TopBP1 functions with 53BP1 in the G1 DNA damage checkpoint
The replication stress protein TopBP1 co-localizes with 53BP1 at sites of DNA damage and mediates its G1 cell cycle checkpoint function.
Keywords: 53BP1, DNA damage, G1 checkpoint, ionizing radiation, TopBP1
Abstract
TopBP1 is a checkpoint protein that colocalizes with ATR at sites of DNA replication stress. In this study, we show that TopBP1 also colocalizes with 53BP1 at sites of DNA double-strand breaks (DSBs), but only in the G1-phase of the cell cycle. Recruitment of TopBP1 to sites of DNA replication stress was dependent on BRCT domains 1–2 and 7–8, whereas recruitment to sites of DNA DSBs was dependent on BRCT domains 1–2 and 4–5. The BRCT domains 4–5 interacted with 53BP1 and recruitment of TopBP1 to sites of DNA DSBs in G1 was dependent on 53BP1. As TopBP1 contains a domain important for ATR activation, we examined whether it contributes to the G1 cell cycle checkpoint. By monitoring the entry of irradiated G1 cells into S-phase, we observed a checkpoint defect after siRNA-mediated depletion of TopBP1, 53BP1 or ATM. Thus, TopBP1 may mediate the checkpoint function of 53BP1 in G1.
Introduction
In eukaryotic cells, the DNA damage checkpoint helps to preserve genomic integrity (Abraham, 2001; Nyberg et al, 2002; Sancar et al, 2004). Key players of this checkpoint are ATM and ATR, members of the phosphoinositide kinase-related family of protein kinases. ATM (ataxia-telangiectasia mutated) responds mainly to DNA double-strand breaks (DSBs), whereas ATR (ATM and Rad3-related) responds primarily to DNA replication stress (Abraham, 2001; Nyberg et al, 2002; Sancar et al, 2004). ATR exists in a stable complex with ATRIP (ATR-interacting protein), a protein that regulates the localization of ATR to stretches of single-stranded DNA at sites of DNA replication stress (Cortez et al, 2001; Zou and Elledge, 2003). Although ATRIP is essential for ATR signalling (Cortez et al, 2001; Zou and Elledge, 2003; Ball and Cortez, 2005), several other proteins participate in ATR activation, including, notably, TopBP1 (Topoisomerase II beta binding protein 1) and Rad9 (higher eukaryote and Schizosaccharomyces pombe terminology; not to be confused with Saccharomyces cerevisiae Rad9) (Kumagai et al, 2006; Majka et al, 2006; Mordes et al, 2008; Lee and Dunphy, 2010).
Mammalian TopBP1 belongs to a family of evolutionarily conserved proteins that includes S. cerevisiae Dbp11, Drosophila melanogaster Mus101 and Xenopus laevis Cut5 (Yamane et al, 1997; Garcia et al, 2005). All these proteins function in the DNA replication checkpoint (Makiniemi et al, 2001; Yamane et al, 2002; Parrilla-Castellar and Karnitz, 2003; Garcia et al, 2005) and have multiple BRCT domains, which usually function in tandem to bind phosphoproteins (Manke et al, 2003; Yu et al, 2003). TopBP1 colocalizes with ATR, ATRIP and RPA (replication protein A) at sites of DNA replication stress (Makiniemi et al, 2001; Garcia et al, 2005). In one study, recruitment to these sites was dependent on BRCT domain 5 (Yamane et al, 2002), whereas in another study on BRCT domains 1–2 (Delacroix et al, 2007; Lee et al, 2007). Once recruited to sites of DNA replication stress, TopBP1 interacts with ATR to stimulate its kinase activity (Kumagai et al, 2006; Mordes et al, 2008). In addition to its role as a checkpoint protein, TopBP1 also functions in the initiation of DNA replication; this latter function of TopBP1 is apparently independent of its checkpoint function (Hashimoto and Takisawa, 2003; Tanaka et al, 2007; Zegerman and Diffley, 2007; Kumagai et al, 2010).
The p53-binding protein 1 (53BP1), such as TopBP1, also responds to DNA damage. However, unlike TopBP1, 53BP1 localizes to sites of DNA DSBs (Schultz et al, 2000; Anderson et al, 2001; Rappold et al, 2001; Xia et al, 2001; Huyen et al, 2004). These sites are physically distinct from the sites of DNA replication stress, as determined by immunofluorescence analysis of irradiated cells with antibodies against 53BP1, ATRIP and RPA (Venere et al, 2007). Depletion of 53BP1 in mammalian cells leads to defects in the G2- and intra-S-phase DNA damage checkpoints, but the magnitude of the defect is limited, and in chicken cells, in which the 53bp1 gene has been deleted, there is no G2- or intra-S-phase checkpoint defect (DiTullio et al, 2002; Wang et al, 2002; Ward et al, 2003; Nakamura et al, 2006). Thus, it is unclear whether 53BP1 has an important role in the DNA damage checkpoint. This is in contrast to what has been observed in S. cerevisiae, in which Rad9, the budding yeast orthologue of human 53BP1, is important for the DNA damage checkpoint and particularly in the G1-phase of the cell cycle (Siede et al, 1993; Wysocki et al, 2005; Hammet et al, 2007).
In this study, we have revisited the recruitment of TopBP1 to sites of DNA damage. We observed that, in addition to its role in the DNA replication stress checkpoint, TopBP1 colocalized with 53BP1 at sites of DNA DSBs in G1-phase. Furthermore, both 53BP1 and TopBP1 were implicated in the activation of the G1 DNA damage checkpoint. Thus, TopBP1 may mediate at least part of the checkpoint activity of 53BP1.
Results
To study the recruitment of TopBP1 to sites of DNA damage, we examined by immunofluorescence the intracellular localization of the endogenous protein in U2OS osteosarcoma cells exposed to ionizing radiation (IR). Consistent with previous studies (Makiniemi et al, 2001; Yamane et al, 2002), in some cells, TopBP1 colocalized with ATRIP and RPA at nuclear foci that we presume are sites of DNA replication stress (stalled or collapsed DNA replication forks ahead of DNA single- or double-stranded breaks) or sites of resected DSBs. However, in other cells, TopBP1 colocalized with 53BP1 (Figure 1A and B; Supplementary Figure 1). TopBP1 recruitment to the latter sites was only evident starting 1–2 h after irradiation and peaked 4 h after irradiation (Figure 1A and B; data not shown).
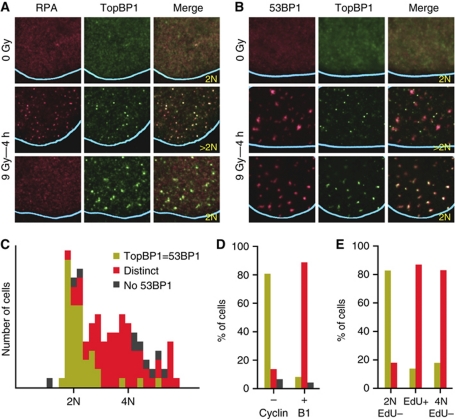
Figure 1.
Endogenous TopBP1 colocalizes with either RPA or 53BP1 in irradiated cells depending on the phase of the cell cycle. (A, B) Localization of endogenous TopBP1, RPA and 53BP1 in U2OS cells 4 h after exposure to IR (9 Gy) or after mock-irradiation (0 Gy), as determined by immunofluorescence. Because the IR-induced RPA foci are much smaller than the 53BP1 foci, only part of the cell nucleus is shown with the blue lines indicating the periphery of the nucleus. In this and all other figures the DNA content of the cells was quantified on the basis of the intensity of DAPI staining over the entire nucleus and shown as 2 or >2N; the DAPI staining intensities of the adjacent cells were used for calibration. (C) Histogram plot of the behaviour of TopBP1 according to genomic DNA content. Endogenous TopBP1 and 53BP1 IR-induced foci were visualized in U2OS cells 4 h after exposure to IR (9 Gy). The behaviour of TopBP1 was scored as: TopBP1=53BP1, colocalized TopBP1 and 53BP1 IR-induced foci; distinct, non-colocalizing TopBP1 and 53BP1 foci; No 53BP1, no 53BP1 foci. (D, E) Colocalization of TopBP1 and 53BP1 IR-induced foci according to (D) Cyclin B1 staining or genomic DNA content and (E) EdU incorporation. The behaviour of TopBP1 is presented using the same colouring scheme as in (C).
To understand why TopBP1 colocalized with RPA in some cells and with 53BP1 in others, we examined whether the behaviour of TopBP1 was cell cycle dependent. Cells, in which TopBP1 colocalized with 53BP1, had a 2N (G1) DNA content, whereas cells, in which the TopBP1 foci were distinct from the 53BP1 foci, had a DNA content greater than 2N (S- or G2-phases of the cell cycle; Figure 1A–C; Supplementary Figure 2A). In a second approach to explore for cell cycle dependency, we repeated the immunofluorescence experiment staining for Cyclin B1, in addition to TopBP1 and 53BP1. The latter two proteins colocalized only in the cells staining negative for Cyclin B1 (Figure 1D; Supplementary Figure 2B). Finally, we monitored colocalization of 53BP1 and TopBP1 in irradiated cells treated with the thymidine analogue 5-ethynyl-2′-deoxyuridine (EdU) to identify S-phase cells. Furthermore, colocalization of these two proteins was only observed in G1 cells (Figure 1E; Supplementary Figure 2C).
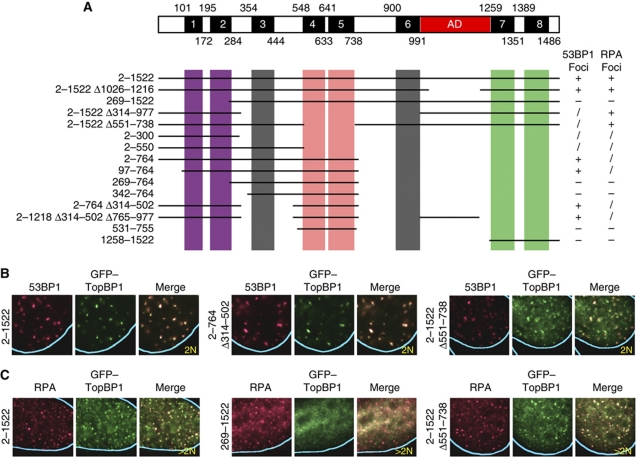
We next examined whether distinct domains of TopBP1 mediated its recruitment to sites of DNA DSBs in G1-phase and to sites of DNA replication stress/resected DNA DSBs in S/G2-phase. TopBP1 contains eight BRCT domains (Yamane et al, 1997; Garcia et al, 2005), of which six are arranged in pairs (BRCT domains 1–2, 4–5 and 7–8) and two are single (BRCT domains 3 and 6). In addition, TopBP1 has an ATR activation domain between BRCT domains 6 and 7 (Kumagai et al, 2006; Mordes et al, 2008). We fused either full-length TopBP1 or TopBP1 deletion mutants to green fluorescent protein (GFP) and monitored the colocalization of these fusion proteins with 53BP1 and RPA in irradiated cells. Efficient recruitment of TopBP1 to 53BP1 foci required BRCT domains 1–2 and 4–5, whereas recruitment to RPA foci required BRCT domains 1–2 and 7–8 (Figure 2; Supplementary Figure 3). By contrast, BRCT domains 3 and 6 and the activation domain of TopBP1 were not required for the recruitment to either 53BP1 or RPA IR-induced foci.
Figure 2.
Mapping the domains that mediate recruitment of TopBP1 to IR-induced 53BP1 or RPA foci. (A) Diagram of human TopBP1 showing the BRCT domains, numbered 1–8, and the ATR activation domain (AD) and summary of the properties of the tested TopBP1 deletion mutants. The mutants were scored according to efficiency of focus formation: +, wild type; /, foci formed, but a significant amount of TopBP1 remained in the nucleoplasm; −, no foci. (B, C) Examples of the localization of selected GFP-tagged TopBP1 mutants in U2OS cells 4 h after exposure to IR (9 Gy). The numbers refer to the residues present in the TopBP1 mutants. Δ, deletion.
To explore the specificity of recruitment mediated by each pair of TopBP1 BRCT domains, we generated GFP-tagged proteins containing single pairs of BRCT domains. These proteins were either not recruited (pairs 4–5 and 7–8) or were recruited inefficiently (pair 1–2) to sites of DNA damage (Figure 3A). To enhance their avidity for their targets, a tetramerization domain (Harbury et al, 1993; Zgheib et al, 2009) was subsequently introduced in these proteins. As homotetramers, all BRCT domain pairs localized efficiently to IR-induced foci. The tetramers of pair 1–2 colocalized with both 53BP1 and RPA foci and recruitment to the 53BP1 foci was cell cycle independent (Figure 3B). The tetramers of pair 4–5 formed foci only in G1 cells and these foci also contained 53BP1, whereas the tetramers of pair 7–8 formed foci only in S/G2 cells and these foci also contained RPA (Figure 3B).
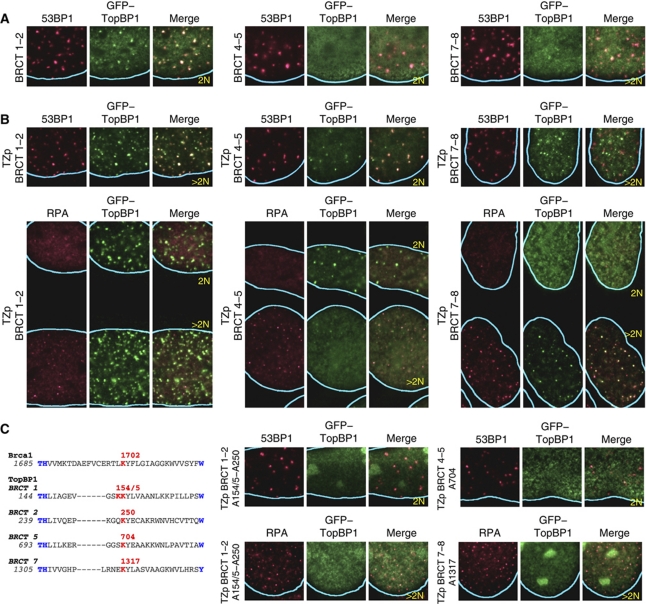
Figure 3.
Different pairs of TopBP1 BRCT domains have distinct properties for recruitment to sites of DNA damage. (A) Recruitment of BRCT domains 1–2 (residues 2–300), 4–5 (residues 531–755) and 7–8 (residues 1258–1522) as monomers to sites of DNA damage. Domains 1–2 formed IR-induced foci with reduced efficiency compared with full-length TopBP1; domains 4–5 and 7–8 did not form IR-induced foci. In this and all subsequent panels, U2OS cells were examined 4 h after irradiation (9 Gy). (B) Recruitment of BRCT domains 1–2 (residues 2–300), 4–5 (residues 531–755) and 7–8 (residues 1258–1522) as tetramers (TZp) to sites of DNA damage. Tetramers of domains 1–2 colocalized with both 53BP1 and RPA IR-induced foci. Tetramers of domains 4–5 colocalized only with 53BP1 foci and then only in G1 cells. Tetramers of domains 7–8 colocalized only with RPA foci. (C) Potential phosphate-binding lysines in BRCT domains 1, 2, 5 and 7 of human TopBP1 and effect of amino acid substitutions targeting these lysines on recruitment of oligomerized TopBP1 BRCT domains to IR-induced foci. The putative phosphate-binding lysines are coloured red; other conserved BRCT domain residues are coloured blue. The corresponding sequence of BRCA1 is shown for reference.
The BRCT domains typically bind phosphoproteins (Manke et al, 2003; Yu et al, 2003). In all examples known to date, the first BRCT domain of a BRCT pair contributes an evolutionarily conserved lysine that interacts with the phosphate group (Shiozaki et al, 2004; Stucki et al, 2005; Kilkenny et al, 2008). We searched for the presence of this lysine in the TopBP1 BRCT domain pairs 1–2, 4–5 and 7–8. Putative phosphate-binding lysines were present in BRCT domains 1 and 7, as expected, but not in domain 4; furthermore, putative phosphate-binding lysines were present in BRCT domains 2 and 5 (Figure 3C). We examined the role of each of these lysines in IR-induced focus formation using alanine mutants. Recruitment of oligomerized BRCT domain pair 1–2 was abolished only when the putative phosphate-binding lysines in both domains 1 and 2 were substituted with alanine; recruitment of oligomerized BRCT domain pair 4–5 was dependent on the lysine in domain 5, whereas recruitment of oligomerized BRCT domain pair 7–8 was dependent on the lysine in domain 7 (Figure 3C; Supplementary Figure 4). Thus, only the pair of BRCT domains 7 and 8 behaves similar to the previously described BRCT domain pairs of BRCA1, MDC1 and Crb2 in having the phosphate-binding site in the N-terminal BRCT domain.
To further validate the different recruitment properties of the various TopBP1 BRCT domains, we examined the localization of the various point mutants described above in cells treated with hydroxyurea (HU). Whereas in S-phase cells, IR induces both DNA replication stress and resected DNA DSBs, HU induces only DNA replication stress. In S-phase, the various TopBP1 mutants behaved identically in response to IR and HU: BRCT domain pairs 1–2 and 7–8 recruited TopBP1 to sites of DNA replication stress, whereas pair 4–5 did not localize to foci (Supplementary Figure 5; data not shown).
The colocalization of TopBP1 and 53BP1 at sites of DNA DSBs in G1 cells raises the question whether recruitment of one of these proteins is dependent on the other. Depletion of TopBP1 did not compromise 53BP1 focus formation (data not shown). In contrast, depletion of 53BP1 abrogated TopBP1 focus formation in G1 cells, but had no effect on the colocalization of TopBP1 with RPA in S and G2 cells (Figure 4A; Supplementary Figures 6A and 7). 53BP1 depletion also abrogated recruitment of oligomerized BRCT domains 4–5 to DNA damage sites, but had no effect on recruitment of BRCT domains 1–2 (Figure 4B; Supplementary Figure 6A). These results suggest that BRCT domains 4–5 may interact with 53BP1 at sites of DNA DSBs. Indeed, it has been reported that endogenous 53BP1 and TopBP1 co-immunoprecipitate (Yamane et al, 2002). In our hands, the interaction between these endogenous proteins was very weak. However, we could detect a specific interaction between tetramers of TopBP1 BRCT domains 4–5 with endogenous 53BP1 (Figure 4C).
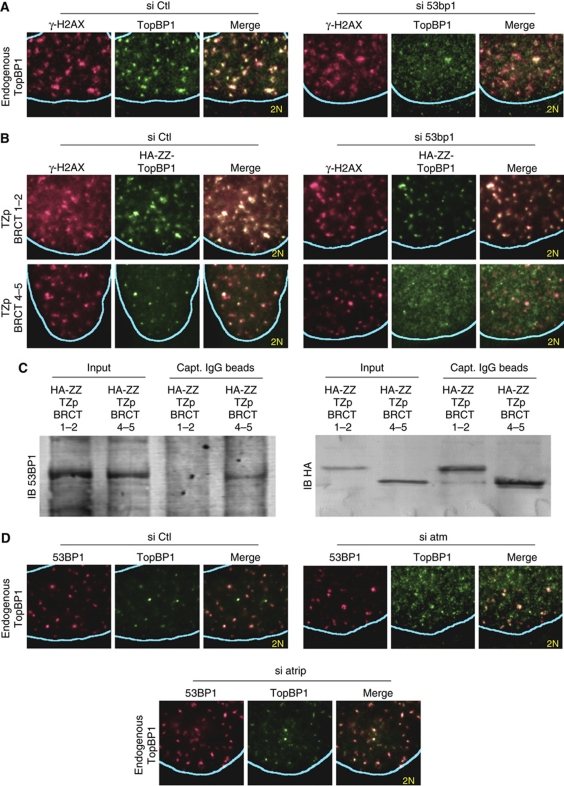
Figure 4.
TopBP1 recruitment to sites of DNA DSBs is 53BP1- and ATM-dependent. (A) Immunofluorescence analysis of U2OS cells for endogenous TopBP1 and phosphorylated histone H2AX (γ-H2AX) 4 h after irradiation (9 Gy). The cells were transfected with control (Ctl) siRNA or siRNA targeting 53bp1. (B) Immunofluorescence analysis of U2OS cells stably expressing haemagglutinin peptide and protein A-tagged (HA-ZZ) oligomerized (TZp) TopBP1 BRCT domains 1–2 (residues 2–300) or 4–5 (residues 531–755). The cells were transfected with control (Ctl) siRNA or siRNA targeting 53bp1 and were monitored for localization of γ-H2AX and the stably expressed TopBP1 proteins 4 h after irradiation (9 Gy). (C) Co-precipitation of oligomerized TopBP1 BRCT domains 4–5 with endogenous 53BP1. U2OS cells stably expressing HA-ZZ-tagged tetramers (TZp) of RCT domains 1–2 (residues 2–300) or 4–5 (residues 531–755) were used to prepare nuclear extracts (Input), which were then incubated with IgG-coated beads. HA-ZZ–TZp–TopBP1 fusion proteins and endogenous 53BP1 bound to the beads were detected by immunoblotting (IB) using anti-HA and anti-53BP1 antibodies, respectively. (D) Immunofluorescence analysis of U2OS cells for endogenous 53BP1 and TopBP1 4 h after irradiation (9 Gy). The cells were transfected with control (Ctl) siRNA or siRNA targeting atm or atrip.
To further validate the 53BP1 dependency of TopBP1 recruitment to sites of DNA DSBs in G1 cells, we took advantage of the fact that 53BP1 recruitment to sites of DNA DSBs is dependent on histone H2AX phosphorylation and the subsequent recruitment of the proteins MDC1 and RNF8 (Fernandez-Capetillo et al, 2002; Stewart et al, 2003; Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007). Indeed, although in G1-phase wild-type mouse embryo fibroblasts (MEFs) TopBP1 and 53BP1 colocalized with foci of phosphorylated histone H2AX (γ-H2AX), in h2ax−/−, mdc1−/− and rnf8−/− MEFs, both TopBP1 and 53BP1 failed to be recruited to sites of DNA DSBs (Supplementary Figure 8A). In contrast, recruitment of TopBP1 to sites of DNA replication stress was not dependent on histone H2AX, MDC1 or RNF8 (Supplementary Figure 8B).
As BRCT domains recognize phosphorylated serines or threonines, we further speculated whether the recruitment of TopBP1 to sites of DNA DSBs in G1 would be ATM or ATR dependent. Depletion of ATM, but not of ATRIP, abrogated the recruitment of endogenous TopBP1 to sites of DNA DSBs in G1 cells (Figure 4D; Supplementary Figure 7). A similar defect in recruitment was also observed in ataxia-telangiectasia cells, which have mutant ATM (Supplementary Figure 6B). However, ATM had no effect on recruitment of TopBP1 to sites of DNA replication stress in S-phase cells (data not shown).
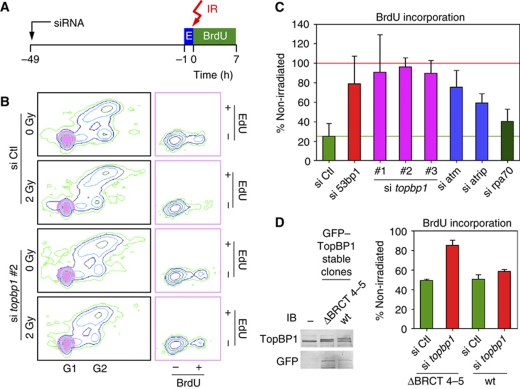
The recruitment of TopBP1 to sites of DNA DSBs in G1 cells suggests that TopBP1 may have a role in the G1 DNA damage checkpoint. To explore this possibility, we developed a G1 checkpoint assay. U2OS osteosarcoma cells were first pulsed for 1 h with the thymidine analogue EdU; EdU was then washed away and the cells were irradiated or mock-irradiated and incubated for 7 h in the presence of 5-bromo-2′-deoxyuridine (BrdU) and nocodazole (Figure 5A). In this assay, cells that were in G1-phase at the time of irradiation would stain negatively for EdU. Furthermore, cells that were in S-, G2- and M-phase would be unable to complete cytokinesis due to the presence of nocodazole, and therefore would remain with a G2 DNA content. Thus, at the time of collection, the Edu-negative cells with a G1-early S-phase DNA content would have been in G1-phase at the time of irradiation. We therefore examined the degree to which these cells incorporated BrdU during the 7-h period after irradiation, indicating entry into S-phase.
Figure 5.
TopBP1 and 53BP1 are required for the G1 DNA damage checkpoint. (A) Design of the G1 checkpoint assay. U2OS cells were transfected with siRNA. After 48 h, the cells were pulsed with EdU (E) for 1 h and then exposed to 2 Gy IR. After irradiation, the cells were incubated with BrdU and nocodazole for 7 h and then examined by flow cytometry. (B) Representative examples of flow cytometry data. Cells having G1 DNA content and staining negatively for EdU (highlighted by the purple circles in the left panels) were plotted on the basis of BrdU versus EdU incorporation (right panels). (C) Statistical analysis of the results of the G1 checkpoint assay performed in quadruplicate. For each sample, the fraction of BrdU-positive cells over the total number of G1/Edu-negative cells was calculated; then the ratio was determined of the fraction for the irradiated cells divided by the fraction for the non-irradiated cells. A ratio of 100% indicates no arrest in G1 after irradiation (marked by a red horizontal line). The green horizontal line indicates the ratio for the cells transfected with control (Ctl) siRNA. Means and s.d. values are shown. (D) A TopBP1 mutant lacking BRCT domains 4–5 is defective in the G1 checkpoint assay. U2OS cells stably transfected with plasmids expressing GFP-tagged wild-type (wt) TopBP1 or a TopBP1 mutant lacking residues 551–738 were treated with control (Ctl) siRNA or siRNA targeting the endogenous topbp1 gene (the 3′ untranslated region; si topbp1 #2) and then subjected to the G1 checkpoint assay. Expression of the endogenous and ectopically expressed TopBP1 proteins was monitored by immunoblotting (IB) with antibodies against TopBP1 and GFP. −; non-transfected cells.
Control cells had a robust G1 checkpoint, as evidenced by the lack of BrdU incorporation in the G1 EdU-negative cell population after irradiation. In contrast, entry of TopBP1-depleted cells into S-phase was essentially unaffected by irradiation (Figure 5B). Depletion of 53BP1, ATM and ATRIP had similar phenotypes as TopBP1 depletion in this assay, although the magnitude of the checkpoint defect was less pronounced than after TopBP1 depletion (Figure 5C). We attribute this to the higher efficiency of siRNA-mediated depletion of TopBP1 (Supplementary Figure 7) and the possibility that ATM and ATR/ATRIP may have redundant roles. In contrast, depletion of RPA, which compromises the intra-S-phase checkpoint (Zou and Elledge, 2003) did not affect the G1 checkpoint (Figure 5C), nor the recruitment of TopBP1 to sites of DNA DSBs in G1 cells (Supplementary Figure 6C).
To establish whether the interaction of 53BP1 and TopBP1 was important for the function of TopBP1 in the G1 checkpoint, we generated stably-transfected clones of U2OS cells expressing either GFP-tagged full-length TopBP1 or a TopBP1 mutant with BRCT domains 4–5 deleted (Δ551–738; Figure 5D). Expression of endogenous TopBP1 was depleted using an siRNA corresponding to the 3 untranslated region of the endogenous topbp1 gene and the G1 checkpoint assay performed. The cells stably expressing full-length TopBP1 retained the G1 checkpoint, whereas the cells expressing the TopBP1 deletion mutant exhibited a checkpoint defect (Figure 5D).
Interestingly, even though TopBP1 also has a role in the initiation of DNA replication, the siRNA-mediated depletion of TopBP1 did not inhibit entry into S-phase in our system (Figure 5B). Presumably, even more efficient depletion of TopBP1 is required to abolish the function of TopBP1 in initiation of DNA replication.
Discussion
The observations that TopBP1 can localize to both sites of DNA DSBs and sites of DNA replication stress and that localization to these sites is cell cycle dependent and mediated by distinct combinations of BRCT domains (1–2 and 4–5 for sites of DNA DSBs in G1-phase and 1–2 and 7–8 for sites of DNA replication stress and resected DNA DSBs in S/G2-phase) provide an explanation for the conflicting data in the literature regarding which domains are important for recruitment (Yamane et al, 2002; Delacroix et al, 2007; Lee et al, 2007). In essence, our studies show that a major part of the TopBP1 protein (BRCT domains 1–2, 4–5 and 7–8) mediates its recruitment to sites of DNA damage.
As BRCT domains are phosphopeptide-binding domains (Manke et al, 2003; Yu et al, 2003), our results suggest that TopBP1 is recruited to sites of DNA damage by interacting with phosphorylated proteins present at these sites. BRCT domains 1–2 are important for the recruitment to sites of both DNA DSBs and DNA replication stress. On the basis of published TopBP1–protein interactions, the recruitment of ToPBP1 to these sites may be mediated by interactions of BRCT domains 1–2 with NBS1 and Rad9, respectively (Delacroix et al, 2007; Lee et al, 2007; Yoo et al, 2009). The interaction between TopBP1 BRCT domains 4–5 and 53BP1 can explain the role of these BRCT domains on the recruitment of TopBP1 to sites of DNA DSBs in G1-phase and its dependency on 53BP1. Interestingly, the homologues of TopBP1 and 53BP1 in fission yeast also interact with each other, suggesting that these recruitment mechanisms may be conserved in evolution (Saka et al, 1997; Du et al, 2006).
TopBP1 has a well-established function in the DNA replication checkpoint, which is conserved in evolution (Makiniemi et al, 2001; Yamane et al, 2002; Parrilla-Castellar and Karnitz, 2003; Garcia et al, 2005). Unexpectedly, we observed that TopBP1 is also critical for the G1 checkpoint and that its function in G1-phase involves 53BP1. A role of 53BP1 as a checkpoint protein in vertebrate cells has been debated (DiTullio et al, 2002; Wang et al, 2002; Ward et al, 2003; Nakamura et al, 2006), even though its homologues in yeast are well-established checkpoint proteins (Weinert and Hartwell, 1988; Saka et al, 1997; Willson et al, 1997). The previous studies on the checkpoint function of 53BP1 in vertebrate cells had focused on the S- and G2-phases of the cell cycle. The results reported here suggest that 53BP1 exerts its checkpoint function predominantly in G1, similar to what has been observed with Rad9, the 53BP1 homologue, in budding yeast (Siede et al, 1993; Wysocki et al, 2005; Hammet et al, 2007). These studies further suggest that the checkpoint function of 53BP1 is mediated at least partially by TopBP1, although direct activation of ATM by 53BP1 may also be a contributor to the checkpoint function of 53BP1 (Lee et al, 2009).
How TopBP1 exerts a checkpoint function in G1-phase remains to be determined (Supplementary Figure 9). One possibility is that TopBP1 activates ATM either through an, as yet unidentified, ATM activation domain or even through its ATR activation domain, as ATM has sequence similarity with the domain of ATR that interacts with TopBP1 (Mordes et al, 2008; Lempiainen and Halazonetis, 2009). Alternatively, TopBP1 may activate ATR. In S- and G2-phase cells, an initial activation of ATM leads to strand resection and RPA-dependent recruitment and activation of ATR (Jazayeri et al, 2006; Myers and Cortez, 2006; Yoo et al, 2007, 2009; Shiotani and Zou, 2009). In G1 cells, we did not detect by immunofluorescence either ATRIP or RPA at sites of DNA DSBs. However, there is a possibility that very limited resection may be occurring, leading to limited recruitment of ATR that is undetectable by immunofluorescence. The two possibilities discussed above, activation of ATM and activation of ATR, are not mutually exclusive. In fact, we favour both, as the G1 checkpoint was dependent on ATM and also, although to a lesser degree, on ATR.
Finally, an unresolved issue is whether the colocalization of 53BP1 and TopBP1 in G1-phase affects DNA repair. The DNA DSBs in G1-phase are repaired mostly by non-homologous end joining, whereas DNA DSBs in S/G2-phase are repaired by homologous recombination. 53BP1 is apparently involved in the choice between these two repair pathways (Nakamura et al, 2006; Difilippantonio et al, 2008; Dimitrova et al, 2008; Bothmer et al, 2010; Bouwman et al, 2010; Bunting et al, 2010). Thus, it will be interesting to determine whether the DNA repair function of 53BP1 is modulated by TopBP1.
Materials and methods
Recombinant plasmids and antibodies
Plasmids encoding TopBP1 polypeptides fused to the C-terminus of GFP were generated from a previously-described mammalian expression plasmid (Huyen et al, 2004). For stable expression the GFP–TopBP1 inserts were transferred from the pSV2 vector to the pIRESN2 bicistronic vector (Clontech Laboratories, Mountain View, CA). For the co-immunoprecipitation experiments, the pIRESN2 vector was used to express a fusion protein consisting from its N-terminus to C-terminus of an HA tag, two tandem IgG-binding domains from protein A (Nilsson et al, 1987), a heterologous tetramerization domain (TZp) fused to a nuclear localization signal (Zgheib et al, 2009) and residues 2–300 or 531–755 of human TopBP1. Antibodies used were specific for 53BP1 (Schultz et al, 2000), ATRIP (Venere et al, 2007), GFP, ATM and TopBP1 (Abcam, Cambridge, UK), RPA70 (Calbiochem, La Jolla, CA, USA), Cyclin B1 (Upstate, Albany, NY, USA) and the HA tag (Covance, Princeton, NJ).
Focus formation assay
U20S cells, obtained from the American type culture collection (Manassas, VA, USA) were either not transfected (to monitor localization of endogenous proteins) or were transiently transfected with plasmids encoding GFP–TopBP1 proteins using Fugene (Roche Diagnostic, Basel, Switzerland) and examined 2 days later. To induce DNA damage, the cells were exposed to IR (9 Gy), and, 30 min to several hours later, fixed and processed for immunofluorescence, as described (Zgheib et al, 2009). Alternatively, cells were treated with 2 mM HU and examined 16 h later. To correlate the behaviour of TopBP1 foci according to genomic DNA content, overlapping images capturing a total of 227 cells were acquired using a × 40 magnification lens. The genomic DNA content of each cell was determined by integrating the density of DAPI staining over the entire nucleus of the cell after the images had been calibrated (via their overlapping part) to one another. Then a histogram plot showing for each cell the genomic DNA content and the behaviour of TopBP1 was prepared (Figure 1C). For all other immunofluorescence assays, the DNA content of the indicated cell was determined to be 2 or >2N based on the integrated DAPI staining value of the cell in comparison to the adjacent cells in the same field. For many experiments, the cells were also treated with EdU for 30 min before fixing and the presence or absence of EdU incorporation, helped distinguish S-phase cells from cells in G1- or G2-phase. The processing of the images and the calculations were performed using Imagevision/IRIX and PERL software (Silicon Graphics, Mountain View, CA, USA). For the correlations of Cyclin B1 staining and EdU incorporation to behaviour of TopBP1 IR-induced foci, more than 200 cells were counted.
Small interfering RNA (siRNA) transfection
U2OS cells were transfected using Oligofectamine (Invitrogen, Carlsbad, CA, USA) or Hyperfectamine (Qiagen, Valencia, CA, USA) with control siRNA (luciferase; Dharmacon, Lafayette, CO) or siRNAs targeting atm, 53bp1, mdc1 (Mochan et al, 2003), atrip (Venere et al, 2007), rpa70 or topbp1. The sequence of the siRNA targeting rpa70 was: ACCACTCTATCCTCTTTCATGdtdt. For topbp1, 3 different siRNAs were used: #1, CAGAAUUGUUGGUCCUCAAdtdt; #2, GAAUUCUGCUUCAGAGUAUAdtdt; and #3, CGAUAGAGGAGACUCAUGAdtdt. The cells were examined 48 h after siRNA transfection.
Preparation of cell extracts, immunoblotting and coprecipitation assays
The preparation of whole cell extracts and immunoblotting were performed as described previously (Venere et al, 2007). For analysis of the interaction of the BRCT domains of TopBP1 with endogenous 53BP1, stable clones of U2OS cells expressing the pIRESN2 vectors described above were selected with G418 as described previously (Venere et al, 2007). Nuclear extracts prepared from these cells (Dignam et al, 1983) were incubated for 1 h at 4°C in buffer consisting of 25 mM BTP (pH 6.8), 10 mM EDTA, 1 mM EGTA, 150 mM KCl, 5% glycerol and 0.75% CHAPS with M-270 Epoxy Dynabeads (Dynal AS, Oslo, Norway) coated with rabbit IgG (Sigma, St Louis, MO, USA). Then the beads were washed extensively in the same buffer and captured HA-tagged TopBP1 and endogenous 53BP1 proteins were detected by immunoblotting.
Checkpoint assay
At 48 h after siRNA transfection, U2OS cells pulse-labelled for 1 h with 10 μM EdU (Invitrogen) were exposed to IR (2 Gy) or were mock-irradiated. Then, the cells were incubated for 7 h with 10 μM BrdU (Sigma) and 0.25 μg/ml nocodazole (Sigma). Afterwards, the cells were collected by trypzinization and fixed in ice-cold 70% ethanol for 20 min. The presence of Edu and BrdU were monitored by Click-It chemistry (Invitrogen) and anti-BrdU antibodies (BD Bioscence, Franklin Lakes, NJ), respectively. After staining of the genomic DNA with propidium iodide, the samples were analysed by flow cytometry.
Supplementary Material
Acknowledgments
This study was supported by grants from the Swiss National Foundation, the NIH, and the European Commission Seventh Framework Programme (GENICA) to TDH.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abraham RT (2001) Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Anderson L, Henderson C, Adachi Y (2001) Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol 21: 1719–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Cortez D (2005) ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J Biol Chem 280: 31390–31396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC (2010) 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med 207: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J (2010) 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17: 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Guntuku S, Qin J, Elledge SJ (2001) ATR and ATRIP: partners in checkpoint signaling. Science 294: 1713–1716 [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM (2007) The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21: 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, Sleckman BP, Nussenzweig A (2008) 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTullio RA Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, Halazonetis TD (2002) 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol 4: 998–1002 [DOI] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Russell P (2006) Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20: 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, Carpenter PB, Bonner WM, Chen J, Nussenzweig A (2002) DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol 4: 993–997 [DOI] [PubMed] [Google Scholar]
- Garcia V, Furuya K, Carr AM (2005) Identification and functional analysis of TopBP1 and its homologs. DNA Repair 4: 1227–1239 [DOI] [PubMed] [Google Scholar]
- Hammet A, Magill C, Heierhorst J, Jackson SP (2007) Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep 8: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T (1993) A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262: 1401–1407 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Takisawa H (2003) Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J 22: 2526–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411 [DOI] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol 8: 37–45 [DOI] [PubMed] [Google Scholar]
- Kilkenny ML, Dore AS, Roe SM, Nestoras K, Ho JC, Watts FZ, Pearl LH (2008) Structural and functional analysis of the Crb2-BRCT2 domain reveals distinct roles in checkpoint signaling and DNA damage repair. Genes Dev 22: 2034–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG (2006) TopBP1 activates the ATR–ATRIP complex. Cell 124: 943–955 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG (2010) Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell 140: 349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Dunphy WG (2010) Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell 21: 926–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Goodarzi AA, Jeggo PA, Paull TT (2009) 53BP1 promotes ATM activity through direct interactions with the MRN complex. EMBO J 29: 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG (2007) The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem 282: 28036–28044 [DOI] [PubMed] [Google Scholar]
- Lempiainen H, Halazonetis TD (2009) Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J 28: 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Majka J, Niedziela-Majka A, Burgers PM (2006) The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell 24: 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiniemi M, Hillukkala T, Tuusa J, Reini K, Vaara M, Huang D, Pospiech H, Majuri I, Westerling T, Makela TP, Syväoja JE (2001) BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J Biol Chem 276: 30399–30406 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB (2003) BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Mochan TA, Venere M, DiTullio RA Jr, Halazonetis TD (2003) 53BP1 and NFBD1/MDC1-Nbs1 function in parallel interacting pathways activating ataxia-telangiectasia mutated (ATM) in response to DNA damage. Cancer Res 63: 8586–8591 [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D (2008) TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev 22: 1478–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JS, Cortez D (2006) Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem 281: 9346–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Sakai W, Kawamoto T, Bree RT, Lowndes NF, Takeda S, Taniguchi Y (2006) Genetic dissection of vertebrate 53BP1: a major role in non-homologous end joining of DNA double strand breaks. DNA Repair (Amst) 5: 741–749 [DOI] [PubMed] [Google Scholar]
- Nilsson B, Moks T, Jansson B, Elmblad A, Holmgren EDF, Henrichson C, Jones A, Uhlen M (1987) A synthetic IgG-binding domain based on Staphylococcal protein A. Protein Eng 1: 107–113 [DOI] [PubMed] [Google Scholar]
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA (2002) Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet 36: 617–656 [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Karnitz LM (2003) Cut5 is required for the binding of Atr and DNA polymerase alpha to genotoxin-damaged chromatin. J Biol Chem 278: 45507–45511 [DOI] [PubMed] [Google Scholar]
- Rappold I, Iwabuchi K, Date T, Chen J (2001) Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol 153: 613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M (1997) Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev 11: 3387–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73: 39–85 [DOI] [PubMed] [Google Scholar]
- Schultz LB, Chehab NH, Malikzay A, Halazonetis TD (2000) p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol 151: 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Zou L (2009) Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell 33: 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki EN, Gu L, Yan N, Shi Y (2004) Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell 14: 405–412 [DOI] [PubMed] [Google Scholar]
- Siede W, Friedberg AS, Friedberg EC (1993) RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 90: 7985–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Venere M, Snyder A, Zgheib O, Halazonetis TD (2007) Phosphorylation of ATR-interacting protein on Ser239 mediates an interaction with breast-ovarian cancer susceptibility 1 and checkpoint function. Cancer Res 67: 6100–6105 [DOI] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Carpenter PB, Elledge SJ (2002) 53BP1, a mediator of the DNA damage checkpoint. Science 298: 1435–1438 [DOI] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, Chen J (2003) p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 23: 2556–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH (1988) The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241: 317–322 [DOI] [PubMed] [Google Scholar]
- Willson J, Wilson S, Warr N, Watts FZ (1997) Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res 25: 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ (2005) Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol 25: 8430–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Morales JC, Dunphy WG, Carpenter PB (2001) Negative cell cycle regulation and DNA damage-inducible phosphorylation of the BRCT protein 53BP1. J Biol Chem 276: 2708–2718 [DOI] [PubMed] [Google Scholar]
- Yamane K, Kawabata M, Tsuruo T (1997) A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur J Biochem 250: 794–799 [DOI] [PubMed] [Google Scholar]
- Yamane K, Wu X, Chen J (2002) A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol Cell Biol 22: 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Dunphy WG (2007) Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J Biol Chem 282: 17501–17506 [DOI] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG (2009) The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol Biol Cell 20: 2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J (2003) The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- Zgheib O, Pataky K, Brugger J, Halazonetis TD (2009) An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks. Mol Cell Biol 29: 1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.