Hen1 is required for oocyte development and piRNA stability in zebrafish
The HEN1 methytransferase adds the 2′-O-methyl modification to piRNAs. Here, it is shown to be present in the nuage of female germ cells, and in its absence piRNAs are selectively uridylated and degraded.
Keywords: germ line, Hen1, piRNA, Piwi, zebrafish
Abstract
Piwi-interacting RNAs (piRNAs) are germ line-specific small RNA molecules that have a function in genome defence and germ cell development. They associate with a specific class of Argonaute proteins, named Piwi, and function through an RNA interference-like mechanism. piRNAs carry a 2′-O-methyl modification at their 3′ end, which is added by the Hen1 enzyme. We show that zebrafish hen1 is specifically expressed in germ cells and is essential for maintaining a female germ line, whereas it is dispensable in the testis. Hen1 protein localizes to nuage through its C-terminal domain, but is not required for nuage formation. In hen1 mutant testes, piRNAs become uridylated and adenylated. Uridylation frequency is highest on retro-transposon-derived piRNAs and is accompanied by decreased piRNA levels and mild derepression of transposon transcripts. Altogether, our data suggest the existence of a uridylation-mediated 3′–5′ exonuclease activity acting on piRNAs in zebrafish germ cells, which is counteracted by nuage-bound Hen1 protein. This system discriminates between piRNA targets and is required for ovary development and fully efficient transposon silencing.
Introduction
Small RNAs are potent regulators of gene expression in many different systems. These RNA co-factors provide sequence specificity to so-called Argonaute proteins that, when targeted to RNA and/or DNA, can initiate a variety of responses (Hutvagner and Simard, 2008). The most basic activity of Argonaute proteins is target cleavage. This is a very specific endonucleolytic activity generating 5′-phosphate and 3′-hydroxyl groups (Martinez and Tuschl, 2004), and it is targeted to the phosphodiester bond opposite bases 10 and 11 of the small RNA molecule (Elbashir et al, 2001). Interestingly, in animal systems, many Argonaute proteins have lost their nucleolytic activity, and have adopted other mechanisms to affect the activities of their targets.
In animal germ cells, a specific subset of Argonaute proteins is expressed: the Piwi proteins (Cox et al, 1998). This subfamily has retained its endonucleolytic activity (Gunawardane et al, 2007) and is guided to their targets by Piwi-interacting RNAs (piRNAs) (Girard et al, 2006; Aravin et al, 2007). In most systems, multiple Piwi paralogues are present (Carmell et al, 2002). In zebrafish, there are two Piwi paralogues: Ziwi and Zili (Houwing et al, 2007, 2008). Together, Piwi proteins and piRNAs are required for a number of different processes, including germ cell differentiation and meiosis (Deng and Lin, 2002; Kuramochi-Miyagawa et al, 2004; Carmell et al, 2007; Houwing et al, 2008). On a molecular level, Piwi proteins are required for transposon control. Whether this relates directly to the germ cell defects observed in Piwi mutants is currently unclear.
In all systems studied a major part of the Piwi-associated piRNAs are derived from repetitive elements (Brennecke et al, 2007; Carmell et al, 2007; Aravin et al, 2008; Houwing et al, 2008). In vertebrates, including zebrafish, retro-elements, or RNA-based transposons, give rise to the most abundant piRNA populations (Houwing et al, 2007; Aravin et al, 2008). As the mode of piRNA biogenesis responds to the actual presence of transposon transcripts (see below), this bias towards retro-elements may reflect the inactivity of DNA transposons in vertebrates (Feschotte and Pritham, 2007).
Biogenesis of piRNAs is distinct from the processing of short-interfering RNAs (siRNAs) and microRNAs (miRNAs): piRNAs are not derived from double-stranded RNA (dsRNA) precursors by Dicer (Vagin et al, 2006; Houwing et al, 2007), but from single-stranded RNA. The current models on piRNA biogenesis involve a central function for the Piwi proteins themselves, with Piwi protein-mediated target cleavage generating the 5′ ends of new piRNAs (Brennecke et al, 2007; Gunawardane et al, 2007; Aravin et al, 2008; Houwing et al, 2008). These 5′ ends are bound by another Piwi protein paralogue, followed by trimming of the 3′ end, resulting in a characteristic piRNA length and 3′-end heterogeneity for each Piwi paralogue. Consequently, different Piwi paralogues tend to bind piRNAs of different polarity; in zebrafish, Ziwi generally binds piRNAs that are anti-sense to transposon coding regions and Zili has a preference for sense piRNAs (Houwing et al, 2008). When both sense and anti-sense transcripts of a piRNA-targeted locus are present, this mechanism creates a cyclic process in which specific piRNA species can be amplified. This is often referred to as the ping-pong cycle (Brennecke et al, 2007).
Apart from the Dicer-independent mode of biogenesis, piRNAs differ in yet another aspect from miRNAs and most siRNAs in animals: they are methylated on the 2′-hydroxyl group at their 3′ end (Vagin et al, 2006; Houwing et al, 2007; Ohara et al, 2007; Kirino and Mourelatos, 2007a). This modification is found on all small RNAs in Arabidopsis (Yu et al, 2005), but in the animal kingdom it is found almost exclusively on piRNAs, with the notable exception of endogenous siRNAs in Drosophila (Horwich et al, 2007). The enzyme responsible for placing this modification onto miRNAs in plants is HEN1 (Yang et al, 2006), an enzyme with a methyltransferase domain and two dsRNA-binding domains. This is consistent with plant HEN1 modifying dsRNA precursors during miRNA biogenesis in Arabidopsis. Recently, a crystal structure of Arabidopsis HEN1 has been reported, showing how the dsRNA-binding domains and the methyltransferase domain come together, resulting in the proper positioning of the 3′ ends of the miRNA precursor in the catalytic site (Huang et al, 2009). In plants lacking HEN1, miRNAs are destabilized and are subject to 3′-end uridylation and degradation, resulting in pleiotropic phenotypes (Chen et al, 2002; Park et al, 2002; Li et al, 2005).
In animals, putative Hen1 homologues have been identified (Tkaczuk et al, 2006), but these lack the dsRNA-binding regions found in the plant HEN1 enzyme, implying that they do not act on double-stranded substrates. Indeed, it has been shown that animal Hen1 can methylate single-stranded RNAs, bind to Piwi proteins, and is required for piRNA accumulation and efficient Piwi-pathway activity (Horwich et al, 2007; Saito et al, 2007; Kirino and Mourelatos, 2007b; Kurth and Mochizuki, 2009). We show that zebrafish Hen1 is specifically expressed in germ cells and is required for oocyte development, and consequently, female development of zebrafish. Hen1 localizes to nuage through interactions with its C-terminal domain (CTD), but is not essential for nuage formation. As expected, Hen1 mediates piRNA methylation, and we reveal that this prevents both adenylation and uridylation. We show that uridylation, but not adenylation, is associated with piRNA destabilization, most likely through a 3′–5′ exonucleolytic pathway. Consequently, in hen1 mutants, transposon transcripts can be mildly up-regulated. The uridylation process discriminates between RNA- and DNA-based transposable elements, possibly reflecting target-dependent uridylation of piRNAs in the absence of Hen1.
Results
Zebrafish hen1
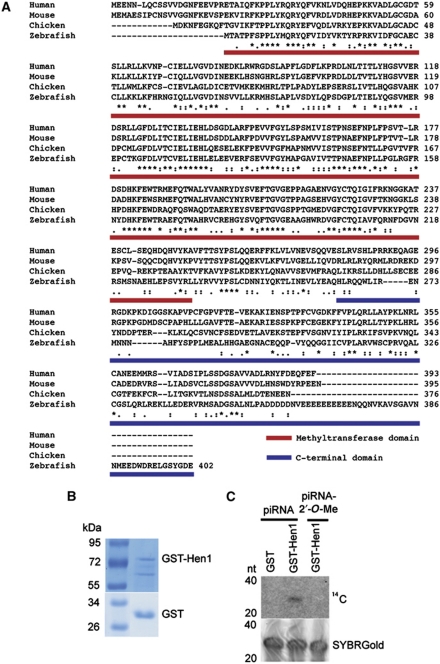
The likely zebrafish homologue of the hen1 gene has previously been identified through bio-informatic analysis as ENSDARG00000018871 (Tkaczuk et al, 2006). Sequence comparison shows that the protein encoded by this locus aligns well with other vertebrate Hen1 homologues (Figure 1A). This homology is mainly observed in the N-terminal part of the protein, which is predicted to harbour the catalytic methyltransferase activity. Indeed, structural analysis has confirmed this hypothesis (Huang et al, 2009). The C-terminal regions of these putative Hen1 homologues differ substantially (Figure 1A).
Figure 1.
Hen1 is a conserved methyltransferase in zebrafish. (A) Alignment of hen1 homologues in different vertebrates. Full-length Hen1 protein of zebrafish was used for experiments in (B) and (C). The red bar depicts the methyltransferase domain and the blue bar shows the C-terminal domain used in Figure 3. Asterisks symbolize conserved residues. (B) Purified GST-Hen1 and GST protein visualized on gel after staining with Page Blue. (C) GST-Hen1 is able to methylate RNA, but cannot do this if the RNA carries a 2′-O-methyl group at its 3′ end. Upper panel shows reaction of purified GST or GST-Hen1 with methylated and unmethylated piRNA. The lower panel shows RNA loading control stained with SYBRGold.
We cloned the cDNA of this putative hen1 homologue into an Escherichia coli expression vector and purified Hen1 using a GST moiety fused to its N-terminus (Figure 1B). This fusion protein shows methyltransferase activity on a single-stranded RNA oligonucleotide, and is inhibited by the presence of a 2′-O-methyl modification on the 3′ most terminal ribose ring of its substrate (Figure 1C). Together, these observations strongly suggest that ENSDARG00000018871 indeed is the zebrafish homologue of HEN1, and we will refer to this gene as hen1.
Gonad-specific expression of hen1
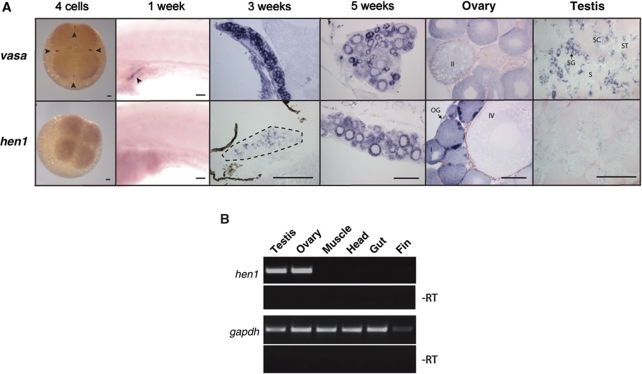
We analysed hen1 mRNA expression during zebrafish development using in situ hybridization (ISH) (Figure 2A). This revealed that hen1 starts to be expressed around 3 weeks of age, specifically in the gonad. Comparison with expression of the germ cell marker vasa strongly suggests germ cell-specific expression. This point in development corresponds to the start of sex determination in the zebrafish (Siegfried and Nusslein-Volhard, 2008), and to the timing of the first phenotypes observed in ziwi mutant zebrafish (Houwing et al, 2007). Hen1 expression remains present in the adult gonads, both in the male as well as in the female, although testis-specific expression is relatively weak. RT–PCR analysis confirms that expression of hen1 in the adult is also restrained to the gonads (Figure 2B).
Figure 2.
Hen1 is specifically expressed in the germ line of the zebrafish. (A) In situ hybridization for vasa and hen1 at different time points of development shows hen1 is not maternally provided and is expressed in the germ line as from 3 weeks of age (dotted line around gonad) and in both testis and ovary. Arrowheads indicate germ plasm. Stages of oogenesis are (OG) oogonia, (I) stage I oocytes, (II) stage II oocytes, (III) stage III oocytes, (IV) stage IV oocytes, and (V, not in picture) stage V oocytes. Stages of spermatogenesis are (SG) spermatogonia, (SC) spermatocytes, (ST) spermatids, and (S) sperm. Scale bars are 100μm. (B) RT–PCR on different organs of the zebrafish shows hen1 is specifically expressed in the germ cells. The gapdh is shown as loading control.
Hen1 subcellular localization
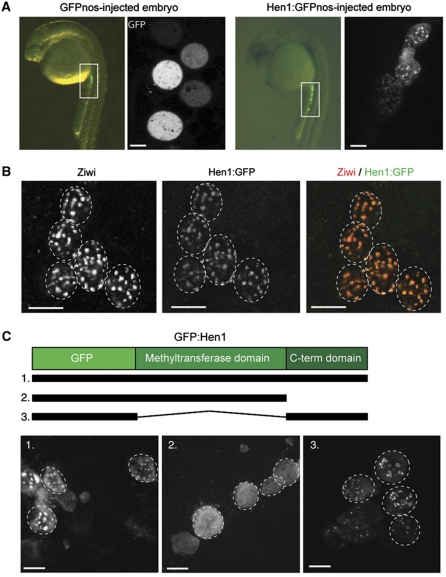
Next, we set out to analyse the subcellular localization of Hen1 protein in germ cells. In the absence of functional anti-sera, we injected embryos with mRNAs encoding GFP-tagged Hen1, flanked by a 3′UTR of the nos1 gene. This 3′UTR destabilizes the mRNA in somatic cells, while keeping it stable in primordial germ cells (PGCs), resulting in PGC-specific expression at 24 h of development (Koprunner et al, 2001). Untagged GFP is localized throughout the PGC cytoplasm. In contrast, GFP:Hen1 localizes to distinct granules around the nucleus (Figure 3A). These structures resemble a germ cell-specific structure named nuage, and we previously described the localization of Piwi protein Ziwi to nuage in PGCs (Houwing et al, 2007). We, therefore, performed a co-localization experiment using antibodies against endogenous Ziwi and the GFP part of the Hen1:GFP fusion protein (Figure 3B). The granules stained by the antibodies overlap perfectly, indicating that Hen1, like Ziwi, localizes to nuage.
Figure 3.
The C-terminal domain of Hen1 controls localization to germ granules. (A) mRNA of hen1 tagged with GFP and the nanos 3′ UTR (lower panel) or only GFPnos (upper panel) was injected into a one-cell embryo. The upper panel shows that GFP is not localized in primordial germ cells (PGCs), whereas Hen1-GFP localized to germ granules at 24 h post-fertilization. mRNA containing a 3′ UTR of nanos is only stable in the germ line. (B) Hen1-GFP co-localizes with Ziwi protein in germ granules (nuage). Hen1-GFP was visualized with a GFP antibody. Scale bar is 10 μm. (C) Schematic representation of Hen1 shows the methyltransferase and C-terminal domain. The three different constructs were injected into one-cell embryos and expression of GFP was assessed at 24hpf. Scale bar is 10 μm.
As described above, the Hen1 protein can roughly be divided into two functional units: the N-terminal region containing the methyltransferase domain and a more divergent CTD with unknown function. We tested both regions (also see Figure 1A) for nuage-targeting properties. As shown in Figure 3C, the CTD of Hen1 alone can direct GFP to nuage, suggesting that this region of the protein is responsible for the observed co-localization of Hen1:GFP with Ziwi, possibly through direct interaction between the CTD and Ziwi (see Discussion). Interestingly, we note that placing a GFP tag on the C-terminus of the CTD fragment results in less efficient expression compared with placing GFP on the N-terminus, suggesting that the CTD is most stable as the true C-terminal end of a protein.
Loss of hen1 affects germ cell development
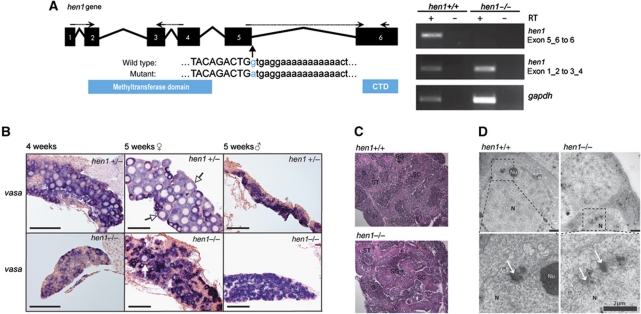
From an ENU-mutagenized library, we obtained a mutant allele of hen1, sa0026, in which the splice-donor site between the last two exons is affected (Figure 4A). Although hen1 mutant transcripts can still be detected, this mutation strongly affects splicing between exons 5 and 6 (Figure 4A), resulting in a stop codon after 108 nucleotides. Therefore, this allele may produce a C-terminally truncated version of the Hen1 protein. Animals homozygous for hen1(sa0026) are viable and fertile. Strikingly, among 297 homozygous adults, we only identified one female animal, whereas heterozygous and homozygous wild-type animals typically showed around 60% females, indicating that hen1(sa0026) homozygosity strongly interferes with the establishment of an adult female gonad (Supplementary Table S1). Unfortunately, the sex of the one female animal only became apparent after dissection of the animal, preventing phenotypic analysis other than RNA analysis. Attempts to generate more female hen1 mutants by decreasing population density and increasing food availability during development only yielded male hen1 mutant animals.
Figure 4.
Germ line development in hen1 mutant fish is delayed. (A) Left panel: schematic representation of the hen1 gene in zebrafish. Arrows indicate the splice-donor mutation in hen1 between exons 5 and 6. Right panel: RT–PCR analysis on specific regions of the hen1 mRNA. The primers used are indicated in the left panel; dashed line indicates primers spanning exon–exon boundary. (B) In situ hybridization for vasa in hen1 heterozygous (upper panels) and mutant (lower panels) zebrafish shows the presence of germ cells. In the heterozygous animal, oocytes (white arrows) are clearly formed at 5 weeks in three out of four animals (also see Supplementary Figure S1). However, in 5-week-old hen1 mutants, in two out of five animals, only some oocytes (white arrow) are present. This suggests sex determination is not yet complete. Scale bar is 100 μm. (C) Heamatoxylin and eosin staining of wild-type (left) and mutant (right) testis. Even though hen1 mutant zebrafish develop into male only, testis development does not seem to be affected, base on morphology and fertility. Stages of spermatogenesis are (SG) spermatogonia, (SC) spermatocytes, (ST) spermatids, and (S) sperm. (D) Electron microscopy of wild-type (left) and hen1 (right) mutant gonads to visualize nuage at 3 weeks of age shows no differences in localization and density of nuage. Nuage is often associated with both the nuclear membrane and mitochondria (m) and is visible as regions of higher density (white arrows). Lower panels show enlargement of boxed areas in upper panels. N: nucleus, Nu: nucleolus.
To analyse germ cell development in closer detail, we looked at gonad and germ cell morphology during development. We dissected gonads from different stages, and visualized germ cells using vasa ISH (Figure 4B). Gonad development is clearly affected in hen1 mutants, as indicated by the slow increase of gonad size compared with wild-type (Supplementary Figure S1). Sectioning of gonads from 4 and 5-week-old animals identified 5-week-old hen1 mutant individuals in which both male and female germ cells are present, indicating that sex determination in these animals has not yet completed as it has in their wild-type siblings. Adult hen1 mutants have testes that are indistinguishable from wild type, based on morphological analysis (Figure 4C).
As Hen1 localizes to nuage, we compared nuage between wild type and hen1 mutant siblings at 3 weeks of age, using electron microscopy (Figure 4D). In these experiments, nuage is visible as an electron dense granular structure, often associated to both the nuclear membrane and mitochondria. No differences in density and localization of these structures between wild type and hen1 mutant fish could be observed. Taken together, these data show that hen1 is dispensable for nuage formation and the initiation of both oogenesis and spermatogenesis, but is essential for maintaining oocyte development.
Reduced piRNA levels in hen1 mutants
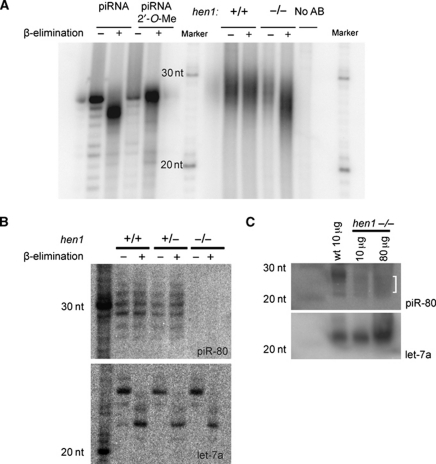
To zoom in to the molecular level, we analysed piRNA biogenesis in adult testes of hen1 mutants and wild-type siblings. First, we checked whether the 2′-O-methyl modification requires Hen1, as would be expected from previously published work (Horwich et al, 2007; Saito et al, 2007; Kurth and Mochizuki, 2009). We used mild oxidative treatment followed by a β-elimination reaction to test this. Unmodified RNA molecules loose their most 3′ base during this procedure and retain a 3′-phosphate group, resulting in an apparent two-base pair size shift, whereas RNA molecules with a 2′-O-methyl modification at their 3′ end are unaffected (Figure 5A, left panel). Consistent with previous findings (Houwing et al, 2007), Ziwi- and Zili-bound piRNAs, isolated from immunoprecipitates from wild-type animals, are resistant to this treatment, reflecting their modified status. In contrast, both types of piRNAs from hen1 mutant animals become sensitive, indicating loss of the modification (Figure 5A; Supplementary Figure S2A, right panels). The fraction of hen1 mutant piRNAs shifting upon oxidative treatment indicates that hen1(sa0026) is a strong loss-of-function allele.
Figure 5.
piRNAs are present at a low level and are not modified in hen1 mutants. (A) The left panel shows β-elimination on synthetic RNA with or without a 2′-O-methyl group. RNA without modification is sensitive to β-elimination, which results in a shift on gel, whereas RNA with a modification is no longer sensitive to oxidation. The right panel shows the same procedure with RNA isolated from Zili immunoprecipitates in wild type and hen1 mutant zebrafish. (B) Northern blot for piRNA-80 in wild type, heterozygous, and mutant hen1 zebrafish before and after β-elimination. piRNA-80 (upper panel) is present and modified in testis RNA derived from wild-type and heterozygous animals. In contrast, piRNA-80 cannot be detected in the hen1 mutant. At the bottom, the same northern blot was probed for let-7a as control for β-elimination and loading. (C) Northern blot on wild-type and mutant hen1 testes for piRNA-80. piRNA-80 is clearly detectable in wild-type testes, but the levels are significantly lower in hen1 mutant, also when increasing amount of RNA were loaded. In addition, piRNA-80 seems to be shorter in the mutant. Let-7a is shown as loading control.
Clearly, this experiment reveals that piRNAs are still present in the hen1 mutant. However, the experimental procedure prevents a quantitative assessment. We, therefore, analysed piRNAs in total RNA preparation of both wild type and hen1 mutant gonads on northern blots, using a probe detecting a single piRNA species (piR80). This revealed strongly reduced piR80 levels (Figure 5B and Supplementary Figure S2B), only just detectable on blots made from small gels in which the usually somewhat broad piR80 signal remains more focussed (Figure 5C).
Loss of Hen1 affects piRNA 3′-end stability
We next deep-sequenced small RNA libraries from adult testes total RNA of wild-type and hen1 mutant animals. Libraries were constructed using adapter ligation to both 5′ and 3′ ends of the small RNAs, allowing accurate assessment of both 5′ and 3′ ends of each cloned RNA species. For wild-type and hen1 mutant samples, we obtained over eight- and nine-million sequence reads, respectively, of which 85% could be mapped to the Zv8 assembly of the zebrafish genome (Supplementary Table S2; Supplementary Figure S3). Sequences annotated as rRNA, snRNA, snoRNA, and tRNA were removed from further analysis. The remaining RNA species displays a bimodal-length distribution reflecting the two major small RNA populations in the adult zebrafish testis: miRNAs and piRNAs.
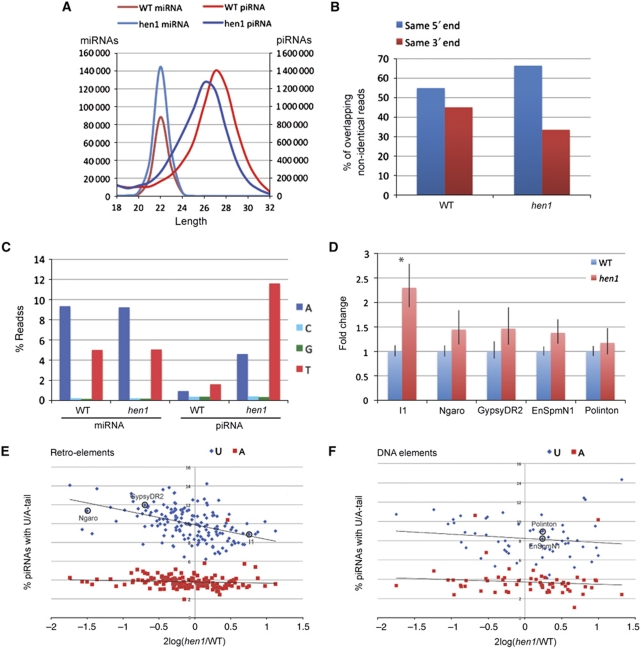
In Figure 6A, we have plotted the length distributions of both these classes. From this it is clear that while the length profile of miRNAs is unaffected by loss of Hen1, the piRNA profile indicates a shortening of this species, mirroring what can be seen on northern blot (Figure 5C). This sequencing approach does not reveal the drop in piRNA levels that we observed on northern blot (Figure 5B and C). This is likely caused by the fact that the 2′-O-methyl modification interferes with the ligation of the cloning adapter at the 3′ end. Thus, upon loss of the modification in the hen1 mutant, ligation becomes more efficient, counteracting the reduced piRNA levels. This effect is even more pronounced when poly-adenylation is used to tag the 3′ end of piRNAs during the cloning procedure (Supplementary Figure S3).
Figure 6.
Deep-sequencing analysis of hen1 mutants. (A) Length distribution of miRNAs and piRNAs in wild type (WT) and mutants. (B) Percentage piRNAs that overlaps with another piRNA, sharing either the same 5′ end or the same 3′ end. Pairs that share both ends or none were not included in the analysis; n=3593068 for the wild type and n=3538814 for the hen1 mutant. (C) Non-templated residues found at the 3′ ends of small RNA species. MiRNAs in wild type and mutant zebrafish show frequent addition of A or T residues. In wild-type testis, piRNA infrequently have non-templated bases, but in hen1 mutant testis, piRNAs show frequent adenylation and uridylation. (D) Quantitative PCR (qPCR) for different retro-elements (I1, Ngaro and GypsyDR2) and DNA elements (EnSpmN1 and Polinton). These are encircled in E) and (F). Shown is the average of three experiments in each of which three different control genes have been analysed (tubulin, gapdh, and ef1a). *P<0.005, unpaired t-test. (E) Uridylation and adenylation frequencies of retro-elements plotted versus their change in cloning frequency. For retro-elements, a negative correlation (R2=0.21) was observed between the frequency of piRNA uridylation and piRNA abundance. No correlation was observed between adenylation and cloning frequency (R2=0.007). (C) qPCR was performed for encircled elements. (F) Uridylation and adenylation frequencies of DNA elements plotted versus their change in cloning frequency. For DNA elements, no correlation (R2=0.02) was observed between the frequency of piRNA uridylation and piRNA abundance. No correlation was observed between adenylation and cloning frequency (R2=0.007). (C) qPCR was performed for encircled elements.
Biogenesis of piRNAs is accompanied by a so-called ping-pong signature: a strong tendency of piRNAs from the two Piwi paralogues to be derived from opposite strands and overlap 10 bases at their 5′ ends (Brennecke et al, 2007). This is likely imposed by the catalytic activities of the Piwi proteins themselves and the presence of this signature in piRNA sequence libraries can be taken as a measure for the activity of the Piwi proteins. This ping-pong signature is very similar in both the wild type and the hen1 mutant-derived piRNA libraries (Supplementary Figure S4), suggesting that piRNA synthesis is not affected by loss of Hen1. The fact that a wild-type ping-pong signature can be observed in hen1 mutants suggests that the observed overall shortening of piRNAs is due to loss of bases at their 3′ end. We also find that piRNAs show increased heterogeneity at their 3′ ends relative to their 5′ ends in hen1 mutants compared with wild type (Figure 6B). Taken together, our data suggest that a 3′–5′ exonuclease activity is responsible for the observed piRNA destabilization in hen1 mutants.
Hen1 inhibits adenylation and uridylation of piRNAs
In Arabidopsis, HEN1 protects miRNAs from uridylation and destabilization (Li et al, 2005), and in mammals, a miRNA has been reported to be adenylated (Katoh et al, 2009). Furthermore, we recently identified an enzyme in Caenorhabditis elegans that destabilizes siRNAs through uridylation (van Wolfswinkel et al, 2009). For these reasons we analysed the small RNA sequence reads for the presence of 3′ non-templated bases. Interestingly, miRNA-reads rather frequently have additional A or T residues, reflecting adenylation and uridylation. This is not affected by loss of Hen1 activity, consistent with the lack of miRNA methylation in zebrafish. In contrast, piRNAs in wild-type testis carry non-templated bases at their 3′-end less frequently. In the hen1 mutant background, adenylation and uridylation both rise significantly: five- and seven-fold, respectively (Figure 6C). The added A and U-tails on both miRNAs and piRNAs are short, with >90% of all the tails being only one or two bases long (Supplementary Figure S5A).
Differential uridylation of DNA and retro-element piRNAs
Roughly 40% of all piRNAs we cloned map to transposable elements, and it has been shown in various organisms, including zebrafish, that piRNAs function in silencing these repeats (Brennecke et al, 2007; Aravin et al, 2008; Houwing et al, 2008). We, therefore, determined transcript levels from five different transposons in wild type and hen1 mutant zebrafish (Figure 6D). This revealed significant up-regulation of transcripts from the I1 retro-element, whereas four other transposon transcripts showed no significant differences, although we note a tendency for them to be slightly up-regulated. These latter four include two DNA transposons, EnSpmN1 and Polinton, and two retro-elements, GypsyDR2 and Ngaro. Our data are consistent with the mild effects of loss of Drosophila hen1 on transposon transcripts (Horwich et al, 2007).
We next looked at transposon-derived piRNAs in more detail. First, we asked whether there are transposons whose piRNA levels are more strongly affected by hen1 mutation than others. We extracted transposon-targeting piRNAs from both the wild type and the hen1 mutant-sequencing data. After scaling the data, we plotted for each element the number of sequence reads obtained from wild type versus that obtained from the hen1 mutant (Supplementary Figure S5B). No individual transposon stood out in this analysis; only some deviations were observed, and these were related to elements with low read counts. Consequently, we restricted ourselves in the rest of the analysis to only those elements for which we retrieved at least 3000 reads in both the wild type and hen1 mutant libraries (Supplementary Table S3).
We then probed whether the observed uridylation and adenylation frequencies of piRNAs in the hen1 mutant differ between piRNAs coming from different transposon types. Upon plotting the frequency of uridylation versus the piRNA-cloning frequency of individual transposons from the hen1 mutant (Supplementary Figure S5C), we observed a statistically highly significant trend for retro-element-derived piRNAs to be more frequently uridylated than DNA element-derived piRNAs (10.0±1.7 versus 8.2±1.9%; P=3 × 10–9). No significant difference was observed for adenylation frequencies (3.7±1.5 versus 3.8±0.8%; P=0.59). Interestingly, this difference in average uridylation frequency is accompanied by a small difference in cloning frequencies between wild type and hen1 mutant as well: piRNAs from retro-elements are slightly underrepresented in the hen1 mutant library (Supplementary Figure S5D). Comparing ratios between mutant and wild-type library read counts yields a small, but significant difference between the two transposon classes (hen1/wt=1.12±0.45 for DNA elements and hen1/wt=0.98±0.32 for retro-elements (P=0.013; paired t-test).
A correlation between piRNA uridylation and piRNA abundance can also be observed at the level of individual transposons. When we plot for each element the uridylation and adenylation frequencies versus their change in cloning frequency, a negative correlation between the frequency of piRNA uridylation and piRNA abundance (R2=0.21) is observed for retro-elements (Figure 6E). Uridylation frequencies did not correlate with decreased cloning frequencies in case of DNA transposons (R2=0.02) (Figure 6F). No correlation was observed between adenylation and cloning frequency (R2=0.007) (Figure 6E and F).
Potential piRNA uridylation enzymes
Although a conclusive identification of the uridylating and adenylating activities acting on piRNAs falls outside the scope of this work, we analysed a number of potential candidate enzymes in terms of their expression. We identified seven candidates based on their homologies to the mammalian non-canonical polyA-polymerases TUTases 1–7, as these include both adenylating and uridylating enzymes (Kwak and Wickens, 2007; Martin and Keller, 2007; Rissland et al, 2007; Wilusz and Wilusz, 2008). For reasons discussed below, we consider drtutase2, drtutase4, and drtutase7 to be the most likely candidates responsible for the observed piRNA modifications.
Complete or nearly complete zebrafish sequences encoding homologues of the mammalian TUTases could be retrieved from public databases (Supplementary Table S4), with the exception of drTUTase4. The complete hypothetical cDNA for this gene was assembled in silico from a number of previously identified gene predictions and verified by RT–PCR and sequencing (Supplementary Figure S6). Using RT–PCR, we determined that all seven dr tutases are expressed in the gonads, but many also in other tissues (Supplementary Figure S7A). ISHs for drtutases 2, 4, and 7 (Supplementary Figure S7B) revealed that they are expressed in the germ cells of the ovary. Interestingly, these genes express most prominently in stages I–II. Early and late stages oocytes appear to be rather devoid of these transcripts.
Discussion
The results we present here describe zebrafish Hen1 as a factor required for ovary development. On a molecular level, we firmly place Hen1 in the zebrafish Piwi pathway: Hen1 can localize to nuage and is required for the 2′-O-methylation of piRNAs in the adult testis. In its absence, terminal transferase activities start to act on piRNAs, leading to adenylation and uridylation, with uridylation correlating with destabilization of piRNAs. Below we discuss the implications of these findings in more detail.
Zebrafish sex determination
Zebrafish sex determination is poorly understood, and no sex determining chromosomes or loci have been identified. Initially, all individuals develop immature non-functional ovaries (Maack and Segner, 2003). In some, oocytes degenerate and male germ cell development initiates (Uchida et al, 2002). The factors that control this process are not well known. Environmental factors play a role; for example, raising zebrafish in low densities with ample food favours female development, whereas crowding and scarcity of food result in mainly male development (Siegfried and Nusslein-Volhard, 2008). In addition, temperature influences the outcome of the sex determination process (Uchida et al, 2002). Finally, one factor that is well known to be essential for female development and is directly related to our study on hen1 is the presence of germ cells (Slanchev et al, 2005; Siegfried and Nusslein-Volhard, 2008).
Zebrafish lacking Piwi proteins develop strictly as infertile males. In ziwi mutants, infertility is due to the loss of germ cells through apoptosis early in development (Houwing et al, 2007). However, animals lacking Zili retain their germ cells, but these fail to differentiate (Houwing et al, 2008). This indicates that the mere presence of any germ cell is not sufficient to support female development.
Hen1 mutants also develop as males, even in conditions in which, by far, most of the heterozygous and homozygous wild-type littermates develop as females (raising embryos in low densities with ample food; data not shown). But in contrast to the piwi mutants, hen1 mutants are fertile, showing that in hen1 mutants germ cells are not lost. Interestingly, we show that hen1 mutant gonads produce early stage oocytes, similar to their wild-type siblings. Apparently, Hen1 is required to maintain early oocytes or to further differentiate them, and failure to do so may trigger the observed switch to male development in hen1 mutant animals.
Interestingly, not all defects in oocyte maturation lead to male development: we have described a missense mutation in zili that triggers meiotic defects, while still permitting female development (Houwing et al, 2008). Taken together, phenotypic analysis of the various piwi-pathway mutants strongly suggests that an event early in oogenesis, but after oocyte specification is important to oocyte maintenance and subsequent female development in zebrafish. Whether this relates to transposon activity is presently not clear.
Piwi pathway in testis and ovary
The onset of hen1 expression in zebrafish, around 3 weeks of development, coincides with the time around which zebrafish sex determination takes place (Maack and Segner, 2003). This is also the time when ziwi mutant zebrafish loose their germ cells (Houwing et al, 2007). As Ziwi, together with piRNAs, is maternally provided, this may indicate that around this time a wave of de novo piRNA synthesis is required to provide the proliferating germ cells with sufficient piRNAs. Obviously, this would require synthesis of additional Ziwi and Hen1 protein. It is likely that these processes are the basis of the germ cell defects observed in both ziwi and hen1 mutants, with the defect in the ziwi mutant being much stronger as it totally blocks the piwi pathway, whereas lack of Hen1 merely makes it less efficient.
However, there are indications that piRNA biosynthesis also occurs earlier. At 3 days of development, zili becomes expressed in the PGCs and Zili protein translocates to the nucleus. In analogy to the mouse Miwi2 data (Aravin et al, 2008), we hypothesize that Zili becomes loaded with newly made piRNAs and is transported to the nucleus to exert its function. Lack of Zili prevents these events and inhibits further PGC differentiation. Yet, zygotic loss of Hen1 does not inhibit PGC development. Potentially, Hen1 protein, but not hen1 RNA, is provided maternally, similar to Ziwi, thus permitting normal piRNA biogenesis and PGC development at these early time points in development.
Hen1 localizes to nuage
We found that a Hen1:GFP fusion protein, when introduced into PGCs localizes to nuage. This would be expected if indeed endogenous Hen1 is provided maternally, as Ziwi localizes strongly to nuage in PGCs as well (Houwing et al, 2007). Furthermore, piRNA biogenesis is presumably occurring in these structures (Aravin et al, 2009). We found that the C-terminal region of Hen1, the CTD, is sufficient for this localization. Interestingly, a model of animal Hen1 binding to a Piwi protein has been proposed, based on structural work on the Arabidopsis HEN1 protein (Huang et al, 2009). This model proposes a direct interaction between the Hen1 CTD and Piwi. However, we have not been able to detect interaction between Hen1, or the Hen1 CTD with any part of Ziwi or Zili in a yeast-two-hybrid experiment (data not shown). This may either indicate that there is no direct interaction between Hen1 and Piwi, or that the Piwi protein has to bind a small RNA to allow the interaction. As Hen1 presumably should work only on loaded Piwi proteins, the latter option seems a realistic possibility.
Interestingly, the hen1(sa0026) allele we describe produces detectable levels of mRNA that may produce a C-terminally truncated protein, unable to localize to nuage. Given the strong effect of hen1(sa0026) on piRNA methylation, it appears that this domain may indeed be critical for Hen1 function. Unfortunately, we have so far been unable to generate Hen1-specific antibodies to further test this idea.
piRNA adenylation and uridylation
We have described a differential response of piRNAs in the hen1 mutant depending on the type of target. Interestingly, we find more frequent uridylation on piRNAs derived from potentially active transposons. This may relate to the fact that the 3′ end of an Argonaute-bound small RNA is bound by the PAZ domain, and only becomes available for modification if the small RNA base pairs extensively with a target. Therefore, the uridylation we observe on unmethylated piRNAs may well require target recognition, an idea that has been proposed before based on work on uridylation of miRNAs and siRNAs in Chlamydomonas (Ibrahim et al, 2010), and has been shown for miRNAs and siRNAs in Drosophila and human cells (Ameres et al, 2010). Such target-dependent uridylation would likely result in the observed difference in uridylation between DNA element-derived piRNAs and RNA element-derived piRNAs, as zebrafish, like most vertebrates (Feschotte and Pritham, 2007), has no known active DNA transposons in its genome. Note that this hypothesis would also imply that the observed adenylation of piRNAs is independent of target recognition, as it does not discriminate between different elements.
The uridylating and adenylating enzymes remain unknown at the moment. Seven non-canonical polyA polymerases, some with uridylating activities have been identified so far (Wilusz and Wilusz, 2008): TUTase1 is the mitochondrial polyA polymerase (Tomecki et al, 2004; Nagaike et al, 2005) and has been implicated, like TUTase3 in Histone mRNA uridylation (Mullen and Marzluff, 2008). U6 TUTase acts specifically on U6 snRNA (Trippe et al, 2006) and TUTase5 has not been well described. Only TUTase2 and 4 have been linked to RNA interference-like pathways before. Mammalian TUTase2 homologues have been shown to polyadenylate their mRNA targets (Kwak et al, 2004), similar to the C. elegans homologue GLD-2 (Wang et al, 2002). However, in mouse, TUTase2 has also been implicated in miRNA stabilization through adenylation of the mature miRNA (Katoh et al, 2009). Furthermore, TUTase4 has in vitro uridylation activity (Kwak and Wickens, 2007) and is involved in uridylation of miRNA precursors (Heo et al, 2009). TUTase7 resembles TUTase4 in domain structure and in vitro activity (Kwak and Wickens, 2007) and is the closest homologue of the C. elegans CDE-1 protein, which is involved in siRNA uridylation (van Wolfswinkel et al, 2009). For these reasons, we consider drTUTases2, 4, and 7 the most likely candidates for the adenylation and uridylation of hen1 mutant piRNAs. However, it should be noted that the in vivo characterization of many of these enzymes is relatively limited, especially for TUTase5.
We found that drtutases2, 4, and 7 are expressed in germ cells, although not specifically in germ cells. Therefore, if these enzymes are indeed involved, their activities are not specific to the Piwi pathway. Consistent with this finding, the previously described miRNA adenylation was found in liver (Katoh et al, 2009) and the target-dependent uridylation activity described by Ameres et al (2010) is also active in somatic cells.
piRNA degradation
We describe that uridylation, but not adenylation, is associated with destablilization of piRNAs. This destabilization likely involves a 3′–5′ exonuclease activity, based on the finding that hen1 mutant piRNAs tend to become shorter at their 3′ ends, not at their 5′ ends. A prominent 3′–5′ exonuclease is the exosome (Houseley et al, 2006), a cytoplasmic activity that has been implicated in specific cases of small RNA biogenesis (Flynt et al, 2010). Whether TUTase4, TUTase7, and the exosome are indeed involved in piRNA homeostasis will have to be determined through further experimental analysis.
Concluding remarks
The zebrafish hen1 mutant described here is a model in which the Piwi pathway is crippled, but still active. It represents the first vertebrate system in which piRNA stability is decreased and offers opportunities to further refine our knowledge of the Piwi pathway in vertebrate germ cells as it allows for proper germ cell function and development, while at the same time it has measurable effects on the Piwi pathway.
Materials and methods
Zebrafish strains and genetics
Zebrafish were kept under standard conditions (Westerfield, 1993) and staged according to Kimmel et al (1995). The hen1 splice-donor mutant (sa0026) was derived from ENU mutagenized libraries using target-selected mutagenesis as described (Wienholds et al, 2002). Fish with mutant alleles were outcrossed against wild-type fish (AB or TL) and subsequently incrossed to obtain homozygous offspring.
Genotyping
DNA was purified from caudial fin tissue, taken from anesthetized zebrafish. A hen1 fragment was amplified with the primers indicated in the Supplementary data, and PCR products were sequenced.
In vitro methyltransferase activity assay
The methyltransferase assay was performed as described previously (Yu et al, 2005). A 100 μl reaction containing 50 mM Tris–HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, 5% glycerol, 80U RNasin, 0.5 μCi S-adenosyl-L-[methyl-14C] methionine (Amersham), 5 μg purified protein (GST or GST-hen1), and 1 nmol of RNA substrate was incubated for 2 h at 37°C. The reaction was stopped with proteinase K for 15 min at 65°C after which it was extracted by phenol/chloroform. Small RNAs were precipitated with ethanol and analysed on a 12% acrylamide gel. The gel was treated with an autoradiography enhancer (En3hance, Perkin Elmer) and exposed to X-ray film at −80°C.
Whole-mount ISH
Embryos and gonads were fixed in 4% PFA overnight at 4°C. Embryos older than 48 h and gonads were treated with proteinase K. ISH was performed as described previously (Houwing et al, 2007). The tissue was embedded in plastic for sectioning.
RT–PCR and qPCR
Total RNA was isolated from testis using FastRNA Pro Green kit (Q-Bio gene). RNA was treated with DNaseI (Promega) and isolated with Trizol. cDNA was synthesized using Oligo dT primers and subsequently used for semi-quantitative PCR or qPCR. Quantitative PCR was performed on a Biorad ICycler system (MiQ5) with SYBR Green. Primers used for quantification are given in the Supplementary data.
mRNA injections
Capped full-length mRNA or mRNA from specific domains were synthesized using mMESSAGE SP6 RNA Polymerase (Ambion) and purified using RNeasy spin columns (Qiagen). Zebrafish embryos were injected with 300 pg of full-length hen1 mRNA, 300 pg mRNA of Hen1 CTD, and 150 pg mRNA of the MT-domain of Hen1. Embryos were fixed for 2 h in MeOH and analysed using confocal fluorescence microscopy.
Histological analysis
Fish were killed in ice-cold water, decapitated, and fixed overnight in 4% PFA in PBS at 4°C. Gonads were isolated from the fish and in gradual steps transferred to 75% ethanol after which they were embedded in paraffin for sectioning. Paraffin sections were stained with haematoxylin and eosin for histological analysis.
Electron microscopy
Trunks of 3-week-old zebrafish were fixed with half-strength Karnovsky fixative (pH 7.4) (Karnovski, 1965) and post-fixed with 1% OsO4 in 0.05 M cacodylate buffer (pH 7.4), dehydrated in acetone series, and embedded in Spurr's low viscosity resin (Spurr, 1969). Semi-thin, 1-μm-thick sections were cut with a histo-Butler diamond knife (Diatome) and mounted on glass slides. After drying, the sections were stained with methylen blue Azur II mixture according to Richardson (Richardson et al, 1960) for light microscopy. Ultrathin sections (70 nm in thickness) were placed on copper grids, stained with uranyl acetate followed by lead citrate, and examined with an electron microscope (ZEISS Libra 120 energy filter electron microscope). Image acquisition and analysis were performed using a 2 k Vario Speed SSCCD camera (Droendle), the iTEM software (TEM imaging platform, Olympus), and Adobe Photoshop.
Northern blotting and β-elimination
Total RNA was isolated using FastRNA Pro Green kit (QBiogene). For regular northern blotting, 5 μg RNA was loaded on a 15% polyacrylamide gel and blotted according to standard procedures. To test the presence of the 3′-end modification of piRNAs, 20 μg total RNA was treated with NaIO4 followed by β-elimination as described previously (Vagin et al, 2006). This was loaded on a 12% polyacrylamide gel and blotted according to standard procedures. Blots were prehybridized in hybridization buffer (Ambion) for 20 min and hybridized overnight at 37°C for DNA probes and 60°C for LNA probes. After washing (20 min at 37°C in 2 × SSC/0.2% SDS and 20 min at 37°C in 1 × SSC/0.1% SDS for DNA probes; 20 min at 50°C in 1 × SSC/0.1% SDS and 20 min at 50°C in 0.5 × SSC/0.1% SDS for LNA probes), blots were exposed to phosphor-imager screens that were scanned on a BAS-2500 imager.
Immunoprecipitation and β-elimination
Immunoprecipitation and 5′-end 32P-labelling of the immunoprecipitated RNA was performed as described previously (Houwing et al, 2008). Half of the RNA sample was treated with NaIO4 followed by β-elimination (Vagin et al, 2006) and phenol/chloroform extraction. The untreated and β-eliminated samples were loaded on a 15% polyacrylamide-sequencing gel. The gel was fixed, dried, and exposed overnight to phosphor-imager screens.
Deep sequencing
Small RNAs in the size range of 19–31 bases were excised from a denaturing gel. Adaptors were ligated to the 5′ and 3′ ends of the isolated RNA, and the product was converted to cDNA using a primer on the 3′ adaptor. After 15 cycles PCR amplification of the library, the product was gel purified and sequenced on a Solexa platform.
Sequence analysis
Adaptor sequences were trimmed from generated data using custom scripts. The resulting inserts were mapped to the zebrafish genome (Zv8 assembly) using the megablast programme (Zhang et al, 2000), allowing mismatches in the reads starting from nucleotide 19 and considering the longest possible matches as true mapping positions. The mismatched 3′ ends of the reads were trimmed, keeping track of the identity of trimmed nucleotides for later counting. Genomic annotations of mapped reads were retrieved from the Ensembl database (release 56) using Perl API provided by Ensembl (Hubbard et al, 2007). Read counts for the different annotated classes are listed in Table S2. Sequences will be made available at GEO upon publication.
Analysis of transposon-derived piRNAs
piRNA reads derived from the various annotated transposable elements were isolated from the sequencing data. For comparison between libraries hen1 mutant read counts were scaled to match the wild-type read count (scaling factor 0.8567; based on total reads from both libraries mapping to transposons). Statistics on global adenylation and uridylation frequencies of different transposon types were performed with a two-tailed t-test. Differences of transposon piRNA characteristics between wild-type and hen1 mutants were analysed using a paired, one-tailed t-test.
Supplementary Material
Acknowledgments
We thank Jeroen Korving, Willemijn Gommans, Annejet Spierenburg, Marc van de Wetering, and Willi Salvenmoser for technical assistance and Bruce Draper for providing unpublished material. We thank Drs Cuppen (Hubrecht Institute) and Stemple (Welcome Trust Sanger Institute) for providing the hen1(sa0026) allele, which was generated as part of the ZF-MODELS Integrated Project in the 6th Framework Programme (Contract No. LSHG-CT-2003-503496) funded by the European Commission. This work was further supported by a grant from the Netherlands Organization for Scientific Research (RFK; ECHO 700.57.006), an ERC Starting Grant from the Ideas Program of the European Union Seventh Framework Program (RFK; Grant 202819) and by the European Union Sixth Framework Program Integrated Project SIROCCO (Contract No. LSHG-CT-2006-037900).
Footnotes
The authors declare that they have no conflict of interest.
References
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD (2010) Target RNA-directed trimming and tailing of small silencing RNAs. Science 328: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J (2007) The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318: 761–764 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A (2009) Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Chen X, Liu J, Cheng Y, Jia D (2002) HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H (2002) miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ (2007) DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet 41: 331–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC (2010) MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell 38: 900–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN (2009) TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708 [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD (2007) The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D (2006) RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF (2008) Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Huang Y, Ji L, Huang Q, Vassylyev DG, Chen X, Ma JB (2009) Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461: 823–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K et al. (2007) Ensembl 2007. Nucleic Acids Res 35 (Database issue): D610–D617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H (2010) Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci USA 107: 3906–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnovski MJ (1965) A formaldehyde—glutaraldehyde fixative of high osmolality for use in electron microscopy. J Cell Biol 27: 137A–138A [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T (2009) Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev 23: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z (2007a) Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14: 347–348 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z (2007b) The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13: 1397–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E (2001) A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev 15: 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T (2004) Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K (2009) 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 15: 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M (2004) Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci USA 101: 4407–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wickens M (2007) A family of poly(U) polymerases. RNA 13: 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X (2005) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maack G, Segner H (2003) Morpholigical development of the gonads in zebrafish. J Fish Biol 62: 895–906 [Google Scholar]
- Martin G, Keller W (2007) RNA-specific ribonucleotidyl transferases. RNA 13: 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T (2004) RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev 18: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF (2008) Degradation of histonee mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 22: 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Katoh T, Ueda T (2005) Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem 280: 19721–19727 [DOI] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T (2007) The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 14: 349–350 [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson KC, Jarret L, Finke EH (1960) Embedding in epoxy resins for ultrathin sectioning in electron microscopy. Stain Technol 35: 313–323 [DOI] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ (2007) Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol 27: 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Siomi H, Siomi MC (2007) Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Nusslein-Volhard C (2008) Germ line control of female sex determination in zebrafish. Dev Biol 324: 277–287 [DOI] [PubMed] [Google Scholar]
- Slanchev K, Stebler J, de la Cueva-Mendez G, Raz E (2005) Development without germ cells: the role of the germ line in zebrafish sex differentiation. Proc Natl Acad Sci USA 102: 4074–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 26: 31–45 [DOI] [PubMed] [Google Scholar]
- Tkaczuk KL, Obarska A, Bujnicki JM (2006) Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis. BMC Evol Biol 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP (2004) Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res 32: 6001–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ (2006) Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida D, Yamashita M, Kitano T, Iguchi T (2002) Oocyte apoptosis during the transition from ovary-like tissue to testes during sex differentiation of juvenile zebrafish. J Exp Biol 205 (Part 6): 711–718 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF (2009) CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139: 135–148 [DOI] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J (2002) A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316 [DOI] [PubMed] [Google Scholar]
- Westerfield (1993) The Zebrafish Book [Google Scholar]
- Wienholds E, Schulte-Merker S, Walderich B, Plasterk RH (2002) Target-selected inactivation of the zebrafish rag1 gene. Science 297: 99–102 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wilusz J (2008) New ways to meet your (3′) end oligouridylation as a step on the path to destruction. Genes Dev 22: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X (2006) HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res 34: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.