Abstract
The Tasmanian devil, a marsupial carnivore, is endangered because of the emergence of a transmissible cancer known as devil facial tumor disease (DFTD). This fatal cancer is clonally derived and is an allograft transmitted between devils by biting. We performed a large-scale genetic analysis of DFTD with microsatellite genotyping, a mitochondrial genome analysis, and deep sequencing of the DFTD transcriptome and microRNAs. These studies confirm that DFTD is a monophyletic clonally transmissible tumor and suggest that the disease is of Schwann cell origin. On the basis of these results, we have generated a diagnostic marker for DFTD and identify a suite of genes relevant to DFTD pathology and transmission. We provide a genomic data set for the Tasmanian devil that is applicable to cancer diagnosis, disease evolution, and conservation biology.
Devil facial tumor disease (DFTD) is a transmissible cancer affecting the Tasmanian devil (Sarcophilus harrisii), an endemic Tasmanian marsupial carnivore. First observed in 1996 in northeastern Tasmania, DFTD has been implicated in devil population collapse (1, 2). DFTD is a rapidly fatal disease that culminates in large tumors, primarily on the face and mouth, which frequently metastasize to internal organs (3). There are no diagnostic tests, treatments, or vaccines available for DFTD, and models predict that the disease could lead to extinction of wild Tasmanian devils within 25 to 35 years (4).
DFTD appears to be a clonal cell line, transmitted (by biting) as an allograft between devils (5, 6) and may be similar in transmission to canine transmissible venereal tumor (CTVT) and a transmissible sarcoma affecting Syrian hamsters (7–9). The prevalence and biology of such somatic cell parasites is generally unknown (10).
Studies of captive Tasmanian devils have suggested that the species is prone to developing tumors, particularly carcinomas (11, 12). However, DFTD does not resemble previously described devil cancers (3, 13), and determining its etiology is critical for developing management strategies for the disease. Cytologically, DFTD appears as a soft tissue neoplasm consisting of undifferentiated round to spindle-shaped cells with few defining ultrastructural features (3, 13). Immunohistochemistry suggests that the tumor is derived from neuroectoderm (13).
Clonal transmission of DFTD was proposed on the basis of karyotypic evidence (5) and was supported by genetic analysis of DFTD tumors at microsatellite and major histocompatibility complex loci (6). We genotyped at 14 micro-satellite loci 25 paired tumor and host samples, as well as 10 samples from DFTD-unaffected devils from 16 locations in Tasmania (14) (figs. S1 and S2 and table S1). All DFTD tumors shared a similar genotype across all loci, regardless of location, sex, or age of the animal (P = 0.18, permutation test) (figs. S1 and S2). Furthermore, the tumor genotype was distinct from that of both the hosts and unaffected devils (P < 0.001, permutation test) (figs. S1 and S2). These data were consistent with previous studies (5, 6) and support the supposition that DFTD is a single clonal cell line propagated as a tumor allograft.
To further assess the clonal origin of DFTD, we sequenced a 1180–base pair fragment of the mitochondrial locus control region (LCR) from 14 tumors, 14 hosts, and 9 DFTD-unaffected devils (table S2). We found that all devils and tumors shared a single LCR haplotype in this region, except for one single-nucleotide polymorphism at position 15,711, which supported the idea that the tumors are clonal. Furthermore, this nucleotide variant was observed only in DFTD-free devils from western Tasmania (figs. S1 and S2 and table S2), a genetically distinct population (15). The karyotypic and genetic consistency between DFTD tumors (figs. S1 and S2) (5, 6) reinforces epidemiological evidence for a recent origin of DFTD (1, 3).
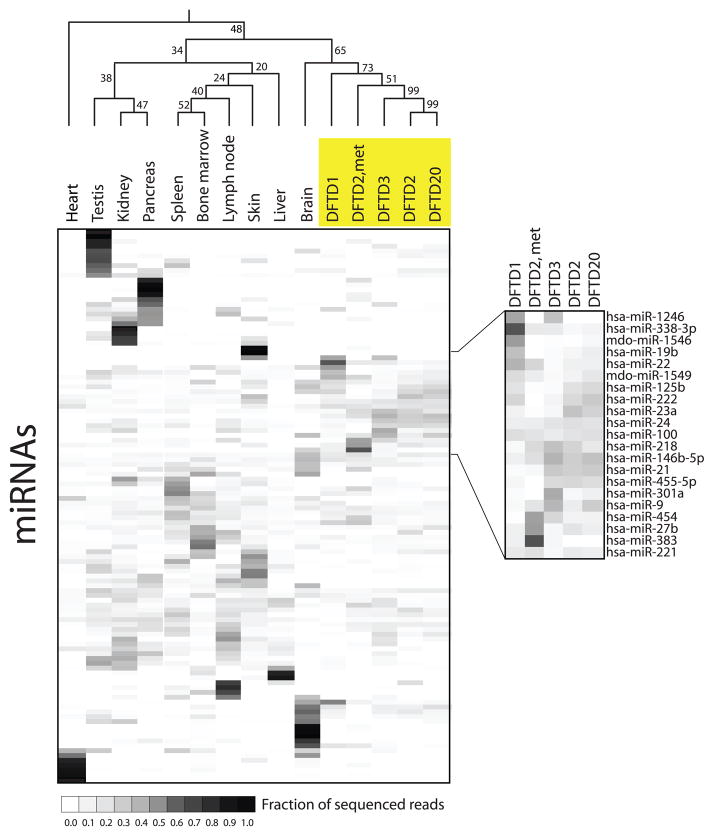
We cloned and deeply sequenced microRNA (miRNA) from 10 devil tissues and five DFTD tumors, including one DFTD mammary metastasis, to gain insight into the histogenesis of DFTD. The 114 Tasmanian devil miRNAs, identified with strict conservation parameters, showed characteristic tissue-specific expression profiles (Fig. 1 and table S3). DFTD had a relatively consistent miRNA profile that was distinct from the other 10 tissues (Fig. 1 and fig. S3). Differential expression of four miRNAs in DFTD relative to a non-DFTD tissue (testis) was confirmed by quantitative polymerase chain reaction (PCR) (fig. S4). Hierarchical clustering based on Pearson’s correlation statistic showed that the DFTD tumors were clustered (Fig. 1 and table S4) and that the non-DFTD miRNA profile most highly correlated with DFTD was that of brain (Fig. 1 and table S4).
Fig. 1.
miRNA profiling of DFTD. Heat map of normalized miRNA reads for 114 miRNAs cloned and sequenced from 10 Tasmanian devil tissues; four DFTD facial tumors (DFTD1, 2, 3, and 20); and one DFTD mammary metastasis (DFTD2, met). miRNAs were clustered on the basis of Pearson’s correlation statistic, and bootstrap values (percentage) are indicated. The DFTD miRNA profile is shown in greater detail. miRNAs were annotated on the basis of conservation with comparison species hsa, human; mdo, opossum.
In cancer, miRNAs can both promote and suppress tumors, as well as regulate processes including cell proliferation, angiogenesis, and metastasis (16). It is noteworthy that the DFTD profile included a number of miRNAs commonly up-regulated in tumors, including miR-21, miR-24, and miR-19b (16), plus a miRNA that has been linked to tumor immune evasion (miR-222) (17) (Fig. 1 and fig. S3). In contrast, DFTD expresses very low levels of miR-29b and miR-126, two miRNAs suggested to suppress tumors (Fig. 1 and fig. S3) (16).
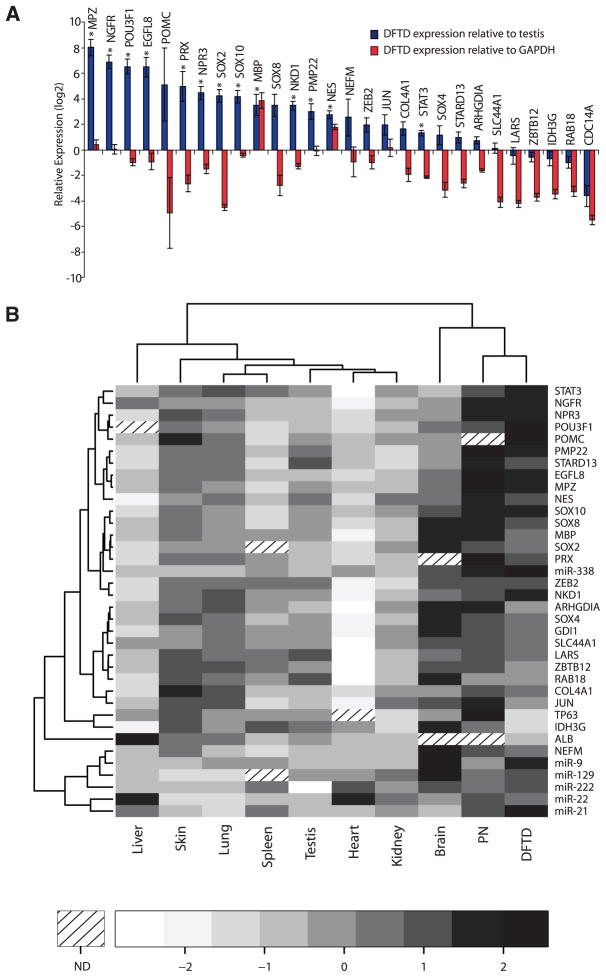
To create a catalog of genes expressed in DFTD, we sequenced the transcriptome of DFTD and testis (chosen because of its expression of a broad range of genes) from an individual Tasmanian devil resulting in 13,665 and 16,438 unique transcripts identified on the basis of similarity with human proteins and opossum transcripts, respectively (see SOM text). Of the annotated transcripts, 0.4% were differentially represented between tumor and testis libraries (fig. S5). The 31 transcripts with the most significant enrichment in the tumor library (tumor-to-testis ratio of at least 2.5, chi-squared test P ≤ 0.05) were validated by semiquantitative PCR (Fig. 2A). Of these, 20 transcripts were at least twice as highly expressed in tumor as in testis (Fig. 2A).
Fig. 2.
DFTD transcriptome. (A) Semiquantitative RT-PCR expression profiling of 31 genes with enriched expression in tumor relative to testis [454–read count fold change ≥2.5, P ≤ 0.05, chi-squared test; (green points in fig. S5)]. Log values of the mean expression difference of DFTD genes relative to testis (blue bars) and relative to GAPDH (red bars) are shown. Error bars represent standard deviation. Significant differences between tumor and testis expression levels (P ≤ 0.05) are indicated by an asterisk (two-sample t test, Holm’s correction for multiple testing). (B) Heat map of semiquantitative PCR expression profiles of 31 genes across a panel of tissues including peripheral nerve (PN), a Schwann cell–enriched tissue. Panel color represents the mean gene expression level, standardized across tissues (z score). Hierarchical clustering based on Pearson’s correlation statistic is indicated by dendrograms. For each tissue three biological replicates were performed (n = 3). ND, not determined.
The gene with the highest expression in DFTD relative to a housekeeping gene, GAPDH (encoding glyceraldehyde-3-phosphate dehydrogenase), was MBP, the gene that encodes myelin basic protein (Fig. 2A). Indeed, 9 of the 20 validated tumor genes (45%) were found to be involved in the myelination pathway (table S6). Myelin, an insulating membranous layer that ensheathes nerve axons, is produced by oligodendrocytes in the central nervous system and by Schwann cells in the peripheral nervous system. Components of a molecular network that controls the differentiation of Schwann cells, including transcription factors (SOX10, SOX2, POU5F1, and JUN) and structural myelin genes (MPZ, PRX, MBP, and PMP22), were apparent in the differentially expressed DFTD transcriptome (Fig. 2A) (18).
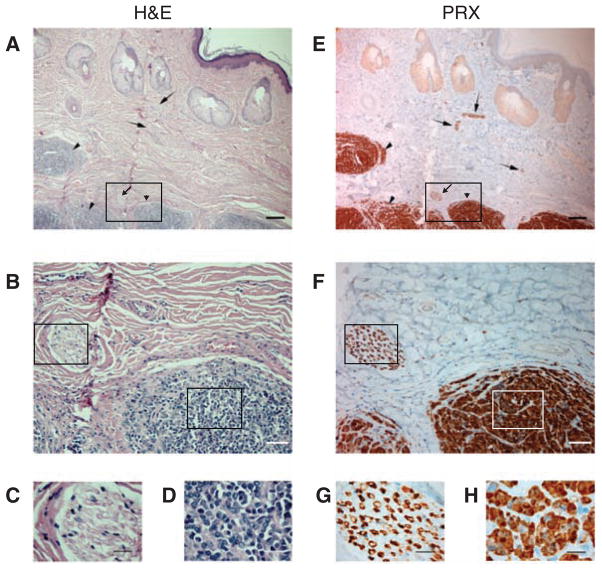
We also measured the expression of 31 tumor-enriched genes from a panel of 10 Tasmanian devil tissues by semiquantitative PCR. Hierarchical clustering of this data set grouped DFTD together with peripheral nerves, a tissue enriched for Schwann cells (Fig. 2B). To confirm the expression of myelin proteins in DFTD, we stained DFTD tumor tissues with the antibody against Schwann cell–specific myelin protein, periaxin (PRX) (Fig. 3). All tumor cells in DFTD lesions, as well as the myelinated sheaths of peripheral nerve bundles, were strongly and specifically positive for PRX (Fig. 3). We also detected protein expression in DFTD of myelin proteins MBP, MPZ, and PMP22, as well as NES, NGFR, and S100 (figs. S6 and S7, and table S7). These experiments confirm that DFTD expresses genes found in myelinating cells, including Schwann cell–specific markers (PRX, PMP22, and MPZ).
Fig. 3.
Identification of a diagnostic marker for DFTD. (A to D) Hematoxylin and eosin (H&E) and (E to H) PRX antibody stains used for DFTD tumor histology. Arrowheads, DFTD tissue; arrows, peripheral nerve bundles (containing Schwann cells). Magnification, 4× (A and E; scale bar, 200 μm); 20× (B and F; scale bar, 50 μm); and 100× (C and G) peripheral nerve bundle (scale bar, 20 μm) and (D and H) DFTD tumor (scale bar, 20 μm). Boxed areas indicate approximate locations of areas magnified in lower panels.
At present, diagnosis of DFTD is based on clinical features and histology. This can pose particular difficulties for the diagnosis of atypical DFTD such as nonfacial DFTD or DFTD metastases. We tested PRX as a potential diagnostic marker for DFTD by staining a panel of both DFTD and non-DFTD Tasmanian devil tumors with PRX. All (n = 20) of the DFTD tumors were positive for PRX, whereas none of the nine non-DFTD tumors tested were positive (table S7). In addition, all (n = 10) of the DFTD metastases collected from a variety of organs were positive for PRX (table S7). Thus, PRX is a strong and specific marker for DFTD and is suitable for diagnostic evaluation.
It is striking that DFTD, a cytologically undifferentiated tumor, expresses markers of highly differentiated Schwann cells because human Schwann cell tumors rarely express a complete set of terminally differentiated myelin genes (19). Although it is possible that Schwann cell genes have been activated in DFTD cells after carcinogenic transformation, it is more likely that the myelin program observed here reflects the DFTD cell of origin. We therefore propose that DFTD is a peripheral nerve sheath tumor that arose from a Schwann cell or Schwann cell precursor; this is supported by the miRNA profile of DFTD (Figs. 1 and 2). Schwann cells participate in nerve repair after injury and also modulate local immune reactions in the peripheral nervous system (20, 21). The plasticity and immunocompetence of Schwann cells may be significant in the evolution of DFTD as a transmissible cancer.
Interestingly, proopiomelanocortin (POMC), a gene encoding a peptide hormone precursor, was expressed in DFTD (Fig. 2). This transcript varies by more than 100-fold between tumors (fig. S8). Technical limitations, due to lack of antibody cross-reactivity with devil, prevented the detection of physiological levels of adrenocorticotropic hormone (ACTH), a labile POMC cleavage product. However, compared with healthy individuals, diseased devils had higher, but not significantly different, levels of serum cortisol (fig. S9, P = 0.06, Student’s t test). Cortisol is a steroid hormone, released by ACTH activity, that is involved in stress responses and has immunosuppressive activity (22). These studies raise the intriguing possibility that DFTD may secrete factors that could alter host physiology and/or behavior, an interesting area for further study.
Our study highlights the activities of two miRNA gene networks that may function in DFTD. PMP22, a myelin gene expressed in DFTD, is a target of miR-29, a miRNA that is down-regulated in DFTD (Figs. 1 and 2 and fig. S3) (23). The expression of transcription factor ZEB2 is negatively correlated with low expression of the miR-200 family, which controls tumor invasiveness in a negative-feedback loop (24)(Figs. 1 and 2 and fig. S3). Furthermore, this work suggests that commonly mutated Schwann cell cancer genes, such as NF1, are interesting candidates for further analysis in DFTD (25).
DFTD is believed to be of neuroendocrine origin on the basis of the expression of the vimentin, S100, neuron-specific enolase, chromogranin A, and synaptophysin markers (13). As Schwann cells and neuroendocrine cells are both derived from the neural crest and overlap in gene expression, it is possible that DFTD has elements of both tissue types.
A Schwann cell origin for DFTD contrasts with that of the canine clonally transmissible cancer, which has been proposed to be of histiocytic origin (26, 27). It will be of interest to define common and unique features of these two cancers and to determine how the histogenesis of transmissible cancers may influence their occurrence, evolution, and biology. Our catalog of genes provides a framework for this work, as well as for use in the effort to develop a DFTD preclinical test and vaccine.
Supplementary Material
Acknowledgments
We thank C. Harmsen and E. Noonan (DPIPWE) and J. Smith (Fort Wayne Children’s Zoo) for assistance with sample collection. We thank K. Claudio Campos, T. Dickson, S. Peck, V. Parameswaran, J. Harris, L. Stimmler, E. Hatchwell, R. Sachidanandam, Z. Xuan, M. Rasmussen, and the students and instructors of the 2006 and 2007 Cold Spring Harbor Laboratory deep-sequencing courses. The 454 sequencing was made possible by a grant from Roche Applied Sciences. E.P.M. was supported by a Sir Keith Murdoch Fellowship from the American Australian Association and an Overseas Postdoctoral Biomedical Fellowship from the Australian National Health and Medical Council (NHMRC). A.T.P., M.B., and A.H. were supported by grants from the NHMRC. A.S. was funded by a fellowship from the Human Frontier Science Program Organization. G.J.H. was supported by grants from the National Institutes of Health. H.S.B. was funded by an Australian Research Council Linkage Grant (LP0562190). E.P.M., C.T., A.K., and G.M.W. were supported by Dr. Eric Guiler Tasmanian Devil Research Grants. Sequences generated by this study have been deposited in GEO, www.ncbi.nlm.nih.gov/geo/ accession GSE18352 (miRNA) and the Short Read Archive http://www.ncbi.nlm.nih.gov/sites/entrez?db=sra accession SRA009772 (cDNA).
Footnotes
References and Notes
- 1.Hawkins CE, et al. Biol Conserv. 2006;131:307. [Google Scholar]
- 2.Lachish S, Jones M, McCallum H. J Anim Ecol. 2007;76:926. doi: 10.1111/j.1365-2656.2007.01272.x. [DOI] [PubMed] [Google Scholar]
- 3.Loh R, et al. Vet Pathol. 2006;43:890. doi: 10.1354/vp.43-6-890. [DOI] [PubMed] [Google Scholar]
- 4.McCallum H, et al. EcoHealth. 2007;4:318. [Google Scholar]
- 5.Pearse AM, Swift K. Nature. 2006;439:549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- 6.Siddle HV, et al. Proc Natl Acad Sci USA. 2007;104:16221. doi: 10.1073/pnas.0704580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper HL, Mackay CM, Banfield WG. J Natl Cancer Inst. 1964;33:691. [PubMed] [Google Scholar]
- 8.Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA. Cell. 2006;126:477. doi: 10.1016/j.cell.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebbeck CA, Thomas R, Breen M, Leroi AM, Burt A. Evolution. 2009;63:2340. doi: 10.1111/j.1558-5646.2009.00724.x. [DOI] [PubMed] [Google Scholar]
- 10.McCallum H. Trends Ecol Evol. 2008;23:631. doi: 10.1016/j.tree.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Canfield PJ, Cunningham AA. J Zoo Wildl Med. 1993;24:158. [Google Scholar]
- 12.Griner LA. J Natl Cancer Inst. 1979;62:589. doi: 10.1093/jnci/62.3.589. [DOI] [PubMed] [Google Scholar]
- 13.Loh R, et al. Vet Pathol. 2006;43:896. doi: 10.1354/vp.43-6-896. [DOI] [PubMed] [Google Scholar]
- 14.Materials and methods are available as supporting material on Science Online.
- 15.Jones ME, Paetkau D, Geffen E, Moritz C. Mol Ecol. 2004;13:2197. doi: 10.1111/j.1365-294X.2004.02239.x. [DOI] [PubMed] [Google Scholar]
- 16.Schickel R, Boyerinas B, Park SM, Peter ME. Oncogene. 2008;27:5959. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 17.Ueda R, et al. Proc Natl Acad Sci USA. 2009;106:10746. doi: 10.1073/pnas.0811817106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svaren J, Meijer D. Glia. 2008;56:1541. doi: 10.1002/glia.20767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark HB, Minesky JJ, Agrawal D, Agrawal HC. Am J Pathol. 1985;121:96. [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlin LB. Scand J Surg. 2008;97:310. doi: 10.1177/145749690809700407. [DOI] [PubMed] [Google Scholar]
- 21.Gold R, Archelos JJ, Hartung HP. Brain Pathol. 1999;9:343. doi: 10.1111/j.1750-3639.1999.tb00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greco D, Stabenfeldt GH. In: Textbook of Veterinary Physiology. Cunningham JG, editor. Saunders; Philadelphia: 1997. [Google Scholar]
- 23.Verrier JD, et al. Glia. 2009;57:1265. doi: 10.1002/glia.20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gregory PA, Bracken CP, Bert AG, Goodall GJ. Cell Cycle. 2008;7:3112. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 25.Carroll SL, Ratner N. Glia. 2008;56:1590. doi: 10.1002/glia.20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mozos E, Méndez A, Gómez-Villamandos JC, Martín De Las Mulas J, Pérez J. Vet Pathol. 1996;33:257. doi: 10.1177/030098589603300301. [DOI] [PubMed] [Google Scholar]
- 27.Marchal T, Chabanne L, Kaplanski C, Rigal D, Magnol JP. Vet Immunol Immunopathol. 1997;57:1. doi: 10.1016/s0165-2427(96)05757-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.