Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion
The methyl-H3K9-binding protein MPP8 is shown to act as repressor of E-cadherin transcription, regulating EMT and metastasis formation.
Keywords: chromodomain, DNA methylation, EMT, H3K9 methylation, transcription
Abstract
H3K9 methylation has been linked to a variety of biological processes including position-effect variegation, heterochromatin formation and transcriptional regulation. To further understand the function of H3K9 methylation, we have identified and characterized MPP8 as a methyl-H3K9-binding protein. MPP8 displays an elevated expression pattern in various human carcinoma cells, whereas knocking-down MPP8 results in the loss of cellular mesenchymal marker as well as the reduction of tumour cell migration and invasiveness, suggesting that MPP8 contributes to tumour progression. Following characterization demonstrates that MPP8 targets the E-cadherin gene promoter and modulates the expression of this key regulator of cell behaviour and tumour progression through its methyl-H3K9 binding. Furthermore, MPP8 interacts with H3K9 methyltransferases GLP and ESET, as well as DNA methyltransferase 3A. MPP8 knockdown decreases DNA methylation on E-cadherin CpG island attended by the loss of DNMT3A localization, indicating MPP8 also directs DNA methylation. Together, our results suggest a model by which MPP8 recognizes methyl-H3K9 marks and directs DNA methylation to repress tumour suppressor gene expression and, in turn, has an important function in epithelial-to-mesenchymal transition and metastasis.
Introduction
Carcinogenesis is a progression of events resulting from not only the accumulation of genetic alterations but also the disruption of epigenetic modifications (Yoo and Jones, 2006). In eukaryotic organisms, the epigenetics network has many layers of complexity that could be summarized in four major modifications: DNA methylation, histone modifications, chromatin remodelling and microRNAs (Esteller, 2006). As one of the key players in the chromatin regulation, histone octamers are wrapped around with 147 bp of DNA to form a nucleosome, which is subjected to at least eight distinct types of post-translational modifications including acetylation and methylation (Kouzarides, 2007). The complex array of these modifications has been proposed to constitute a ‘histone code' that can be recognized by different chromatin regulatory proteins, which in turn affect the chromatin structure or regulate the accessibility of DNA to various machineries (Jenuwein and Allis, 2001). In normal cells, these modifications are delicately balanced, and small changes in a given parameter can lead to major consequences and ultimately result in cellular transformation and malignant outgrowth (Ting et al, 2006).
Epigenetic silencing of tumour suppressor genes is a common event during carcinogenesis and often involves aberrant DNA methylation and histone modifications (Wang et al, 2007). One well-studied example is the transcriptional repression of the pleiotropic cell behaviour regulator E-cadherin. Emerging evidences indicate that epigenetic silencing of E-cadherin during tumour progression is achieved by a combination of different mechanisms, including transcription factors, promoter hypermethylation, histone deacetylation and methylation (Peinado et al, 2007). For example, zinc-finger transcription factor SNAIL recruits H3K27 HMTase polycomb repressive complex 2 (PRC2) and Sin3A/HDAC complexes to repress E-cadherin expression (Peinado et al, 2004; Herranz et al, 2008). DNA-binding proteins ZEB1/2 and several chromatin modifying enzymes co-exist in the transcription co-repressor CtBP-1 complex to downregulate E-cadherin expression (Shi et al, 2003). In addition, promoter hypermethylation has been associated with E-cadherin gene silencing in various carcinoma cells (Grady et al, 2000). Although the interplay between DNA methylation and histone modifications has been well-documented, the molecular details of how these mechanisms cooperate for E-cadherin gene repression remain unclear.
As one of the best-studied histone modifications, histone methylation occurs on both arginine and lysine residues and can be recognized by effector proteins harbouring different methyl-histone-binding domains (Kouzarides, 2007). So far, six distinct motifs including chromodomain, Tudor domain, WD40 repeat domain, MBT domain, PHD domain and ankyrin-repeats domain, have been shown to be recruited by different methylated lysines on histone tails and this recruitment is a critical step for the functional consequences associated with different methylation events (Taverna et al, 2007; Collins et al, 2008). The first identified example of methyl-lysine-dependent protein–protein interaction was between H3K9 methylation and heterochromatin protein 1 (HP1). HP1 proteins recognize methyl-H3K9 through its N-terminal chromodomain and form a protein dimer with a wide range of chromosomal proteins through its C-terminal chromoshadow domain for various functions including heterochromatin formation, telomere capping and transcriptional regulation (Kwon and Workman, 2008). Although methyl-H3K9 is one of the major repressive marks and HP1 proteins have been co-purified with several repressive protein complexes (Ogawa et al, 2002; Shi et al, 2003), both HP1γ and H3K9me3 also associate with the coding regions of a number of active genes and their presence relies on elongation by RNA pol II (Vakoc et al, 2005). Therefore, the functional outcomes of H3K9 methylation are determined by their localization in chromatin context and accessibility to different methyl-histone-binding proteins.
To determine whether other effector proteins are also involved in mediating biological functions of H3K9 methylation, we performed an in vitro screen using a protein chromodomain microarray. We have identified an uncharacterized protein MPP8, which is capable of recognizing methylated H3K9 marks through its chromodomain in vitro and in cells. We also demonstrate that MPP8 represses E-cadherin gene expression and is involved in regulation of tumour cell growth and epithelial-to-mesenchymal transition (EMT) through methyl-H3K9 binding. Biochemical analyses reveal that MPP8 associates with H3K9 methylation and DNA methylation machineries and co-localizes in E-cadherin promoter region. Importantly, MPP8 also directs DNA methylation by recruiting DNMT3A to the 5′-regulatory regions of E-cadherin gene. Together, our work not only characterized MPP8 as a methyl-H3K9-binding protein, but also revealed a novel molecular mechanism by which MPP8 couples histone H3K9 methylation and DNA methylation for tumour suppressor gene silencing and metastasis.
Results
MPP8 chromodomain is a methyl-H3K9-binding motif
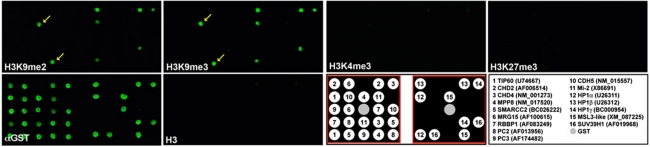
The fact that different methyl-binding proteins can be recruited to same or different methylated lysines to mediate functional outcomes indicates that the translation of the ‘histone code' is more complex. For example, methyl-H3K9 recruits chromodomain containing proteins HP1 and CDY (Fischle et al, 2008; Kwon and Workman, 2008), as well as the ankyrin-repeats containing protein G9a (Collins et al, 2008), whereas the WD40 domain containing protein EED recognizes both H3K9me3 and H3K27me3 marks (Margueron et al, 2009). To explore the possibility that additional chromodomain containing proteins possess the ability to ‘read' the H3K9me marks, we generated a protein chromodomain array, which contains 16 representative chromodomains fused with GST and screened their binding ability to different methyl-H3 peptides as described previously (Kim et al, 2006). As expected, chromodomains from HP1 proteins recognize H3K9me2 and H3K9me3 peptides, but not the unmethylated control (Figure 1). Furthermore, we observed that H3K9me2 and me3 peptides also bind to the chromodomain of an uncharacterized protein MPP8 (arrows in Figure 1), which was initially isolated in an expression-cloning screen using an antibody against M-phase phosphoproteins (Matsumoto-Taniura et al, 1996). To further determine the specificity of these interactions, we re-probed the same array with H3K4me3 and H3K27me3 peptides. None of the arrayed chromodomains showed obvious binding to these peptides (Figure 1).
Figure 1.
The MPP8 chromodomain is a methyl-H3K9-binding motif. The chromodomain array was probed with Cy3 labelled H3K9me2, H3K9me3, H3K4me3, H3K27me3 and H3 (1–18) peptides and α-GST, as indicated. A key to the arrayed chromodomains is given together with accession numbers. The middle position (grey circle) contains GST alone as a background indicator. Arrows indicate binding of MPP8 chromodomain to H3K9me2 and me3 peptides on the array.
MPP8 binds to methylated H3K9 in vitro and in vivo
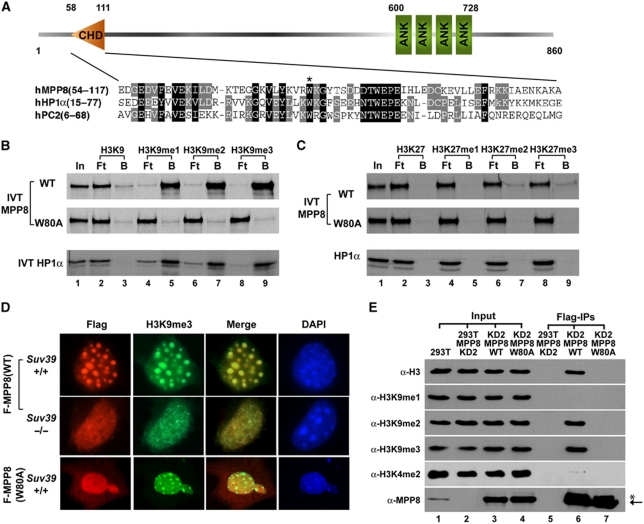
Different from HP1 proteins, MPP8 is a relatively large protein (860 aa) and does not have homologs in Drosophila melanogaster and Caenorhabditis elegans, suggesting that MPP8 could function through different mechanisms. Domain structure analysis reveals that MPP8 contains one chromodomain near the N-terminal, with four copies of ankyrin-repeats near the C-terminal (Figure 2A). In vitro translation coupled peptide pull-down assays demonstrate that full-length (FL)-MPP8 is still capable of binding to immobilized methyl-H3K9 peptides (1–20 aa) but not the unmethylated form (Figure 2B, upper panel). Furthermore, we mutated a conserved aromatic residue in MPP8 chromodomain (W80A) for same binding assays. As indicated in Figure 2B, this single amino acid substitution completely abolished MPP8 binding ability to same methyl-H3K9 peptides (middle panel). As control, HP1α showed the similar methyl-H3K9-binding profile, which is consistent with previous reports. To better mimic the in vivo situation, we also chemically installed different methyl-H3K9 analogs on H3 (Simon et al, 2007) for the in vitro pull-down assays and observed a similar methyl-H3K9-binding profile of MPP8 (Supplementary Figure S1A). Together, these results indicate that the chromodomain is critical for methyl-H3K9 binding of MPP8 in vitro even though the ankyrin-repeats domain has been shown as another methyl-lysine-binding module (Collins et al, 2008). In addition, we examined whether MPP8 recognizes H3K27me marks because of the high sequence similarity between K9 site and K27 site (Min et al, 2003). In the same assays, MPP8 displayed no binding to either H3 peptides harbouring methylated or unmethylated K27 (18–37 aa) (Figure 2C) or H3 chemically installed with different methyl-K27 analogs (Supplementary Figure S1B). Collectively, these data clearly demonstrate that MPP8 specifically binds to methyl-H3K9 in vitro and its binding ability requires an intact chromodomain.
Figure 2.
MPP8 binds to methylated histone H3K9 through chromodomain. (A) Diagram of full-length MPP8 and chromodomain alignment among human MPP8, HP1α and PC2. Conserved amino acids are shaded. The conserved W80 is indicated with a star. (B, C) MPP8 specifically recognizes methyl-H3K9 in vitro. 35S-labelled in vitro translated MPP8-wt or W80A mutant were tested for binding with immobilized biotin-H3 peptides with or without methylation at K9 (B) or K27 (C). Methylation states are indicated. 35S-labelled in vitro translated HP1α serves as controls in the parallel. ‘In' represents 10% of total input, ‘B' and ‘Ft' represent bound and flow through fractions, respectively. (D) H3K9me3 marks recruit MPP8 onto heterochromatin in cells. Immortalized MEF cells derived from wild-type or Suv39h1/h2 knockout mice were transfected with vectors expressing Flag-MPP8 or W80A mutant. Cells were fixed and stained with mouse anti-Flag (red) and chicken anti-H3K9me3 (green) antibodies. Blue colour (DAPI) shows the nuclei. (E) MPP8 preferentially binds to chromatin containing H3K9me2 and me3 marks in cells. MPP8 stable knockdown 293T cells (KD2) were transfected with vectors expressing shRNA resistant Flag-MPP8 or W80A mutant. MPP8 and bound chromatins were IPed by anti-Flag antibody and analysed by western blot. Antibodies are indicated and a non-specific cross-react band is labelled with a star.
H3K9me3 is enriched in pericentric heterochromatin regions where DAPI heavily stains (Rice et al, 2003). We thus expect MPP8 to be localized in these regions if MPP8 recognizes H3K9me3 marks in cells. In this regards, we expressed Flag-MPP8-FL or its W80A mutant in immortalized MEF cells. Immunostaining results indicate that Flag-MPP8 wild-type (wt) localizes on heterochromatin where H3K9me3 marks are enriched, whereas the W80A mutant uniformly distributes in the nucleus (Figure 2D, upper and bottom panels). These results in combination with our peptide pull-down data suggest that MPP8 localizes on heterochromatin through H3K9me3 binding. To test this possibility, we expressed Flag-MPP8-wt in immortalized MEF cells derived from Suv39h1/h2 double knockout mice (Peters et al, 2001). Similar to the previous reports, depletion of Suv39h1/h2 led to the loss of enrichment of H3K9me3 in heterochromatin regions (Figure 2D, column 2). Importantly, loss of heterochromatin localization of H3K9me3 is concomitant with a loss of MPP8 localization on heterochromatin (Figure 2D, middle panels). Given that the expression of Flag-MPP8-W80A mutant in MEF cells does not affect distribution of H3K9me3 on heterochromatin (Figure 2D, bottom panels), these results demonstrate that MPP8 recognizes H3K9me3 marks in cells.
To further explore MPP8 functions, we generated MPP8 antibody that specifically recognizes a protein band with molecular weight about 110 kDa from whole cell lysates on western blot (Supplementary Figure S2A). To validate that this protein band indeed represents the endogenous MPP8, we generated two MPP8 stable knockdown 293T cell lines using vector-based short hairpin RNA (shRNA) targeting to different regions of human MPP8. Western blot analysis not only confirmed the antibody specificity but also indicates that over 90% knockdown efficiency has been achieved with both shRNAs at the protein level (KD1 and KD2; Supplementary Figure S2B). Next, we rescue expressed shRNA resistant Flag-MPP8-wt or W80A mutant in MPP8-KD2 293T cells. Flag-MPP8 was then immunoprecipitated (IPed) and the MPP8 bound nucleosome was analysed by western blot using specific antibodies against different H3K9 methylation states to determine the methyl-H3K9-binding preference of MPP8 in cells. As indicated in Figure 2E, Flag-MPP8-wt was able to pull-down H3 harbouring K9me2 and me3 marks but not K9me1 or K4me2 marks (lane 6), whereas the W80A mutant was not co-purified with nucleosome (lane 7). These results are consistent with our in vitro binding data as well as immunostaining results, and indicate that MPP8 preferentially recognizes di- and tri-methylated H3K9 through its chromodomain in cells. We thus conclude that MPP8 is a methyl-H3K9-binding protein.
MPP8 is important for tumour cell proliferation
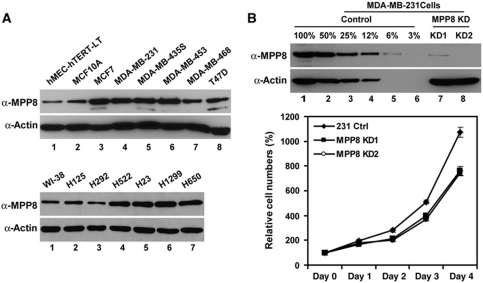
Given that H3K9 methylation is critical for transcriptional regulation and genomic stability (Peters et al, 2001), we speculate that methyl-H3K9-binding protein MPP8 could have a function in cancer. To test this possibility, we examined MPP8 expression in several human carcinoma cell lines. As shown in Figure 3A, four of six breast cancer cell lines showed an increased MPP8 expression compared with immortalized human mammary epithelial cells hEMCs-hTERT-LT and MCF10A (lanes 1–6, top panels). Additionally, this elevated expression pattern was also observed in four of six human non-small cell lung cancer cells (lanes 4–7, bottom panels) and most of commonly cultured human tumour cell lines including HeLa and U2OS compared with the normal human fibroblast cell WI-38 (data not shown), implicating that MPP8 may contribute to tumour cell maintenance. To test this idea, we established two stable MPP8 knockdown MDA-MB-231 cell lines using the same shRNA vectors we described above. Western blot analysis indicates that over 90% knockdown efficiency has been achieved in both cell lines (Figure 3B, top panels). Furthermore, MPP8 knockdown leads to a moderate growth reduction in MDA-MB-231 cells (Figure 3B, bottom panel), indicating MPP8 has a function in tumour cell proliferation.
Figure 3.
MPP8 is important for tumour cell proliferation. (A) Western blot analysis of two immortalized human mammary epithelial cells (MCF10A and hMEC-hTERT-LT) and six breast cancer cell lines (top panels); human normal fibroblast cells (WI-38) and six non-small cell lung cancer cells (bottom panels). (B) MPP8 knockdown results in decreased cell proliferation. Top panels are western blot analysis of control and two stable MPP8 knockdown MDA-MB-231 cell lines. Bottom panel shows the growth curve of different MPP8 knockdown cells. Viable cells were quantified every 24 h for 4 days after initial seeding. Cell numbers from six independent experiments were averaged, and the variations are presented with error bars.
MPP8 promotes tumour cell motility and invasion
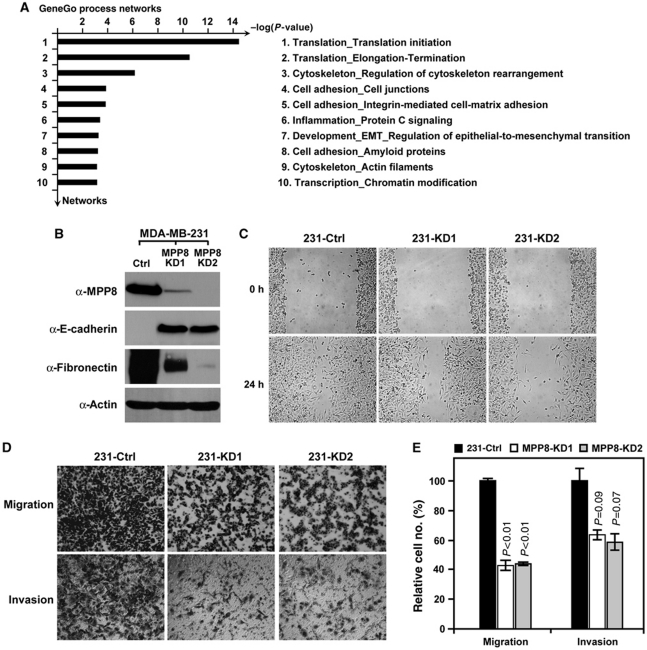
To further evaluate the functional outcomes caused by MPP8 knockdown, we performed the cDNA microarray analysis using RNA generated from control or MPP8-KD2 MDA-MB-231 cells. The raw data were analysed using SAM software with R package as two class unpaired samples and the genes with statistical significance between control and MPP8 knockdown samples were further analysed on GeneGo MetaCoreTM platform. Data mining and pathway analysis reveal that a large group of dysregulated genes in response to MPP8 knockdown are involved in cell adhesion and EMT, implicating MPP8 has important functions in this process (Figure 4A). In addition, pathway analysis also indicates that MPP8 could be involved in translation and transcription regulations.
Figure 4.
MPP8 promotes tumour cell migration and invasion. (A) GeneGo statistical process analysis of DNA microarray data derived from control and MPP8-KD2 MDA-MB-231 cells. Ten dysregulated GeneGo processes with best significance (lowest P-value) in response to MPP8 knockdown are listed. (B) Western blot analysis using whole cell lysates derived from control and MPP8 knockdown MDA-MB-231 cells. Specific antibodies as indicated. (C) Scratch wounding assays on the confluent layers of control and MPP8 knockdown MDA-MB-231 cells. Images were acquired at 0 and 24 h after wounding. (D, E) Migration and invasion assays of control and MPP8 knockdown MDA-MB-231 cells. Cells were induced to move or invade through uncoated or Matrigel-coated membranes for 12 or 24 h, respectively. Membranes were then fixed, photographed (D) or quantitated (E). Columns represent the mean of triplicate assays and the variations are presented with error bars. Control cell numbers were normalized as 100%. P-values are indicated.
EMT is a highly conserved cellular process that has pivotal functions in diverse processes during embryonic development, chronic inflammation, fibrosis, and tumour progression (Thiery, 2002). To further investigate whether MPP8 is indeed involved in EMT, we examined the epithelial and mesenchymal cell markers in control and MPP8 knockdown cells by western blot. MDA-MB-231 cells are typical metastatic mesenchymal cells that express a high level of mesenchymal marker Fibronectin, but not epithelial protein E-cadherin. However, MPP8 knockdown results in a significant loss of the Fibronectin expression and a marked increase of E-cadherin expression (Figure 4B). In addition, differential interference contrast microscopy analysis indicates that cells changed their shapes to a cuboidal form in response to MPP8 knockdown (Supplementary Figure S3). These results together suggest that MPP8-KD2 MDA-MB-231 cells undergo a mesenchymal-to-epithelial like transition. We further applied several in vitro assays to determine whether the functional changes in cell behaviour occurred following altered protein expression patterns. As indicated in Figure 4C, MPP8-KD2 cells displayed a notably slower recovery compared with control cells in the monolayer wound-healing assays, indicating MPP8 is important for cell mobility. More specifically, the motility and invasiveness of these cells were independently assessed using the Boyden chamber assays. Whereas the control cells showed great motility and invasiveness, MPP8 knockdown MDA-MB-231 cells displayed a 50–60% reduction on migratory ability and a 50% reduction on invasive ability to move through trans-well membranes with or without Matrigel coating, respectively (Figure 4D and E). Collectively, these results demonstrate that MPP8 promotes tumour cell migration and invasion and could therefore contribute to maintaining the metastatic status of human breast carcinoma cell MDA-MB-231.
MPP8 represses E-cadherin gene expression
It has been widely acknowledged that the hallmark of EMT is the functional loss of E-cadherin (Yang and Weinberg, 2008). As a pleiotropic regulator of cell behaviour, E-cadherin controls a complex transcriptional network and the loss of E-cadherin itself is sufficient to cause EMT or to afford functional traits that allow completion of the later steps of metastasis (Onder et al, 2008). In addition, E-cadherin also negatively regulates cell growth by modulating proliferation-dependent β-catenin transcriptional activity (Stockinger et al, 2001). These observations prompted us to investigate whether MPP8 regulates E-cadherin expression and in turn mediates cellular behaviour changes. Although E-cadherin mRNA level is extremely low in MDA-MB-231 cells, MPP8 knockdown significantly increases E-cadherin transcription (Figure 5A). The similar E-cadherin de-repression was also observed after knocking-down MPP8 in the invasive H23 (non-small cell lung cancer) cells and HeLa (cervical cancer) cells (Supplementary Figure S4A and B), suggesting that MPP8 represses E-cadherin expression in different cancer cells. To test this possibility, we carried out reporter assays in 293T cells using a vector in which the luciferase gene is driven by the E-cadherin promoter (−420∼+23) (Shi et al, 2003). As indicated in Figure 5B, overexpression of MPP8-wt but not W80A mutant causes a moderate (∼30%) repression on reporter gene activity (lanes 1–3). As 293T cells express a high level of endogenous MPP8, we used MPP8 knockdown cells for same assays. As expected, we observed two- to three-fold increases on luciferase activity in MPP8 knockdown cells and these increases are MPP8 dosage dependent (lanes 1, 4 and 5). To assess the role of methyl-H3K9 binding in MPP8-mediated repression, we next rescue expressed MPP8-wt or W80A in MPP8-KD2 cells. Expression of MPP8-wt significantly decreased reporter activity, whereas W80A mutant showed a moderate repressive effect (lanes 5–7). In addition, ChIP analysis reveals that exogenous E-cadherin promoter is partially assembled into nucleosome with H3K9me3 marks and MPP8 knockdown or rescue expression does not affect H3K9 methylation pattern (Figure 5C; Supplementary Figure S5). These data suggest that MPP8 could repress E-cadherin expression through methyl-H3K9 binding.
Figure 5.
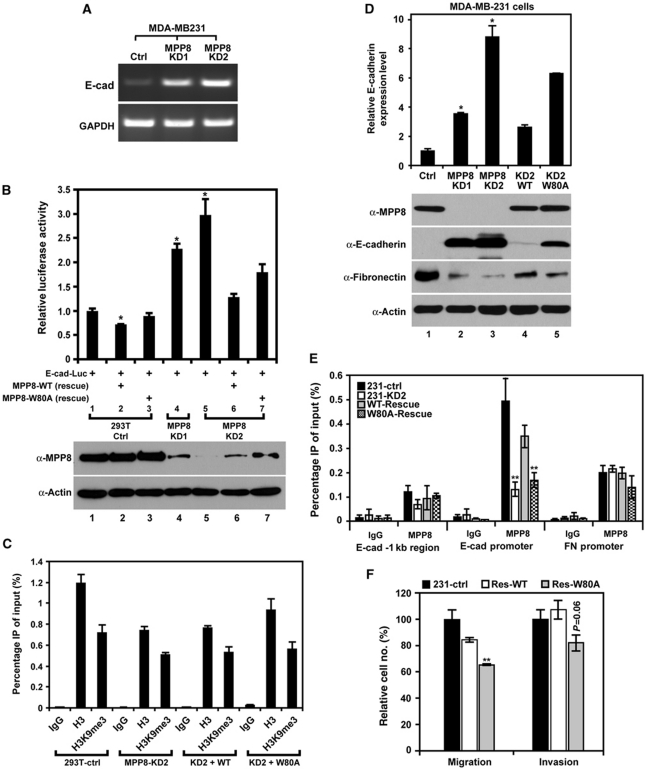
MPP8 represses E-cadherin gene expression. (A) E-cadherin mRNA levels in control and two MPP8 stable knockdown MDA-MB-231 cells were assayed with RT–PCR. GAPDH serves as controls. (B) MPP8 represses transcription from E-cadherin promoter. As indicated below each column, mock, and MPP8 knockdown 293T cells were transfected with E-cad-Luc (−420∼+23) luciferase reporter with or without MPP8-wt or W80A rescue construct. The luciferase activity of reporter alone was normalized as 1. Results are the mean of three independent experiments with s.d. (error bars). MPP8 expression was analysed by western blot. (C) ChIP analysis using H3K9me3- and H3-specific antibodies. qPCR was conducted using primers specific for exogenous E-cad-Luc promoter and chromatin derived from control, MPP8 knockdown and rescue 293T cells. Graphs show the mean of ChIP enrichment values (n=3) with s.d. (error bars). (D) RT–qPCR (top) and western blot (bottom) analysis of control, MPP8 knockdown and rescue MDA-MB-231 cells. Antibodies are indicated, and qPCR results were derived from three independent samples (±s.d.) and normalized to GAPDH. (E) ChIP–qPCR analysis using anti-MPP8 antibody and primers specific for E-cadherin (E-cad) promoter or Fibronectin (FN) promoter using chromatin derived from control, MPP8 knockdown and rescue MDA-MB-231 cells. Graphs show the mean of ChIP enrichment values (n=3) with s.d. (error bars). (F) Migration and invasion assays of control and MPP8 rescue MDA-MB-231 cells. Columns represent the mean of triplicate assays with s.d. (error bars). Control cells were normalized as 100%. In all panels, ‘*' represents P-values <0.01 and ‘**' represents P-values <0.05.
To further assess the importance of MPP8 methyl-H3K9 binding in endogenous E-cadherin gene silencing, we stably rescue expressed Flag-MPP8-wt or W80A mutant in MPP8-KD2 MDA-MB-231 cells. As indicated in Figure 5D, E-cadherin expression increased significantly in MPP8 knockdown cells. However, this elevated E-cadherin expression is severely repressed at both mRNA and protein levels by rescue expressed MPP8-wt in MPP8-KD2 cells, whereas MPP8-W80A only showed a mild effect (lanes 1, 3–5), suggesting that methyl-H3K9 binding of MPP8 is crucial for E-cadherin repression. Therefore, we examined MPP8 localization on the E-cadherin promoter by ChIP assays in different MDA-MB-231 cells which we generated. As shown in Figure 5E, MPP8 targets the E-cadherin promoter endogenously but not the −1 kb upstream region. In MPP8-KD2 cells, MPP8 presence on E-cadherin promoter is severely impaired; however, this localization can be greatly restored by rescue expression of MPP8-wt but not W80A mutant (Figure 5E). These results together demonstrate that MPP8 directly targets E-cadherin promoter for gene silencing and methyl-H3K9 binding of MPP8 is critical for E-cadherin promoter targeting as well as transcriptional repression. In addition, the observations that MPP8-W80A mutant has a mild repressive effect on exogenous and endogenous E-cadherin expression indicate that MPP8 may function through other mechanisms.
Having established the role of MPP8 in E-cadherin repression, we next examined the role of MPP8 methyl-H3K9 binding in modulating cell behaviour. Consistent with their E-cadherin level (Figure 5D), MPP8-wt rescue cells displayed the similar migratory and invasive ability compared with control MDA-MD-231 cells in the Boyden chamber assays, whereas MPP8-W80A rescue cells still showed reduced motility and invasiveness (Figure 5F). In addition, immunostaining results indicate that re-expressed E-cadherin in MPP8 knockdown MDA-MB-231 cells and H23 cells localizes at the cell membrane and enables the cells to establish cell–cell contacts (Supplementary Figures S4E and S6). These results together suggest that MPP8-mediated E-cadherin repression through methyl-H3K9 binding results in functional changes in cell behaviour. Although MPP8 knockdown or rescue expression also affects Fibronectin expression (Figure 5D), MPP8 does not target Fibronectin promoter specifically (Figure 5E), indicating it could be an indirect effect caused by altered E-cadherin expression (Onder et al, 2008). Furthermore, knocking-down MPP8 in invasive lung cancer H23 cells also results in a severe reduction of migration and invasion in the Boyden chamber assays (Supplementary Figure S4C and D) in addition to the de-repression of E-cadherin (Supplementary Figure S4A). Collectively, we conclude that MPP8 represses E-cadherin through methyl-H3K9 binding and has pivotal functions in promoting tumour cell motility, invasiveness and EMT.
MPP8 interacts with H3K9 HMTases and DNMT3A
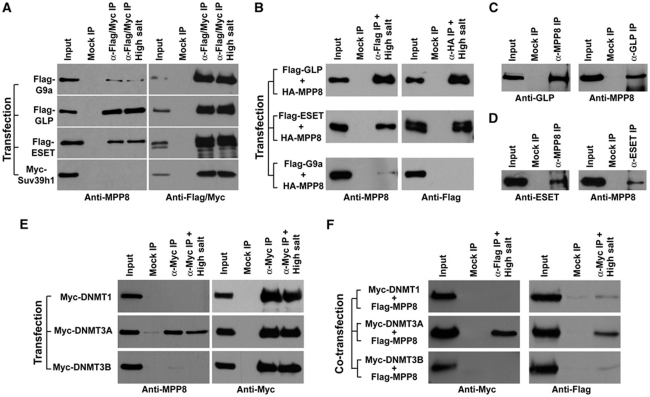
It has been demonstrated that H3K9 HMTases G9a and GLP cooperate with HDAC1/2 and LSD1 in CtBP-1 complex for E-cadherin repression (Shi et al, 2003). We thus asked whether MPP8 interacts with H3K9 HMTase for E-cadherin silencing. To this end, we expressed Flag-tagged H3K9 HMTases G9a, GLP, ESET and Myc-SUV39H1 in 293T cells. Western blot analysis after co-IP experiments indicates that GLP and ESET, but not SUV39H1, interact with endogenous MPP8 and the MPP8–GLP and MPP8–ESET interactions remained stable under high salt washing conditions (Figure 6A). We also observed a weak interaction between Flag-G9a and MPP8, which could be attributed to the strong interactions between G9a and GLP (Tachibana et al, 2005). In addition, we co-expressed HA-MPP8 with various Flag-tagged H3K9 HMTases in 293T cells for IP-western analysis. As indicated in Figure 6B, HA-MPP8 specifically captures the Flag-GLP or ESET whereas Flag-GLP or ESET co-isolates the HA-MPP8 from cell extracts when both were expressed. We also carried out endogenous IP using antibodies against GLP, ESET or MPP8. Western blot analysis demonstrates that these endogenous proteins formed specific complexes as well (Figure 6C and D).
Figure 6.
MPP8 interacts with HMTase GLP, ESET and DNMT3A. (A) 293T cells were transfected with each of H3K9 HMTase expression vectors. Antibodies used for IP and following western blot are indicated. High salt represents washing buffer containing 600 mM KCl compared with normal (300 mM KCl). (B) Similar IP-western analysis after co-transfecting with expression vectors for different H3K9 HMTases together with HA-MPP8. Antibodies used for IPs and western blot are indicated. (C, D) MPP8 interacts with GLP and ESET endogenously. Cell extracts derived from 293T cells were incubated with anti-MPP8 and anti-GLP antibodies (C) or anti-MPP8 and anti-ESET antibodies (D) for IP. Endogenous protein complexes were analysed by western blot with antibodies indicated below each panel. (E) 293T cells were transfected with each of Myc-DNMT1, DNMT3A and DNMT3B expression vector and the similar IP-western analysis were carried out using indicated antibodies. High salt represents washing buffer containing 300 mM KCl compared with normal (150 mM KCl). (F) Co-transfections were performed with each of Myc-tagged DNMT vectors and Flag-MPP8 vector. IPs were carried out with under high salt condition and analysed by western blot using indicated antibodies. In all IP-western experiments, normal mouse or rabbit IgG was used for mock IPs and ‘Input' represents 5% of total cell extract.
As another major repressive mark, DNA methylation has been associated with E-cadherin gene silencing in various human tumours (Yoshiura et al, 1995). We thus examined whether MPP8 also interacts with DNMTs using similar IP-western approach. As indicated in Figure 6E, exogenously expressed Myc-DNMT3A but not DNMT1 or DNMT3B interacts with endogenous MPP8 and this protein–protein interaction remains stable under more stringent washing conditions (Figure 6E). We also co-expressed Flag-MPP8 together with different Myc-tagged DNMTs in 293T cells. Western blot analysis after co-IP experiments further confirmed this specific MPP8–DNMT3A interaction (Figure 6F). Together, we conclude that MPP8 specifically interacts with two euchromatic H3K9 HMTases GLP and ESET, as well as de novo DNA methyltransferase 3A.
MPP8 recruits DNMT3A to E-cadherin promoter and directs DNA methylation
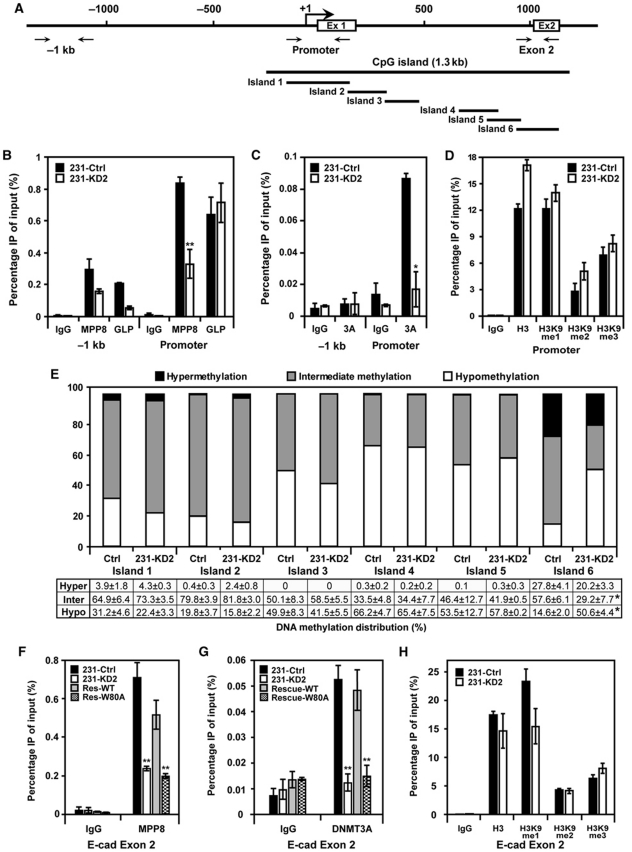
The observation that MPP8 associates with H3K9 HMTases and DNMT3A prompted us to further investigate whether MPP8 functions together with these enzymes for E-cadherin repression. ChIP analysis indicates that MPP8, GLP and DNMT3A all localize on E-cadherin promoter in MDA-MB-231 cells but not in the −1 kb upstream region. Interestingly, MPP8 knockdown severely reduces the E-cadherin promoter occupancy of DNMT3A in addition to MPP8. GLP localization in the same region is not affected (Figure 7B and C). These results suggest that the E-cadherin promoter binding by MPP8 is critical for DNMT3A targeting but not for GLP. Additional ChIP analysis reveals that all three H3K9 methylation forms exist on E-cadherin promoter and this methylation pattern remains similar after MPP8 knockdown (Figure 7D; Supplementary Figure S7A). In addition, we did not observe any significant changes of several other histone modifications on E-cadherin promoter in MPP8-KD2 cells, including H3K4me2, K36me2, H3ac and H4K20me3 (Supplementary Figure S7C–G). These results suggest that the de-repression of E-cadherin in MPP8 knockdown cells is mainly caused by the loss of MPP8 protein and its promoter localization but not the altered histone methylation and acetylation states.
Figure 7.
MPP8 recruits DNMT3A to direct DNA methylation. (A) Diagram of the 5′ region of E-cadherin gene with exon 1, exon 2 and transcription start site indicated. Solid arrows indicate primer sites used for ChIP assays. Bold line indicates the entire CpG island and islands 1–6 represent qPCR products to quantify CpG methylation using MethylScreen approach. (B–D) DSS/Formaldehyde-fixed chromatin was isolated from control and MPP8-KD2 MDA-MB-231 cells and ChIP–qPCR analysis was conducted using GLP, MPP8 (B), DNMT3A (C) and H3K9, me1, me2 and me3 (D) specific antibodies and primers specific for the promoter and −1 kb region of E-cadherin. Graphs show the mean ChIP enrichment values (n=3) with s.d. (error bars). (E) MethylScreen analysis of six small CpG islands obtained from genomic DNA derived from control and MPP8-KD2 MDA-MB-231 cells. White boxes reflect the unmethylated molecular population, whereas black represents the proportion uniformly methylated. The grey boxes represent the portion of molecules that were partially but not completely methylated. Quantitative results represent mean of triplicate assays (±s.d.). (F–H) ChIP–qPCR analysis using MPP8 (F), DNMT3A (G) and H3K9, me1, me2 and me3 (H) specific antibodies and chromatin derived from control, MPP8 knockdown and rescue MDA-MB-231 cells. qPCR was conducted using primers specific for E-cadherin exon2 (island 6). Graphs show the mean ChIP enrichment values (n=3) with s.d. (error bars). In all panels, ‘*' represents P-values <0.01 and ‘**' represents P-values <0.05.
Our ChIP data suggest a possibility that MPP8 directs DNA methylation through DNMT3A recruitment, we thus analysed the DNA methylation patterns in the 5′-regulatory region of E-cadherin gene in control and MPP8-KD2 MDA-MB-231 cells using the recently developed MethylScreen approach (Holemon et al, 2007). The entire 1.3 kb CpG island was first divided into six small CpG islands based on methylation-dependent restriction enzymes (MDRE) and methylation-sensitive restriction enzymes (MSRE) recognition sites (Figure 7A). Genomic DNA from control and MPP8-KD2 cells were digested by different sets of enzymes and the DNA methylation status was determined by qPCR and calculated as previously described (Holemon et al, 2007). As shown in Figure 7E, most of CpG sites in E-cadherin promoter, exon 1 and intron 1 regions (islands 1–5) are either intermediately methylated or hypomethylated in MDA-MB-231 cells, which is consistent with the previous report (Reinhold et al, 2007). MPP8 knockdown does not result in any obvious changes of DNA methylation patterns in these regions. On the contrary, most of CpG sites in the exon 2 region (island 6) are either hypermethylated (28%) or intermediately methylated (58%). Inhibition of MPP8 expression leads to about 3.5-fold increase of DNA hypomethylation (15–51%) and a significant decrease of intermediate DNA methylation (58–29%) in this region. DNA hypermethylation levels also decreased slightly in MPP8-KD2 cells. Furthermore, ChIP analysis revealed a decreased MPP8 and DNMT3A occupancy on exon 2 in MPP8-KD2 cells (Figure 7F and G). We thus conclude that MPP8 recruits DNMT3A to direct CpG methylation in this region. Interestingly, rescue expression of MPP8-wt, but not MPP8-W80A in MPP8-KD2 cells restored the localization of both MPP8 and DNMT3A on exon 2, suggesting that methyl-H3K9 binding by MPP8 is critical for the DNMT3A recruitment in this region (Figure 7F and G). Similar to the promoter region, H3K9 methylation states in the exon 2 region is not affected by MPP8 levels (Figure 7H; Supplementary Figure S7B). These results together suggest a possibility that MPP8-directed DNA methylation is crucial for E-cadherin repression.
MPP8-directed DNA methylation is critical for E-cadherin repression
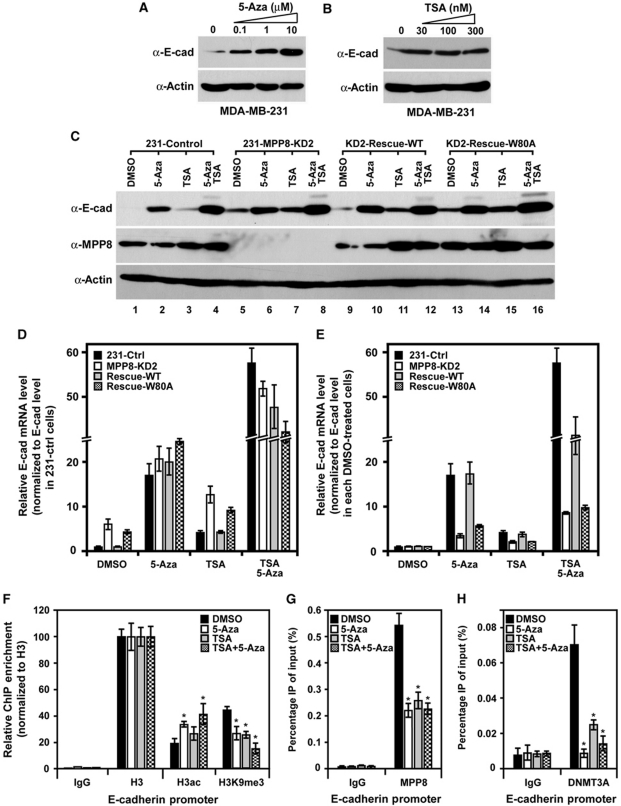
Given that E-cadherin expression can be repressed by multiple epigenetic mechanisms, we next assessed the importance of MPP8-directed DNA methylation in E-cadherin repression using a pharmacologic approach. Consistent with previous reports (Pruitt et al, 2006; Liu et al, 2008), HDAC inhibitor TSA or DNA methylation inhibitor 5-Aza treatment could cause the re-expression of E-cadherin in MDA-MB-231 cells (Figure 8A and B). However, 5-Aza showed a more significant effect on E-cadherin re-expression compared with TSA, and the double treatment has an additive effect (Figure 8C). qRT–PCR analysis further reveals that E-cadherin mRNA increased 4- or 17-fold with TSA or 5-Aza treatment, respectively, whereas double treatment displayed a 57-fold increase (Figure 8D). These data indicate that E-cadherin repression is mainly mediated by DNA methylation in MDA-MB-231 cells and histone de-acetylation serves as another independent mechanism. In MPP8-KD2 cells, E-cadherin expression increased ∼7-fold compared with control cells (Figure 8D, DMSO). Although MPP8 knockdown apparently increased TSA-induced E-cadherin re-expression, 5-Aza or double treatment still displayed a similar E-cadherin re-expression compared with control cells (Figure 8C and D), indicating that MPP8-mediated E-cadherin repression is through DNA methylation but not histone deacetylation. Furthermore, when normalized to control treatment, 5-Aza, TSA and double treatment only displayed three-, two- and eight-fold additive increases on E-cadherin re-expression in MPP8-KD2 MDA-MB-231 cells (Figure 8E, white bars). These results together suggest that MPP8-directed DNA methylation is the major mechanism for E-cadherin repression. Additionally, MPP8-wt and W80A rescue cells displayed similar E-cadherin de-repression patterns compared with control and MPP8-KD2 cells, respectively, when treated with different inhibitors (Figure 8C–E). These data are consistent with our previous results and suggest that MPP8-directed DNMT3A recruitment and DNA methylation in E-cadherin 5′-regulatory region require methyl-H3K9 binding of MPP8.
Figure 8.
MPP8 represses E-cadherin expression through DNA methylation. (A, B) Western blot analysis of MDA-MB-231 cells treated with different amount of 5-Aza (A) for 96 h or TSA (B) for 24 h. (C) Western blot analysis of control, MPP8 knockdown and rescue MDA-MB-231 cells treated with 5-Aza (10 μM, 96 h), TSA (100 nM, 24 h) or both inhibitors. (D) Real-time RT–qPCR analysis of E-cadherin mRNA level in control, MPP8 knockdown and rescue MDA-MB-231 cells treated with 5-Aza, TSA or both inhibitors. E-cadherin expression was normalized to GAPDH expression and E-cadherin mRNA level in control MDA-MB-231 cells with DMSO treatment was normalized as 1. Graphs show the mean of relative E-cadherin mRNA level (n=3) with s.d. (error bars). (E) The same RT–qPCR results as (D), but E-cadherin expression in each DMSO-treated cells was normalized as 1 to assess the effect of different inhibitors in each assayed cells. (F–H) ChIP–qPCR analysis using H3ac, H3K9me3 (F), MPP8 (G) and DNMT3A (H) specific antibodies and chromatin derived from MDA-MB-231 cells treated with DMSO, 5-Aza, TSA or both inhibitors. qPCR was conducted using primers specific for E-cadherin promoter. Graphs show the mean ChIP enrichment values (n=3) with s.d. (error bars). In all panels, ‘*' represents P-values <0.01.
Although TSA and 5-Aza have been widely used to re-activate epigenetically silenced genes, these inhibitors have both direct and indirect effects on transcriptional regulation. For example, 5-Aza treatment could decrease H3K9me2 and me3 levels in the regulatory region of different genes in various cancer cells (Lakshmikuttyamma et al, 2010). Similarly, ChIP analysis reveals that TSA or 5-Aza treatment increases H3 acetylation level moderately on E-cadherin promoter and this increased H3 acetylation is attended by a moderate reduction of H3K9me3 in MDA-MB-231 cells (Figure 8F). In addition, we observed a significant reduction of MPP8 localization on E-cadherin promoter (Figure 8G) after cells were treated with different inhibitors. Furthermore, decreased MPP8 enrichment is concomitant with a severe reduction of DNMT3A localization in the same region (Figure 8H). And the similar localization patterns were also observed in E-cadherin exon 2 region (Supplementary Figure S8). Although the molecular details of how 5-Aza decreases H3K9 methylation remain unclear, these results further demonstrate the importance of MPP8 methyl-H3K9 binding in DNA methylation and E-cadherin repression.
Discussion
MPP8 recognizes methyl-H3K9 through chromodomain
In mammals, chromodomain is conserved in a wide range of chromatin regulatory proteins and some of them have been characterized as methyl-histone-binding proteins including HP1, PC and CDY. Here, we demonstrated MPP8 as another methyl-H3K9-binding protein. Although MPP8 contains two methyl-histone-binding motifs—chromodomain and ankyrin- repeats, ankyrin-repeats of MPP8 do not have any conserved residues that exist in G9a and GLP ankyrin-repeats for methyl-H3K9 binding (Collins et al, 2008). On the contrary, the chromodomain of MPP8 is similar to HP1 (Figure 2A) and contains critical residues for methyl-H3K9 recognition (Jacobs and Khorasanizadeh, 2002; Nielsen et al, 2002). These observations are consistent with our results, which indicate that MPP8 binds to methyl-H3K9 only through its chromodomain. Furthermore, MPP8 does not have the conserved residues that exist in PC2 proteins for chromodomain dimerization, which is a key determinant for the methyl-H3K27 recognition (Min et al, 2003). Consistent with our results, two recent studies showed that MPP8 chromodomain alone is capable of binding to methylated H3K9 peptides on the human epigenome peptide array (Bua et al, 2009) or in the AlphaScreen assays (Quinn et al, 2010). Although MPP8 chromodomain has been predicted and later demonstrated binding to methyl-H3K27 peptides in vitro (Fischle et al, 2008; Bua et al, 2009), our results do not support this when chromodomain is in the context of FL MPP8 protein. Interestingly, we also noticed that the binding of MPP8 with methyl-H3K9 peptides is stronger and more stable compared with hHP1α in our in vitro assays (Figure 2B and unpublished results).
MPP8, EMT and tumour metastasis
The fact that MPP8 expression is elevated in various tumour cells indicates MPP8 has a role in cancer. Our results revealed that MPP8 expression level not only affects tumour cell proliferation but also regulates cell mobility and invasiveness (Figures 3B and 4C–E). The observation that MPP8 directly regulates E-cadherin gene expression (Figure 5) together with the fact that E-cadherin is a central modulator governing EMT and metastatic dissemination (Onder et al, 2008) suggest that MPP8 could have critical functions to promote tumour progression and metastasis. Given that metastasis is the most common cause of death in many cancer patients and is a major obstacle to successful anti-cancer treatment, our studies provide a possibility that blocking MPP8-H3K9me and/or MPP8–DNMT3A interactions could have potential therapeutic value and the levels of MPP8 could serve as a metastatic diagnostic marker in the future.
In addition, HP1 proteins have been shown to modulate invasive potential of breast cancer cells as well (Norwood et al, 2006). Different from MPP8, HP1 expression is downregulated in invasive metastatic breast cancer cells and overexpression of HP1 in MDA-MB-231 cells led to a reduction of cell invasiveness. Mutagenesis studies revealed that the dimerization of HP1 proteins is critical for these functions, although HP1 proteins have been co-purified with several repressor protein complexes (Ogawa et al, 2002; Shi et al, 2003). Therefore, it will be intriguing to further dissect molecular details of how H3K9 ‘methylation code' is translated to completely opposite cellular behaviours by different methyl-H3K9-binding proteins.
MPP8, H3K9 methylation, DNA methylation and gene repression
In most carcinomas, epigenetic silencing of E-cadherin ex-pression requires multiple regulatory machineries such as transcription factors including SNAIL, ZEB1/2 and bHLH factors (Peinado et al, 2007) as well as histone modifying protein complexes including SUZ12/PRC2, Sin3A/HDAC and CtBP-1 (Shi et al, 2003; Peinado et al, 2007; Herranz et al, 2008). Here, our results revealed a novel mechanism by which the methyl-H3K9-binding protein MPP8 could couple H3K9 methylation and DNA methylation for E-cadherin repression. Our working model predicts that HMTase GLP first methylates H3K9 on E-cadherin promoter. MPP8 is then recruited through methyl-H3K9 binding and protein–protein interactions for transcriptional repression. MPP8 next recruits DNMT3A to introduce de novo CpG methylation on exon 2 and finally turns off E-cadherin gene expression. Given that MPP8 knockdown does not result in a rearrangement of ‘histone code', we speculate that MPP8-mediated E-cadherin repression is downstream of histone methylation and acetylation. However, altered H3K9 methylation could significantly regulate MPP8-mediated E-cadherin repression as methyl-H3K9 binding by MPP8 is critical for DNMT3A recruitment. Furthermore, although the possible cross-talk between MPP8 and other repressive protein machineries is under investigation, we have been unable to detect obvious interactions between MPP8 and SNAIL, TWIST or CtBP-1 (data not shown), indicating that MPP8 may function as another major player in EMT. In addition, the importance of MPP8–ESET interaction still remains unclear. As GLP and ESET target to different chromatic context (Dodge et al, 2004; Tachibana et al, 2005), one possibility is that different H3K9 HMTases can recruit MPP8 to different genes, such as amyloid P component serum, another MPP8 target gene we identified (Supplementary Figure S9).
The correlation between DNA methylation status and E-cadherin expression has been established in various epithelial tumours (Yoshiura et al, 1995; Graff et al, 2000). However, inactive chromatin at the 5′-end of E-cadherin gene is heterogeneously modified and the hypermethylation of CpG island is not a prerequisite for transcriptional repression, indicating that multiple epigenetic pathways could coordinate in this process (Koizume et al, 2002). In mammalian cells, interplays between two major repressive epigenetic marks, H3K9 methylation and DNA methylation have been extensively studied (Brenner and Fuks, 2007). For example, G9a and Glp control DNA methylation for transcriptional silencing, although neither enzymatic activities of G9a/GLP nor deposited H3K9 methylation is required in this regulation (Dong et al, 2008; Epsztejn-Litman et al, 2008; Tachibana et al, 2008). For the MPP8-directed DNA methylation we uncovered, H3K9 methylation as well as recognition of H3K9me marks by MPP8 are critical for the recruitment of DNMT3A, suggesting that MPP8 could bridge H3K9 methylation and DNA methylation for transcriptional repression. However, the observations that MPP8 knockdown only affects CpG methylation in E-cadherin exon 2 region and 5-Aza treatment has a moderate additive effect on E-cadherin de-repression in MPP8-KD2 cells indicate the regulation of E-cadherin DNA methylation is more complicated. For example, DNMT1 has been reported to localize to the E-cadherin promoter although this region lacks CpG methylation (Herranz et al, 2008). Alternatively, MPP8 could also regulate transcription through ESET and DNMT3A using different molecular mechanisms as ESET not only interacts with DNMT3A/3B and targets several tumour suppressor gene promoters (Li et al, 2006), but also directs DNA methylation on retrovirus-like elements in ES cells (Matsui et al, 2010).
Materials and methods
Cell culture and antibodies
All cells were cultured in DMEM supplemented with 10% FBS and antibiotics (Invitrogen). MPP8 stable knockdown and rescue cells were maintained in medium containing 1–2 μg/ml puromycin. MPP8 antibody was generated in rabbit using purified recombinant N-terminal protein (1–188 aa) as antigen and affinity purified from serum. Other antibodies used for the study are described in the Supplementary data.
Protein and DNA microarrays
Chromodomain microarray preparation and hybridization were performed as described previously (Kim et al, 2006). cDNA microarray analysis was carried out using GeneChip Human Genome U133 Plus 2.0 array (Affymetrix) according to the manufacturer's protocols. Data sets were analysed using SAM software (3.05) with R package (2.8.1) for individual genes and Gene Set Enrichment Analysis. For individual genes, tests of statistical significance between wt and MPP8 knockdown samples were conducted as two-classes unpaired samples using Wilcoxon tests. Permutation number was set to no less than 1000 and FDR was strictly control under 5%. Generated gene lists were then analysed using GeneGo MetaCore software (version 5.3, build 18499) for data mining and pathway analysis.
MPP8 knockdown, rescue, and cell growth, migration and invasion assays
For knockdown, two desired oligonucleotides targeting to human MPP8 (NM_017520, 445–463 for KD1 and 1095–1113 for KD2) were ligated into pTYF-H1-PGK-puro vector and the lentivirus was generated with three helper vectors using standard protocols. Stable cell lines were generated by virus infection followed by puromycin selection (2 μg/ml). For rescue, retroviral vector expressing shRNA resistant Flag-MPP8-wt or W80A (1102-AACCAG-1107 of NM_017520) was transfected into Linx cells for virus production. MPP8-KD2 cells were then infected with the retroviruses and selected by neomycin (1.5 mg/ml) and puromycin (2 μg/ml). For cell growth analysis, 3.6 × 103 cells were seeded in 96-well plates. Relative cell numbers were measured every 24 h by using Non-Radioactive cell proliferation assay kit (Promega). All experiments were performed in sextuplicates. For cell mobility, a scratch wound was generated using a 200 μl pipette tip on confluent cell monolayers in six-well plate in normal culture medium with 10% FBS. Cells were then washed with fresh medium to remove floating cells and microphotographs were taken at different time point. The trans-well migration and invasion assays were carried out using 24-well Cell Migration and Invasion Assay kit (Cell Biolabs). Cells were incubated in trans-wells 12 h for migration and 24 h for invasion and all experiments were done in triplicates.
In vitro and in vivo binding assays
In vitro translation coupled peptide pull-down assays were performed as previously described (Cao et al, 2002) with modifications (Supplementary data). For the in vivo pull-down experiments, MPP8-KD2 293T cells were transfected with vector expressing shRNA resistant Flag-MPP8-wt or W80A. Cells were lysated with 600 μl of IPH buffer (50 mM Tris–HCl, 150 mM NaCl, 0.5% NP40, pH 8.0) with brief sonication and then incubated with 0.6 μl of 1 M CaCl2 and 120 units micrococcal nuclease (Worthington) at room temperature for 30 min with gentle mixing. Digestion was stopped by adding 6 μl of 0.5 M EDTA, and the cell lysates were cleared and applied for immunoprecipitation using anti-FLAG-Agarose (Sigma). The beads were washed four times with IPH buffer containing 5 mM EDTA and 750 mM NaCl and beads-bound proteins were resolved by SDS–PAGE. Western blotting was carried out with various ChIP-grade antibodies from Abcam.
ChIP and DNA methylation assay
For ChIP assays, formaldehyde or DSS-formaldehyde fixed chromatin was sheared by MNase digestion followed by sonication and incubated with various antibodies as previously described (Cao and Zhang, 2004) with modifications (Supplementary data). MethylScreen DNA methylation assays were carried out as described previously (Holemon et al, 2007). Briefly, genomic DNA was first prepared using Genomic DNA kit (Zymo research). For CpG islands 1, 2, 3 and 6, genomic DNA was digested by MDRE, MSRE and two sets of enzymes together using Methyl-Profiler DNA Methylation Enzyme kit (SA Biosciences). For CpG islands 4 and 5, genomic DNA was digested by McrBC (NEB) as MDRE, a mixture of HhaI, HpyCH4IV and AciI (NEB) as MSRE and a two sets of enzymes together (double digestion). All digestions were carried out for 12 h at 37°C and qPCR was performed in triplicates using SYBR Green SuperMix (Quanta Biosciences). To improve amplification efficiency, DMSO, Betain (Sigma) and 7-deaza dGTP (Roche) were added to master mix (Musso et al, 2006). Standard curves were generated using serial diluted genomic DNA for same PCR reactions. Results from qPCR were calculated according to the manufacturer's instruction (SA Biosciences). Primer pairs used for qPCR are described in the Supplementary data.
Statistical analysis
Statistical analyses (t-test) were carried out by using Microsoft Excel. Two-tailed distribution and homoscedastic parameters were used.
Supplementary Material
Acknowledgments
We thank Thomas Jenuwein and Yi Zhang for the Suv39h1/h2 null cells and other reagents. David Allis for histone H3K9 and K27 peptides. Ken Wright, Gerd Pfeifer, G Chinnadurai and Yang Shi for various constructs. Alexandra Espejo for assistance with the protein microarrays, Chunxiang Liu for assistance with DNA methylation assays. Research in the laboratory of JF was supported by Florida Department of Health (08BN-01-17192) grant, ACS-IRG and Moffitt Lung Cancer SPORE program. MTB is supported by NIH (DK62248, ES07784, ES01104) grants.
Footnotes
The authors declare that they have no conflict of interest.
References
- Brenner C, Fuks F (2007) A methylation rendezvous: reader meets writers. Dev Cell 12: 843–844 [DOI] [PubMed] [Google Scholar]
- Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT, Guccione E, Gozani O (2009) Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PloS one 4: e6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science (New York, NY) 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y (2004) SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 15: 57–67 [DOI] [PubMed] [Google Scholar]
- Collins RE, Northrop JP, Horton JR, Lee DY, Zhang X, Stallcup MR, Cheng X (2008) The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol 15: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E (2004) Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 24: 2478–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schubeler D, Tachibana M, Shinkai Y, Lorincz MC (2008) DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 27: 2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, Bergman Y (2008) De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 15: 1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M (2006) The necessity of a human epigenome project. Carcinogenesis 27: 1121–1125 [DOI] [PubMed] [Google Scholar]
- Fischle W, Franz H, Jacobs SA, Allis CD, Khorasanizadeh S (2008) Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J Biol Chem 283: 19626–19635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady WM, Willis J, Guilford PJ, Dunbier AK, Toro TT, Lynch H, Wiesner G, Ferguson K, Eng C, Park JG, Kim SJ, Markowitz S (2000) Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet 26: 16–17 [DOI] [PubMed] [Google Scholar]
- Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG (2000) Methylation patterns of the E-cadherin 5′ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem 275: 2727–2732 [DOI] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, Garcia de Herreros A, Peiro S (2008) Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol 28: 4772–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holemon H, Korshunova Y, Ordway JM, Bedell JA, Citek RW, Lakey N, Leon J, Finney M, McPherson JD, Jeddeloh JA (2007) MethylScreen: DNA methylation density monitoring using quantitative PCR. BioTechniques 43: 683–693 [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S (2002) Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science (New York, NY) 295: 2080–2083 [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science (New York, NY) 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT (2006) Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep 7: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizume S, Tachibana K, Sekiya T, Hirohashi S, Shiraishi M (2002) Heterogeneity in the modification and involvement of chromatin components of the CpG island of the silenced human CDH1 gene in cancer cells. Nucleic Acids Res 30: 4770–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kwon SH, Workman JL (2008) The heterochromatin protein 1 (HP1) family: put away a bias toward HP1. Mol Cells 26: 217–227 [PubMed] [Google Scholar]
- Lakshmikuttyamma A, Scott SA, DeCoteau JF, Geyer CR (2010) Reexpression of epigenetically silenced AML tumor suppressor genes by SUV39H1 inhibition. Oncogene 29: 576–588 [DOI] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD, Pfeifer GP (2006) The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem 281: 19489–19500 [DOI] [PubMed] [Google Scholar]
- Liu YN, Liu Y, Lee HJ, Hsu YH, Chen JH (2008) Activated androgen receptor downregulates E-cadherin gene expression and promotes tumor metastasis. Mol Cell Biol 28: 7096–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ (2009) Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Matsumoto-Taniura N, Pirollet F, Monroe R, Gerace L, Westendorf JM (1996) Identification of novel M phase phosphoproteins by expression cloning. Mol Biol Cell 7: 1455–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM (2003) Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso M, Bocciardi R, Parodi S, Ravazzolo R, Ceccherini I (2006) Betaine, dimethyl sulfoxide, and 7-deaza-dGTP, a powerful mixture for amplification of GC-rich DNA sequences. J Mol Diagn 8: 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED (2002) Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416: 103–107 [DOI] [PubMed] [Google Scholar]
- Norwood LE, Moss TJ, Margaryan NV, Cook SL, Wright L, Seftor EA, Hendrix MJ, Kirschmann DA, Wallrath LL (2006) A requirement for dimerization of HP1Hsalpha in suppression of breast cancer invasion. J Biol Chem 281: 18668–18676 [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science (New York, NY) 296: 1132–1136 [DOI] [PubMed] [Google Scholar]
- Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA (2008) Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res 68: 3645–3654 [DOI] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A (2004) Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 24: 306–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7: 415–428 [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T (2001) Loss of the Suv39 h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107: 323–337 [DOI] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB (2006) Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet 2: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn AM, Bedford MT, Espejo A, Spannhoff A, Austin CP, Oppermann U, Simeonov A (2010) homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics. Nucleic Acids Res 38: e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold WC, Reimers MA, Maunakea AK, Kim S, Lababidi S, Scherf U, Shankavaram UT, Ziegler MS, Stewart C, Kouros-Mehr H, Cui H, Dolginow D, Scudiero DA, Pommier YG, Munroe DJ, Feinberg AP, Weinstein JN (2007) Detailed DNA methylation profiles of the E-cadherin promoter in the NCI-60 cancer cells. Mol Cancer Ther 6: 391–403 [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12: 1591–1598 [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422: 735–738 [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM (2007) The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128: 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger A, Eger A, Wolf J, Beug H, Foisner R (2001) E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J Cell Biol 154: 1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y (2008) G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J 27: 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y (2005) Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 19: 815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP (2002) Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2: 442–454 [DOI] [PubMed] [Google Scholar]
- Ting AH, McGarvey KM, Baylin SB (2006) The cancer epigenome--components and functional correlates. Genes Dev 20: 3215–3231 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA (2005) Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell 19: 381–391 [DOI] [PubMed] [Google Scholar]
- Wang GG, Allis CD, Chi P (2007) Chromatin remodeling and cancer, Part I: Covalent histone modifications. Trends Mol Med 13: 363–372 [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA (2008) Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14: 818–829 [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jones PA (2006) Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov 5: 37–50 [DOI] [PubMed] [Google Scholar]
- Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S (1995) Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci USA 92: 7416–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.