-type ΔNTD-PTB, the intensity of many of these cleavage bands was reduced as shown by the black arrows in Figure 5C. The cuts at nt 467–470 and nt 536–543 made by Fe(II)-T829C were significantly weaker (Figure 6A–C), and the weak cleavages generated by Fe(II)-D928DC at nt 525–528 in the absence of PTB were diminished even further when PTB was present (Figure 6C). On the other hand, PTB had no consistent effect on the cleavage band intensity at nt 455–456 (Figure 6A–C), indicating no reduction in the actual amount of eIF4G p50 bound to Domain V.
Figure 6.
The binding position and orientation of eIF4G p50 on Domain V is modified in the presence of PTB. The figure shows representative primer extension analysis of directed hydroxyl-radical probing of PV IRES nucleotides 410–480 with PVpr5 (A), nt 450–550 with PVpr6 (B) and nt 460–540 with PVpr7 (C), using the designated Fe(II)-BABE p50 mutants in the presence or absence of ΔNTD-PTB. Hydroxyl-radical probing was performed as in Figure 2, using the specified derivatised eIF4G p50 mutants at 350 or 700 nM in the presence or absence of equimolar ΔNTD-PTB (A), 700 nM or 1 μM each p50 mutant in the presence or absence of 700 nM ΔNTD-PTB (B) and 700 nM each p50 mutant with or without 700 nM ΔNTD-PTB (C). Sites of hydroxyl-radical cleavage are indicated by the vertical lines to the left of each lane. The cleavage sites reported by de Breyne et al (2009) are indicated at the right margin of panel C. Lanes T, A, C, G depict sequencing ladders generated from the same primers. The changes in band intensity at different sites are summarised in Figure 5C.
The most striking change induced by PTB, however, was that the cleavages at nt 467–470 were slightly displaced to nt 467–474 and thus became somewhat more dispersed (Figure 6A). This observation carries the strong implication that the presence of ΔNTD-PTB results in a subtle repositioning or reorientation of the interaction between eIF4G p50 and Domain V. We conclude that PTB and eIF4G p50 (or the native eIF4F complex) reciprocally modify each other's binding to Domain V, and that the effect of PTB on the position and orientation of p50 binding is greater than that of p50 on PTB binding.
Discussion
In our previous tethered hydroxyl-radical probing analysis of the orientation of PTB interaction with the EMCV IRES, we found that the core IRES bound a single PTB at widely dispersed sites in the generally accepted two-dimensional structure of the IRES with RBDs1 and 2 binding Domain K near the 3′-end, and RBDs3 and 4 binding to Domain H and the basal part of Domain I near the 5′-end of the core IRES (Kafasla et al, 2009). Moreover, the presence of eIF4G p50 had no influence on these interactions, as shown by no change in the intensity or positions of the cleavages. This suggested a model in which the binding of PTB to widely dispersed sites constrains the three-dimensional flexibility of the IRES, favouring the attainment of the optimal structure for efficient internal initiation.
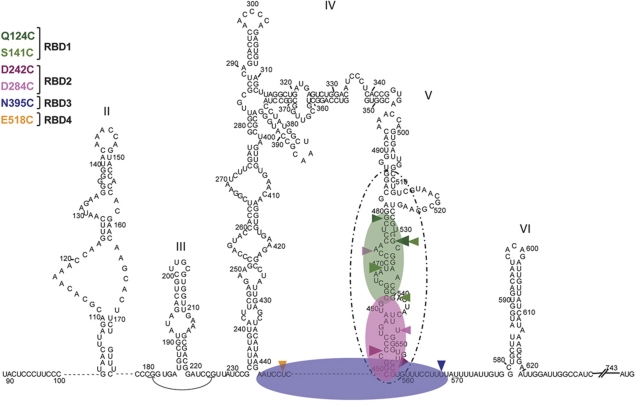
The results reported here show a very different scenario for PTB interaction with the poliovirus IRES. The outcome of the tethered hydroxyl-radical probing assays (particularly the pattern of strong and medium cleavages), coupled with the mass spectrometry data, lead unambiguously to the model depicted in Figure 7, in which the PV-1(M) IRES binds a single PTB in a unique orientation, with RBDs1 and 2 interacting with the basal part of Domain V, and RBDs3 and 4 binding the putatively single-stranded linkers flanking Domain V. Thus, the interactions with the PV-1(M) IRES are much more localised and far less widely dispersed than with the EMCV IRES.
Figure 7.
Docking PTB onto the PV IRES. Strong and medium cleavages caused by the designated Fe(II)-PTB derivatives are shown as in Figure 3C. The deduced orientation of PTB binding is also shown in the same colour code: transparent green denotes RBD1, RBD2 is pink and the RBD3+4 di-domain is transparent blue. The proposed position of the central domain of eIF4G (de Breyne et al, 2009) is shown by the dashed oval line.
This model for PTB binding to the PV-1(M) IRES is highly consistent with the recent suggestion that RBDs1 and 2 bind preferentially to short pyrimidine-rich motifs displayed in loops and bulges, whereas the RBDs3+4 di-domain preferentially binds longer flexible RNA structures (Clerte and Hall, 2009). It is also consistent with earlier low resolution data mapping the PTB interaction with the PV IRES. In UV-crosslinking assays using various segments of the PV-1(M) 5′-UTR (Hellen et al, 1994) and either HeLa cell cytoplasmic extracts or purified PTB, the strongest crosslinking signals were seen with all the probes that included the whole of Domain V (plus the short Domain IV–V linker and the first part of the pyrimidine-rich tract), and all probes, which lacked the complete Domain V gave either a much weaker signal, or no detectable crosslinking whatsoever. Similarly, in UV-crosslinking assays with a HeLa cell extract and individual domains of the structurally similar human rhinovirus-2 IRES, the strongest signals (which were as strong as those seen with the complete 5′-UTR) were observed with a Domain V probe, which included the downstream oligopyrimidine tract (Anderson et al, 2007). In addition, PTB binding to the poliovirus type 3 (PV-3) IRES has been shown to protect residues in the basal half of Domain V and the oligopyrimidine tract against ribonucleases (Guest et al, 2004). These previous results, taken together with the hydroxyl-radical probing data reported here, leave no doubt that Domain V, specifically the basal half of Domain V (plus its neighbouring flanking regions), is the main PTB-binding site on the PV IRES, in which the strongest and closest PTB–RNA interactions occur.
Moreover, comparisons between the IRESs of PV-3 Sabin and Leon strains have shown that this PTB-Domain V interaction strongly influences IRES activity (Gutierrez et al, 1997; Guest et al, 2004). These two IRESs differ in Domain V by a single point mutation at the equivalent position to PV-1(M) nt 470, which is a region in which PTB binding occurred in our assays (Figures 3C, 5C and 7). In a one-step growth assay in SH-SY5Y neuroblastoma cells, the yield of virus with the PV-3 (Sabin) 5′-UTR was much reduced as compared with one with the PV-3 (Leon) IRES, although no striking difference was seen in HeLa cells (Gutierrez et al, 1997). The low yield with SH-SY5Y cells was largely, if not entirely, because of defective IRES-dependent translation when the PV-3 (Sabin) IRES was present (Gutierrez et al, 1997; Guest et al, 2004), which correlated with a lower UV-crosslinking of endogenous PTB in SH-SY5Y cell extracts (but not in HeLa cell extracts) to a probe encompassing the PV-3 (Sabin) Domain V as compared with the equivalent probe from PV-3 (Leon). This reduced crosslinking to the PV-3 (Sabin) Domain V appears to be partly due to a lower affinity of PTB for the PV-3 (Sabin) Domain V than PV-3 (Leon), as shown by ribonuclease protection assays (Guest et al, 2004), and partly due to the fact that undifferentiated SH-SY5Y cells have very little PTB in comparison with HeLa cells (Mitchell et al, 2001).
Moreover, when reporter constructs with these IRESes were introduced by electroporation into developing chick embryo spinal cords, expression was much lower with the PV-3 (Sabin) IRES than PV-3 (Leon), but the defect could be rescued by co-electroporation of a PTB expression construct, though, somewhat surprisingly, such rescue could only be achieved with canonical PTB and not with nPTB (Guest et al, 2004). Taken together, all these results indicate that the Sabin3 Domain V has lower affinity for PTB than the Leon3 Domain V, which is the direct cause of a lower IRES activity especially in cells with a low endogenous PTB concentration such as SH-SY5Y and CNS cells.
How might PTB binding to Domain V influence IRES activity? It is widely agreed that an important feature of internal initiation on both type I and type II picornavirus IRESs is the binding of eIF4G (or its C-terminal two-thirds p100 fragment or the central domain p50 fragment) to a site just upstream of the oligopyrimidine tract (Jackson et al, 2010). The binding sites have been identified as Domain V in type I IRESs (de Breyne et al, 2009) and the A-rich bulge at the three-way junction between Domains J and K in the type II EMCV IRES (Kolupaeva et al, 1998, 2003). As the eIF4G p50 domain has the potential to interact with the eIF3 associated with the 43S pre-initiation complex (a 40S subunit with associated eIF1, eIF1A, eIF3 and an eIF2/GTP/Met-tRNAi ternary complex), the binding of eIF4G p50 (or the intact eIF4F complex) to these sites can promote delivery of the 43S complex to the internal ribosome entry site AUG, which is located just downstream of the oligopyrimidine tract, at nt 586 in PV-1(M) and at nt 834 in EMCV (Jackson et al, 2010).
Although this cannot be the complete explanation of the mechanism of internal initiation, because it does not account for the fact that IRES function requires the upstream Domains II–IV in type I IRESs and EMCV Domains H and I, nevertheless the binding of the p50 domain to these IRESs is indispensable for internal initiation, and efficient delivery of the 43S complex to the entry site presumably requires a precise positioning and orientation of the bound p50 domain. Consequently, our finding that PTB and eIF4G p50 each reciprocally induce subtle changes in the binding of the other protein on PV-1 Domain V is extremely interesting. It suggests that the stimulatory effect of PTB could well be due to the fact that it causes eIF4G p50 (or the intact eIF4F complex) to bind in the optimal position and orientation for efficient 43S pre-initiation complex loading. At present, we cannot distinguish whether this subtle repositioning of eIF4G p50 is due to direct steric hindrance, with PTB occluding certain sites from interacting with p50, or whether it is a more indirect effect in which PTB binding remodels the RNA structure in a way that causes a small shift in the position and orientation of p50 binding.
Although this model raises the question of why the poliovirus IRES has not evolved to function efficiently in the absence of PTB, the explanation may simply be that if the virus has always replicated in environments, which have sufficient cytoplasmic PTB, there would be no selective pressure to evolve towards a PTB-independent sequence or structure. Certainly, there is extraordinarily high sequence conservation of the basal part of Domain V in type I IRESs, not only among all enteroviruses (de Breyne et al, 2009), but also including all human rhinoviruses, and this exceptionally strong conservation extends to the short Domain IV–V linker and the 5′-part of the oligopyrimidine tract.
It will be interesting to see that those cellular mRNA IRESs that are strongly stimulated by PTB (Sawicka et al, 2008) follow the poliovirus or the EMCV model.
Materials and methods
Plasmids and primers
RNA for use in probing and translation assays was generated from plasmid pPV-CAT, which comprises the PV-1(Mahoney) 5′-UTR (nt 1–747) linked in-frame to CAT-coding sequences and cloned between the XhoI and SmaI sites of pSP73 (Promega). PCR was used to generate a slightly shorter derivative of pPV-CAT lacking nt 1–93 (the cloverleaf Domain I) of the PV-1 5′-UTR. The constructs used for expression of PTB mutant proteins were as described by Kafasla et al (2009).
The oligonucleotides used for primer extension analysis of cleavage products were as follows: PVpr1 5′ CTT GGT TTT GTG CGT CTA AG 3′; PVpr2 5′ ATC CGT CGC TTT CAA CCA CG 3′, PVpr3 5′ TCA TCA GCC TAA GCT ACA CTC 3′; PVpr4 5′ GGC TCT TCA CAC CTT GTT CAC 3′; PVpr5 5′ TTG CGC GTT ACG ACA GGC C 3′; PVpr6 5′ CCA ATT CGC TTT ATG ATA AC 3′; PVpr7 5′ GCT CCA TTA TGA TAC AAT TGT CTG 3′; PVpr8 5′ GGT TAT AGG TTA CAT TGA GC 3′.
In vitro transcription and translation
Uncapped monocistronic mRNA for use in translation assays was generated by T7 RNA polymerase transcription of the plasmid pPV-CAT linearised with SmaI. PV-1(M) IRES RNAs for use in probing assays were generated by T7 RNA polymerase transcription of the plasmid pPV-CAT linearised with MscI (to produce RNA probes consisting of PV nt 1–630 (or 94–630), or with PvuII (which cleaves in the CAT-coding sequence at position +114) to produce a full-length PV 5′-UTR RNA. Transcription reactions and the translation assays were carried out as described previously (Kaminski and Jackson, 1998; Hunt and Jackson, 1999).
Directed hydroxyl-radical probing
The PTB mutant proteins were expressed, purified and derivatised exactly as described by Kafasla et al (2009). For the directed hydroxyl-radical probing assays, 5 pmol of PV-IRES RNA was heated briefly at 95°C, then slowly cooled at room temperature after the addition of binding buffer (80 mM HEPES-KOH pH 7.4, 70 mM KCl and 2.5 mM MgCl2) and incubated (in a total volume of 50 μl) for 30 min at 37°C with the appropriate amount of Fe(II)-PTB′ mutant (usually 35 pmol). The reaction was then put on ice, and the Fenton reaction was initiated by addition of 1 μl 250 mM freshly prepared ascorbic acid and 1 μl 1.25% H2O2. After 15 min incubation on ice, the RNA was reisolated by phenol extraction and ethanol precipitation, and the resulting RNA fragments were analysed by primer extension using AMV-RT. Usually, one-eighth of the reisolated RNA was heated to 95°C for 90 s with 1 pmol of 32P-end labelled oligonucleotide primer, and then incubated further at 42°C for 30 min. Then, reverse transcription was performed at 42°C for 45 min in a buffer containing 50 mM Tris–HCl pH 8.0, 40 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, 1.25 mM of each of dATP, dCTP, dGTP, dTTP, 1 mM DTT, 1U RNase inhibitor (Dundee Cell Products), and 5U of AMV-RT (Promega) in a volume of 10 μl. The reaction was stopped by the addition of 10 μl of formamide dyes. The RT results were analysed by urea-acrylamide (6%) gel electrophoresis followed by autoradiography. Constructs for the expression of His-tagged eIF4G p50 single cysteine derivatives, described by de Breyne et al (2009), were a gift of CUT Hellen and TV Pestova. The p50 proteins were purified and used for hydroxyl-radical probing exactly as for the PTB′ derivatives.
Nanospray mass spectrometry of PTB/IRES complexes
The IRES RNA was heated to 95°C for 3 min and immediately diluted with assembly buffer (88 mM ammonium acetate, 2 mM magnesium acetate, pH 7.4). After slowly cooling at room temperature for 30 min, the appropriate amount of PTB was added and the mix was incubated at 37°C for 30 min, then put on ice. The concentrations were either 7.4 μM RNA and 7.4 μM PTB (for equimolar PTB and RNA), or 6.1 μM RNA and 12.2 μM PTB (for a PTB/RNA molar ratio of 2). For mass spectrometry measurements, the complexes were buffer exchanged into 200 mM ammonium acetate using micro-Bio-Spin Chromatography Columns (Bio-Rad, UK) with a 6 kDa cutoff, and diluted 1:1 with the same buffer to give an RNA concentration of 3.7 or 3.1 μM. In the case of the RNA alone, the biospin step could be omitted. NanoESI spectra were acquired on a Q star (MDS Analytical Technologies) modified for optimised performance at high masses (Sobott et al, 2002; Chernushevich and Thomson, 2004).
Supplementary Material
Acknowledgments
We thank Christopher Hellen and Tatyana Pestova for their gift of constructs for expression of the single cysteine eIF4G p50 derivatives, Tuija Pöyry and Tatyana Pestova for the native reticulocyte eIF4F complex preparation, Stephen Curry for helpful advice and Jenny Reed for technical support. This work was supported by BBSRC Project Grant BB/E004857/1 (to RJJ) and Eurocores Programme 06-RNAQuality-FP-004 (to CVR).
Footnotes
The authors declare that they have no conflict of interest.
References
- Alexander L, Lu HH, Wimmer E (1994) Polioviruses containing picornavirus type 1 and/or type 2 internal ribosomal entry site elements: genetic hybrids and the expression of a foreign gene. Proc Natl Acad Sci USA 91: 1406–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali IK, Jackson RJ (2001) The translation of capped mRNAs has an absolute requirement for the central domain of eIF4G but not for the cap-binding initiation factor eIF4E. Cold Spring Harb Symp Quant Biol 66: 377–387 [DOI] [PubMed] [Google Scholar]
- Anderson EC, Hunt SL, Jackson RJ (2007) Internal initiation of translation from the human rhinovirus-2 internal ribosome entry site requires the binding of Unr to two distinct sites on the 5′ untranslated region. J Gen Virol 88: 3043–3052 [DOI] [PubMed] [Google Scholar]
- Balvay L, Soto Rifo R, Ricci EP, Decimo E, Ohlmann T (2009) Structural and functional diversity of viral IRESs. Biochim Biophys Acta 1789: 542–557 [DOI] [PubMed] [Google Scholar]
- Blyn LB, Towner JS, Semler BL, Ehrenfeld E (1997) Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J Virol 71: 6243–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussadia O, Niepmann M, Créancier L, Prats AC, Dautry F, Jacquemin-Sablon H (2003) Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J Virol 77: 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernushevich IV, Thomson BA (2004) Collisional cooling of large ions in electrospray mass spectrometry. Anal Chem 76: 1754–1760 [DOI] [PubMed] [Google Scholar]
- Clerte C, Hall KB (2009) The domains of polypyrimidine tract binding protein have distinct RNA structural preferences. Biochemistry 48: 2063–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver GM, Noller HF (2000) Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol 318: 461–475 [DOI] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB (2005) SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU (2009) Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci USA 106: 9197–9202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti A, Pinol Roma S, Michael WM, Morandi C, Dreyfuss G (1992) hnRNP-I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res 20: 3671–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest S, Pilipenko E, Sharma K, Chumakov K, Roos RP (2004) Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3. J Virol 78: 11097–11107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez AL, Denova-Ocampo M, Racaniello VR, del Angel RM (1997) Attenuating mutations in the poliovirus 5′ untranslated region alter its interaction with polypyrimidine tract-binding protein. J Virol 71: 3826–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CUT, Pestova TV, Litterst M, Wimmer E (1994) The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J Virol 68: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SL, Hsuan JJ, Totty N, Jackson RJ (1999) Unr, a cellular cytoplasmic RNA-binding protein with five cold shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev 13: 437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SL, Jackson RJ (1999) Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA 5: 344–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka N, Kohara M, Hagino-Yamagishi K, Abe S, Komatsu T, Tago K, Arita M, Nomoto A. (1989) Construction of less neurovirulent polioviruses by introducing deletions into the 5′ noncoding sequence of the genome. J Virol 63: 5354–5363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Kaminski A (1995) Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1: 985–1000 [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nature Rev Mol Cell Biol 10: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafasla P, Morgner N, Pöyry TAA, Curry S, Robinson CV, Jackson RJ (2009) Polypyrimidine tract binding protein stabilizes the encephalomyocarditis virus IRES structure via binding multiple sites in a unique orientation. Mol Cell 34: 556–568 [DOI] [PubMed] [Google Scholar]
- Kaminski A, Howell MT, Jackson RJ (1990) Initiation of encephalomyocarditis virus RNA translation: the authentic initiation site is not selected by a scanning mechanism. EMBO J 9: 3753–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Belsham GJ, Jackson RJ (1994) Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J 13: 1673–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Jackson RJ (1998) The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU, Shatsky IN. (1998) Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem 273: 18599–18604 [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU. (2003) Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol 23: 687–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Nomoto A (1987) Construction of viable deletion and insertion mutants of the Sabin strain of type 1 poliovirus: function of the 5′noncoding sequence in viral replication. J Virol 1: 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laederach A, Das R, Vicens Q, Pearlman SM, Brenowitz M, Herschlag D, Altman RB (2008) Semi-automated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat Protoc 3: 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L, Kiel MC, Kaji A, Noller HF (2002) Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell 111: 129–140 [DOI] [PubMed] [Google Scholar]
- Meerovitch K, Nicholson R, Sonenberg N (1991) In vitro mutational analysis of cis-acting RNA translational elements within the poliovirus type 2 5′ untranslated region. J Virol 65: 5895–5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SA, Brown EC, Coldwell MJ, Jackson RJ, Willis AE (2001) Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol Cell Biol 21: 3364–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monie TP, Hernandez H, Robinson CV, Simpson P, Matthews S, Curry S (2005) The polypyrimidine tract binding protein is a monomer. RNA 11: 1803–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natan E, Hirschberg D, Morgner N, Robinson CV, Fersht AR (2009) Ultraslow oligomerization equilibria of p53 and its implications. Proc Natl Acad Sci USA 106: 14327–14332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson R, Pelletier J, Le S-Y, Sonenberg N (1991) Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. J Virol 65: 5886–5894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FHT (2005) Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309: 2054–2057 [DOI] [PubMed] [Google Scholar]
- Pelletier J, Flynn ME, Kaplan G, Racaniello V, Sonenberg N (1988) Mutational analysis of upstream AUG codons of poliovirus RNA. J Virol 62: 4486–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Hellen CUT (1996) Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol 16: 6870–6878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko EV, Gmyl AP, Maslova SV, Svitkin YV, Sinyakov AN, Agol VI (1992) Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell 68: 119–131 [DOI] [PubMed] [Google Scholar]
- Sawicka K, Bushell M, Spriggs KA, Willis AE (2008) Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans 36: 641–647 [DOI] [PubMed] [Google Scholar]
- Simpson PJ, Monie TP, Szendroi A, Davydova N, Tyzack JK, Conte MR, Read CM, Cary PD, Svergun DI, Konarev PV, Curry S, Matthews S (2004) Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure (Camb) 12: 1631–1643 [DOI] [PubMed] [Google Scholar]
- Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV (2002) A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem 74: 1402–1407 [DOI] [PubMed] [Google Scholar]
- Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW (2005) Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans 33: 457–460 [DOI] [PubMed] [Google Scholar]
- Valcarcel J, Gebauer F (1997) Post-transcriptional regulation: the dawn of PTB. Curr Biol 7: R705–R708 [DOI] [PubMed] [Google Scholar]
- Walter BL, Nguyen JH, Ehrenfeld E, Semler BL (1999) Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5: 1570–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.