Abstract
RNA folding thermodynamics are crucial for structure prediction, which requires characterization of both enthalpic and entropic contributions of tertiary motifs to conformational stability. We explore the temperature dependence of RNA folding due to the ubiquitous GAAA tetraloop–receptor docking interaction, exploiting immobilized and freely diffusing single-molecule fluorescence resonance energy transfer (smFRET) methods. The equilibrium constant for intramolecular docking is obtained as a function of temperature (T = 21–47 °C), from which a van’t Hoff analysis yields the enthalpy (ΔH°) and entropy (ΔS°) of docking. Tetraloop–receptor docking is significantly exothermic and entropically unfavorable in 1 mM MgCl2 and 100 mM NaCl, with excellent agreement between immobilized (ΔH° = −17.4 ± 1.6 kcal/mol, and ΔS° = −56.2 ± 5.4 cal mol−1 K−1) and freely diffusing (ΔH° = −17.2 ± 1.6 kcal/mol, and ΔS° = −55.9 ± 5.2 cal mol−1 K−1) species. Kinetic heterogeneity in the tetraloop–receptor construct is unaffected over the temperature range investigated, indicating a large energy barrier for interconversion between the actively docking and nondocking subpopulations. Formation of the tetraloop–receptor interaction can account for ~60% of the ΔH° and ΔS° of P4–P6 domain folding in the Tetrahymena ribozyme, suggesting that it may act as a thermodynamic clamp for the domain. Comparison of the isolated tetraloop–receptor and other tertiary folding thermodynamics supports a theme that enthalpy- versus entropy-driven folding is determined by the number of hydrogen bonding and base stacking interactions.
RNA folding is generally hierarchical, with tertiary structure occurring through interactions of preformed secondary elements (1–3). As a result, the kinetics and thermodynamics of tertiary interactions are crucial to understanding RNA folding and functionality as well for accurate structural predictions (1, 4). Toward this end, individual folding motifs must be characterized both in isolation and in combination for a unifying thermodynamic description of RNA folding to emerge. Among the most widespread long-range RNA interactions are A-minor motifs (5–7), which are commonly manifested in GNRA tetraloop–receptor binding, where N is any nucleotide and R is a purine (5, 8). The ubiquitous GAAA tetraloop–receptor interaction has been of particular interest because it properly forms outside of the ribozyme’s framework (9, 10). The GAAA tetraloop docks into an 11-nucleotide internal receptor (11), the bound and unbound structures of which have been extensively studied (10–13). There have been previous thermodynamic investigations of the tetraloop–receptor interaction in a variety of RNA folding contexts (8, 14–18). However, none of these studies has been able to isolate the enthalpy and entropy changes associated with tetraloop–receptor tertiary docking.
Enthalpies and entropies of tertiary structure formation can be obtained from the temperature dependence of equilibrium constants and/or precision calorimetry measurements. Differential scanning calorimetry has revealed enthalpy and free energy changes for pseudoknot tertiary folding (19). However, thermodynamics for tertiary structure formation can be challenging to isolate by methods such as isothermal titration calorimetry (ITC),1 since studies are performed as a titration of two species, for which resolution of secondary and tertiary contributions is achieved through clever experimental design (20–24). Other ensemble methods for dissecting thermodynamic contributions to folding, such as temperature gradient gel electrophoresis, have had restricted application because experimental conditions are constrained by the limited resolution of gel shifts (25). Time-resolved ensemble FRET methods have been elegantly employed to probe tertiary folding thermodynamics, though data interpretation often requires preconceived models of fluorophore distance distributions, which potentially mask rare subpopulations (26).
Single-molecule fluorescence resonance energy transfer (smFRET) methods have been widely applied to RNA folding (for recent reviews, see refs 27 and 28). Such methods allow isolation and direct characterization of RNA tertiary conformational dynamics with subpopulations that exhibit different rate constants and conformations readily distinguished under a wide range of experimental conditions, e.g., varying Mg2+ concentrations (16, 29, 30). Despite the potential for temperature-dependent smFRET measurements of tertiary RNA folding thermodynamics, there have been surprisingly few such studies, specifically only P1 helix docking into the prefolded core of the Tetrahymena ribozyme and four-way helical junction folding (31, 32). Additionally, both of these smFRET studies have utilized surface-immobilized constructs.
In this work, we combine a microfluidic temperature-controlled stage with time-correlated single-photon counting methods in a confocal smFRET microscope to extract thermodynamic parameters for RNA folding under both freely diffusing and surface-immobilized conditions. We determine the standard state free energy (ΔG°), enthalpy (ΔH°), and entropy (ΔS°) changes for intramolecular GAAA tetraloop–receptor docking under physiologically relevant cation conditions and identify the folding process as enthalpically driven but at a large entropic cost. We extract tertiary folding enthalpies and entropies by freely diffusing smFRET methods, which are advantageous because they require only trace amounts of sample while avoiding possible perturbative effects of surface tethering. Such freely diffusing methods have already yielded great insight into the secondary folding of RNA hairpins (33). We interpret the GAAA–tetraloop tertiary interaction in the context of previous thermodynamics studies to illuminate a possible enthalpic and entropic paradigm for RNA folding.
EXPERIMENTAL PROCEDURES
RNA Preparation
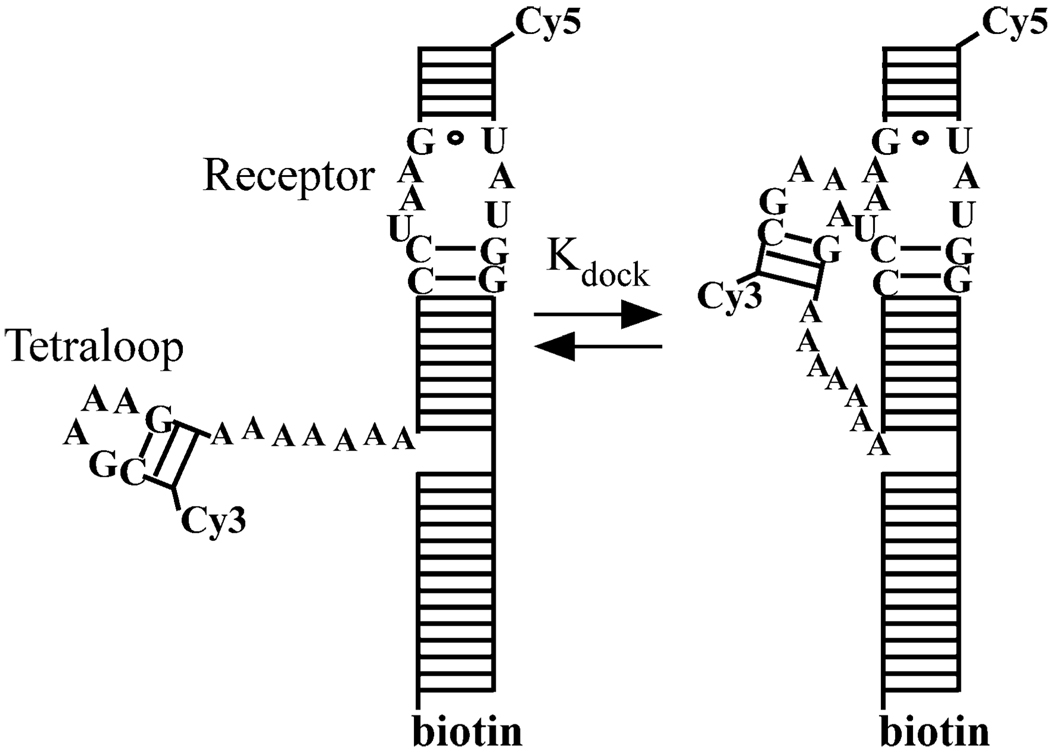
Cy3-Cy5-labeled tetraloop–receptor constructs depicted in Figure 1 were prepared as previously described (15, 16). Briefly, synthetic 5′-amino-modified RNA oligomers (Dharmacon, Lafayette, CO) are labeled with Cy3 and Cy5 N-succinimidyl esters (Amersham Biosciences, Piscataway, NJ) and HPLC-purified. The sequences of the RNA oligonucleotides are 5′ Cy5-GCC GAU AUG GAC GAC ACG CCC UCA GAC GAG UGC G 3′ and 5′ Cy3-GGC GAA AGC CAA AAA AAC GUG UCG UCC UAA GUC GGC 3′. We formed the complete construct (Figure 1) by annealing the Cy3 (1 µM) and Cy5 (1.5 µM) RNA oligomers with 2 µM biotinylated DNA oligomer (5′ biotin-CGC ACT CGT CTG AG 3′, Integrated DNA Technologies, Coralville, IA), heating the sample to 70 °C, and slowly cooling the sample to room temperature in an annealing buffer of 50 mM HEPES, 100 mM NaCl, and 100 µM EDTA (pH 7.5). The secondary structure of the Cy3 strand forms the tetraloop with an A7 linker connecting it to the receptor domain created by the hybridized Cy3 and Cy5 strands. Molecules can be tethered to streptavidin-coated glass surfaces with the biotinylated extension formed by base pairing of the DNA and Cy5 strands. The micromolar stock of annealed RNA is diluted in working buffer that differs from the annealing buffer by the addition of 1 mM MgCl2.
FIGURE 1.
Secondary structure depiction of the tetraloop–receptor RNA docking–undocking equilibrium that can be observed by Cy3–Cy5 FRET. The undocked (left) GAAA tetraloop docks (right) into the receptor via a flexible A7 linker, resulting in an increased FRET efficiency. Biotinylation of the RNA allows for immobilization on streptavidin-coated coverglass.
Single-Molecule Fluorescence Measurements
Immobilized or freely diffusing molecules are observed using a time-resolved confocal microscope system (Microtime 200, PicoQuant, GmbH) based on an inverted microscope (Olympus IX-71) equipped with a time-correlating single-photon counting module and objective scanning mode (P-721 PIFOC objective nanopositioner with x-y scanner P-733.2CL, Physik Instrumente, Karlsruhe, Germany). A 1.2 NA water-immersion objective (Olympus UPLSAPO 60XW) is used to focus a frequency-doubled pulsed picosecond (40 MHz) semiconductor laser at 532 nm (PicoTA, PicoQuant, GmbH) onto a glass surface for immobilized studies (1 µW at the microscope back plane, 1.7 kW/cm2 at the focus) and 15 µm above the glass surface for freely diffusing studies (100 µW at the back plane, 170 kW/cm2 at the focus). Fluorescence collected through the same objective is separated from the excitation source by a dichroic beamsplitter (Z532/635, Chroma Technology, Rockingham, VT) and focused through a 50 µm pinhole. Donor and acceptor emission are separated by a dichroic beamsplitter (645DCXR, Chroma Technology) and transmitted through bandpass filters (HQ585/70 M and HQ700/75M, Chroma Technology) for detection by single-photon counting avalanche photodiodes (SPCM-AQR-14, Perkin-Elmer). Data acquisition is achieved with a photon counting module (PicoHarp 300, PicoQuant, GmbH) in the time-tagged time-resolved mode, enabling the recording of every detected photon with its individual timing and detection channel information and processing with SymPhoTime (PicoQuant, GmbH).
Precise temperature control of the sample is achieved by implementation of a heated flow cell (FCS2, Bioptechs, Butler, PA) with an internal volume of 31 µL defined by a gasket spacer (0.1 mm × 14 mm × 27 mm) between the coverglass and a resistively heated, indium–tin oxide-coated slide (Bioptechs). The objective is heated to the same temperature as the flow cell to prevent a temperature gradient with respect to the immersion optic, with the objective thermally isolated from the microscope turret by a thermal spacer (Bioptechs). The reported temperatures and standard deviations are from an average over four point measurements: the top of the heated slide, on the coverglass in an immersion droplet, on the objective in an immersion droplet, and on the side of the objective. Temperatures are stable within ±0.2 °C during a given measurement, which are performed from ambient temperature to 47.4 °C.
Round No. 1.5 coverslips for the flow cell assembly are cleaned by rinsing with acetone, ethanol, HPLC-grade water, and then methanol followed by baking at 500 °C for 5 h. All experiments are performed in a buffer of 50 mM hemisodium HEPES (pH 7.5 at 25 °C), 100 mM NaCl, 100 µM ETDA, and 1 mM MgCl2. HEPES is a temperature stable buffer with only minor changes in pH with temperature (ΔpKa = −0.014 °C−1), indicating a pH reduction to 7.22 at 45 °C (34). Immobilized samples are prepared in the FCS2 holder using a biotinylated BSA–streptavidin–biotinylated RNA tethering scheme (35). An enzymatic oxygen scavenging solution of glucose (9 mg/mL), glucose oxidase (0.43 mg/mL), catalase (0.072 mg/mL), and 2 mM Trolox is added to the buffer to reduce fluorophore photobleaching and photophysics (36). For freely diffusing measurements, the glass surfaces are passivated with BSA and experiments are performed with a 200 pM RNA solution in the holder (37). The oxygen scavenging system is not used in the freely diffusing experiments as it was found to offer no advantage at the powers used (37).
Single-Molecule FRET Efficiency Analysis
Immobilized single-molecule trajectories are analyzed with 10 ms data binning, which clearly resolves the undocked and docked events (16, 37). Freely diffusing time traces are analyzed with 1 ms integration times, which is on the order of a molecule’s dwell time in the laser focus (16, 37). As previously derived, the corrected intensity-based FRET efficiency (EFRET) is calculated from the background-subtracted signals on the two channels, ΔI1 and ΔI2, designed primarily for donor and acceptor detection, respectively. Corrections are implemented for (i) collection efficiencies and cross talk of the donor and acceptor emission on channels 1 and 2 (β1A, β2A, β1D, and β2D), (ii) differential quantum yields of the donor and acceptor (QD and QA), and (iii) fractional direct excitation of the acceptor versus donor (αA, where 1 – αD = αA)
| (1) |
Quantum yield ratios and collection efficiencies are determined in independent measurements of singly labeled constructs: QA/QD = 1.2 ± 0.3, β1A = 0.00000 ± 0.00003, β2A = 0.0242 ± 0.0018, β1D = 0.0269 ± 0.0024, and β2D = 0.00211 ± 0.00018 (37). Fractional direct laser excitation of the acceptor and donor is calculated from the extinction coefficients at 532 nm (αA = 0.07 ± 0.01, and αD = 0.93 ± 0.01) (37). In contrast to the immobilized studies, donor-only species in the freely diffusing studies are necessarily included and appear at negative EFRET values due to correction for acceptor direct excitation (37).
RESULTS
Temperature Dependence of Tetraloop–Receptor Docking Equilibrium Revealed in Single-Molecule Trajectories
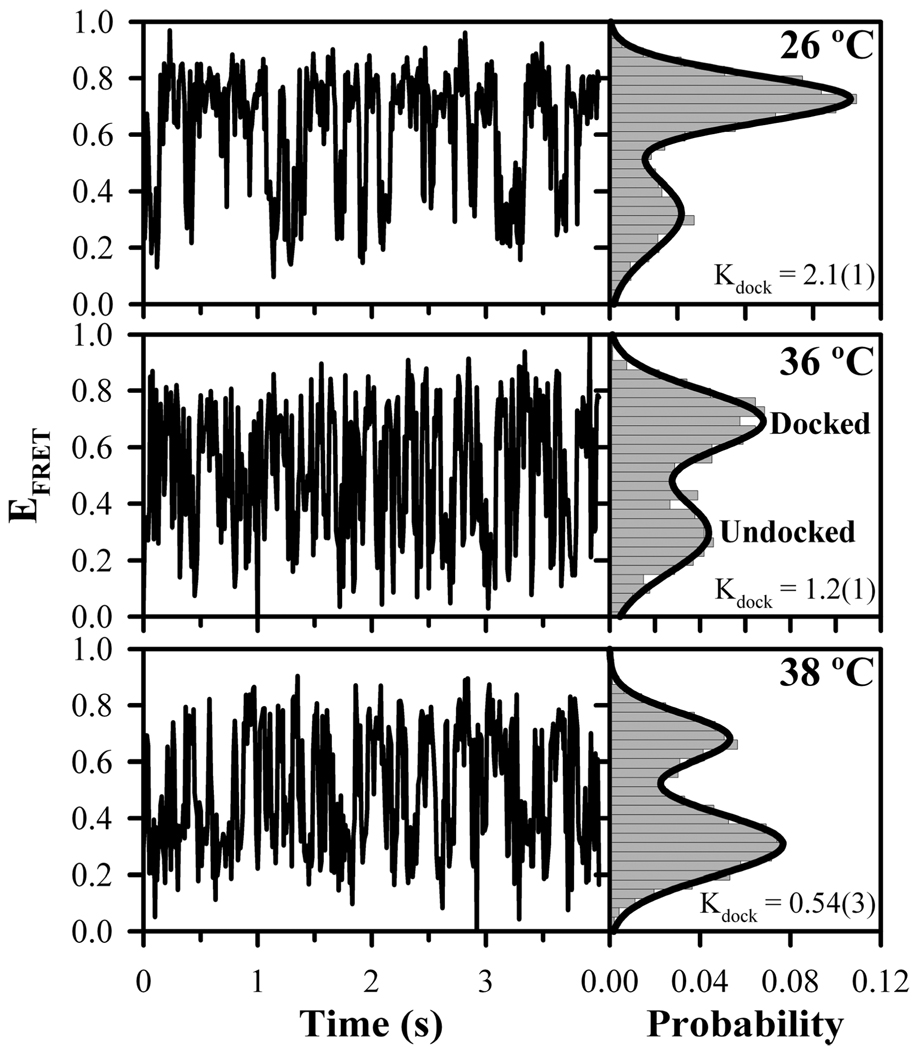
The docking–undocking equilibrium of the GAAA tetraloop and receptor is readily monitored by FRET between the donor (Cy3) and acceptor (Cy5) in the RNA construct (Figure 1), as described elsewhere (16). The tetraloop is flexibly attached to the receptor domain with a single-stranded A7 linker, which was previously shown to allow for an isolated thermodynamic study of a tertiary interaction (15, 16). Temperature-dependent conformational dynamics in single-immobilized molecules under physiological conditions [100 mM NaCl, 1 mM MgCl2, and 50 mM HEPES (pH 7.5)] are observed in EFRET trajectories calculated ratiometrically from real-time donor and acceptor fluorescence signals (see Experimental Procedures) (16, 37). Sample trajectories portraying the temperature dependence of the tetraloop–receptor docking equilibrium are shown in Figure 2. Fluctuations between undocked (low EFRET) and docked (high EFRET) conformations are visible in each of the time traces (16). Probability distributions from the EFRET trajectories allow two states to be distinguished (right panel, Figure 2).
FIGURE 2.
Immobilized single-molecule EFRET trajectories and the corresponding probability distributions at 26 (top), 36 (middle), and 38 °C (bottom). The low and high EFRET peaks correspond to the undocked and docked states, respectively. Integrated areas of the undocked and docked peaks are determined from the superimposed two-Gaussian fits with the equilibrium constant for docking, Kdock, calculated as the ratio of the docked to undocked area. Increasing temperature shifts the equilibrium to the undocked state.
The EFRET probability histograms are described well by Gaussian distributions (37); fitting the histograms to a sum of Gaussians permits quantitation of relative docked versus undocked populations. From cumulative probability distributions of many molecules, we determine 〈EFRET〉 = 0.29 ± 0.02 and 0.70 ± 0.02 and width σ = 0.14 ± 0.01 and 0.094 ± 0.003 for the undocked and docked states, respectively, in excellent agreement with previous studies of this construct (37). The EFRET centers (〈EFRET〉) and Gaussian widths (σ) for the molecules shown in Figure 2 at 26, 36, and 38 °C show no systematic shifts as the temperature is increased, indicating that (i) obscuring of docked and undocked peaks due to finite binning is negligible and (ii) the tetraloop and receptor secondary structure are maintained upon heating, since disruption of these regions would lead to shifts in the EFRET peaks. Secondary structure is expected be unaffected in this study as both the tetraloop and receptor domains are extremely stable; the melting temperature (Tm) of the tetraloop domain was experimentally determined at ~64 °C (38), and the Tm of the receptor domain is predicted to be ~62 °C from the DINAMelt server (39). Furthermore, the robustness of the EFRET peak position suggests fluorophore quantum yields are sufficiently independent of temperature over the range measured.
The effect of temperature on fluorophore quantum yields is independently assessed on singly labeled constructs at the experimental extremes. Comparison of relative quantum yields at 21 and 45 °C shows a decrease of ~20% for both Cy3 and Cy5 as the RNA is heated over this range; the quantum yield effect is reversible as observed by temperature cycling. A similar diminution of Cy3 quantum yield has also been observed by Levitus and co-workers for Cy3–DNA conjugates (40). Quantum yield changes induce a 2-fold effect on the observed EFRET, where EFRET = R06/(R06 + R6), both through the Förster radius (R0) and in the EFRET calculation from emission intensities (see eq 1) for a defined distance (R). The latter contribution is negligible in the ratiometric determination of EFRET as the relative ratio QA/QD does not change (see eq 1). The R0 effect is also subtle because R0 is proportional to QD1/6 (41). For the observed decrease of ~20% in donor quantum yield, we predict an EFRET center decrease of only ~0.03 for undocked and docked peaks over the entire temperature range investigated (see Figure S1 of the Supporting Information). Such a peak shift may be present but is on the order of EFRET center reproducibility (±0.02) and not relevant within experimental uncertainty. Furthermore, absolute positions of these peaks have no effect on the analysis presented here as we seek only the relative areas of the docked and undocked peaks, which are populated on the basis of the fractional dwell time of the molecule in each configuration.
The calculated fluorophore distance from the corrected EFRET in the docked conformation is consistent with proper formation of the tetraloop–receptor interaction as observed in the X-ray crystal structure of the P4–P6 domain of the Tetrahymena ribozyme (11). The interphosphate distance between the corresponding nucleotides to which the fluoro-phores would be attached in the P4–P6 domain is 31.6 Å. Our observed EFRET of 0.29 corresponds to an R of 46 Å in the docked state for an R0 of 53.4 Å [R6 = R06(1/EFRET – 1)]. The functionalized fluorophores are attached to aminomodified RNA with a three-carbon linker, which places 12 atoms between each fluorophore and its nucleotide phosphate. This added distance can easily account for the additional 14.4 Å between Cy3 and Cy5 as compared to the interphosphate distance in P4–P6, though the exact position of the fluorophores in this construct is not structurally known. The observed EFRET for the undocked conformation also indicates a distance (R = 62 Å) that is consistent with a right angle extension of the tetraloop A7 arm with respect to the receptor helix (R ~ 70 Å). However, inferring absolute distances from FRET efficiencies is challenging because fluorophore rotation is potentially hindered upon covalent attachment to biomolecules. Hindered rotation affects R0, which is calculated assuming free rotation (i.e., κ2 is assumed to be ⅔), though this assumption is spurious as shown in recent studies by Lilley and co-workers (42). Nevertheless, observed EFRET values are in agreement with the anticipated unfolded and folded conformations of the tetraloop–receptor construct.
The undocked and docked conformations of the tetraloop receptor are clearly assigned, allowing determination of equilibrium constants from the ratios of integrated peak areas (Kdock = area docked/area undocked), which is the fractional dwell time in the docked versus undocked state (18). An enhanced tendency for tetraloop–receptor constructs to be in the undocked conformation with an increase in temperature is apparent from the single-molecule traces, with Kdock decreasing from 2.1 ± 0.1 at 26 °C to 0.54 ± 0.03 at 38 °C (Figure 2). Cumulative Kdock values for many molecules are in good agreement with the individual molecules (see Figure 2) and summarized over the full temperature range (21.0–47.4 °C) in Table 1 [Kdock(immobilized)], indicating a strongly exothermic folding process.
Table 1.
Temperature Dependence of Kdock and Thermodynamic Parameters for Tetraloop–Receptor Docking from Immobilized and Freely Diffusing (free) Single-Molecule Methods
| T (°C) | Kdock(immobilized) | Kdock(free) | ΔG° (kcal/mol) = −RT ln Kdock |
|---|---|---|---|
| 21 ± 0.2 | 4.18 ± 0.18 | 3.7 ± 1.8 | −0.80 ± 0.05 |
| 26 ± 0.2 | 2.3 ± 0.1 | − | −0.50 ± 0.03 |
| 29 ± 0.2 | 2.2 ± 0.2 | 1.7 ± 0.3 | −0.4 ± 0.1 |
| 34 ± 2 | 1.32 ± 0.08 | 0.87 ± 0.16 | −0.05 ± 0.16 |
| 36 ± 2 | 1.17 ± 0.09 | − | −0.10 ± 0.05 |
| 38 ± 2 | 0.64 ± 0.03 | 0.60 ± 0.11 | 0.30 ± 0.03 |
| 43 ± 3 | − | 0.55 ± 0.10 | 0.43 ± 0.12 |
| 47.4 ± 3.6 | − | 0.42 ± 0.09 | 0.58 ± 0.12 |
| immobilized | free | ||
| ΔH° (kcal/mol) | −17.4 ± 1.6 | −17.2 ± 1.6 | |
| ΔS° (cal mol−1 K−1) | −56.2 ± 5.4 | −55.9 ± 5.2 | |
Significant kinetic heterogeneity exists in the tetraloop–receptor system, as discussed previously (16). This heterogeneity can be adequately described as two non-interconverting populations: (i) 68% of species that actively dock and undock, and (ii) a 32% minority population of molecules that exhibit no folding events. The presence of “nondocking” molecules was also confirmed under freely diffusing conditions, yielding a 32% subpopulation in quantitative agreement with the tethered studies (37). Since these molecules show no docking events over the range of time scales and temperatures sampled, they are excluded from the equilibrium analysis described above. The molecular origin of the species is not yet known, e.g., whether the kinetic trap is of secondary or tertiary origin. These molecules never achieve the bound form of the tetraloop receptor so cannot be included in assessing tetraloop–receptor thermodynamics. Studies have suggested that the nondockers can be removed from the sample by native gel electrophoresis (15); however, we retain them in the study, to assess whether they can interconvert at increased temperatures. Temperatures greater than investigated may allow the nondockers to fold, which would be consistent with the remarkable robustness of subpopulations in the hairpin ribozyme (43). Further confirmation of the temperature insensitivity of the nondocking RNA subpopulation is provided by studies under freely diffusing conditions as described below.
Temperature Dependence of the Docking Equilibrium in Freely Diffusing RNA
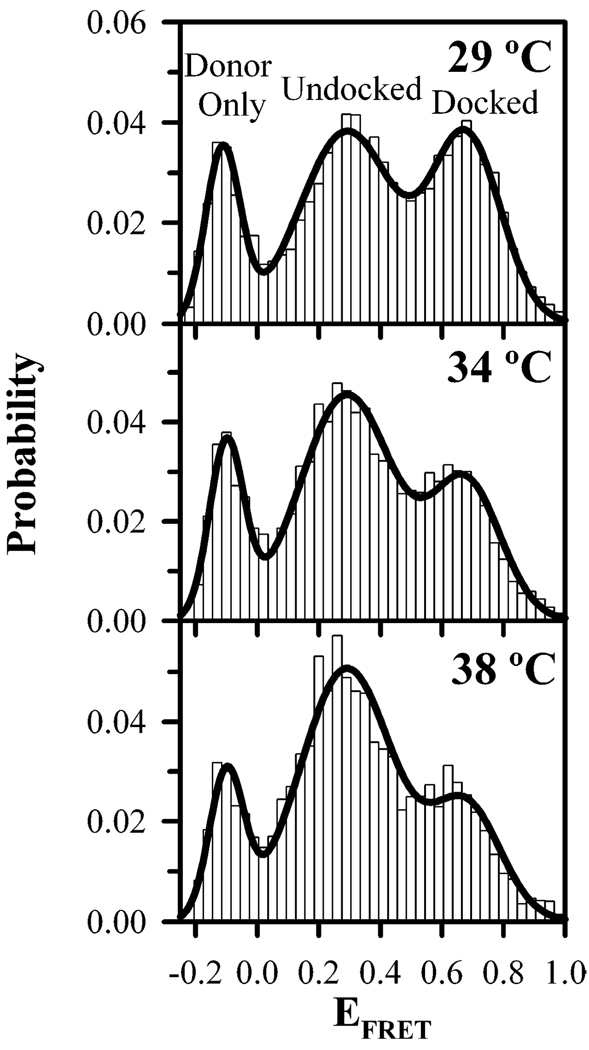
For temperature-dependent measurements of the tetraloop–-receptor interaction without surface immobilization, the equilibrium conformational distribution of the tetraloop–receptor construct is acquired from individual molecules freely diffusing through the confocal detection volume. This method allows for rapid sampling of many molecules, but with the ability to still discern conformational states (37). Low concentrations (200 pM) of RNA ensure that we are in the single-molecule detection regime, which is confirmed by cross correlation analysis of the donor and acceptor channels for freely diffusing time traces (see Figure S2 of the Supporting Information), yielding an average occupancy of the focal volume of 0.71 ± 0.09 molecule. Cross correlations also indicate the concentration of donor- and acceptor-labeled species is maintained upon heating, indicating that RNA construct remains intact. Time traces are binned at 1 ms integration for analysis, which is on the order of the dwell time of a molecule in the detection volume (37). This bin time is much shorter than the typical duration of a docking or undocking event; thus, a molecule can be cleanly identified in either the undocked or docked conformation (16). Fluorescence emission from labeled RNA molecules is distinguished from background via an intensity threshold (Ithresh = 10σbackground = 30 kHz) of a minimum sum of photons above background levels with typically >104 events acquired to ensure high-quality statistics (44). In freely diffusing studies, all fluorescent species in the solution are sampled. Therefore, signals from donor-only molecules give rise to a peak at negative EFRET due to direct excitation correction for the missing or photobleached acceptor (see Experimental Procedures) (37). As illustrated in Figure 3, the donor-only population shows no change with temperature, consistent with the RNA constructs remaining hybridized throughout temperature cycling.
FIGURE 3.
Temperature dependence of tetraloop–receptor docking shown in EFRET histograms generated from photon bursts occurring when freely diffusing RNAs traverse the laser focal volume (see Experimental Procedures). Three distinct populations with peaks corresponding to the donor-only (EFRET < 0), undocked (center EFRET = 0.291 ± 0.003), and docked RNA (center EFRET = 0.679 ± 0.004) exist at 29, 34, and 38 °C. There is a marked increase in the relative fraction of the undocked population with temperature, indicating disruption of the tertiary interaction, which is quantified by fitting the histograms to a sum of Gaussian distributions (solid black lines).
The freely diffusing EFRET histogram data can be globally fit over all temperatures by a sum of Gaussian distributions with common widths and centers, confirming that the docked and undocked peak shapes and positions are not significantly affected by heating. Such an analysis yields an 〈EFRET〉undocked of 0.291 ± 0.003 and a σundocked of 0.153 ± 0.004, and an 〈EFRET〉docked of 0.679 ± 0.004 and a σdocked of 0.112 ± 0.003, in agreement with the above immobilized results and previous freely diffusing studies at room temperature (〈EFRET〉 = 0.28 ± 0.01 and 0.687 ± 0.005 and σ = 0.165 ± 0.004 and 0.106 ± 0.002 for undocked and docked states, respectively) (37). Temperature and immobilization, therefore, do not introduce any observable changes in FRET centers and widths for both the undocked and docked RNA conformations, indicating that the system is described well by a two-state reaction scheme under all conditions in these studies. As demonstrated in Figure 3, the docked and undocked EFRET amplitudes do display temperature sensitivity. Specifically, increased temperature shifts the tetraloop–receptor construct to the undocked conformation, which is quantified by extracting Kdock from the equilibrium distributions. Extracting equilibrium constants from the threshold event distribution is potentially only an estimate of Kdock because of preferential detection of molecules dominantly emitting photons from the fluorophore with higher net collection efficiency. However, systematic variation of the threshold (15–75 kHz) reveals a negligible effect on the results presented. Such a result is expected as the collection efficiencies and quantum yields of the donor and acceptor are nearly equivalent in our experimental apparatus (see Experimental Procedures).
The freely diffusing detection method samples an equilibrium conformational distribution of all tetraloop–receptor constructs, including both actively docking (68%) and nondocking (32%) subpopulations. The nondocking sub-population can be incorporated in the analysis by noting that the fractional population in the docked state (Ffree) must scale linearly (i.e., Ffree = αFimmobilized) with the fractional docked state populations under immobilized conditions, where nondocking molecules are excluded (37). This fraction can be written explicitly as
| (2) |
where Ndocked and Nundocked + Nnondock are integrated areas of docked and undocked peaks in freely diffusing studies, respectively. The actively docking fraction, α, has been previously measured for both immobilized (α = 0.68 ± 0.01) (37) and freely diffusing (α = 0.66 ± 0.02) species (37). As a result, Kdock is readily extracted from Ffree by
| (3) |
where the nondocking fraction is identified by immobilized studies to be independent of temperature. As summarized in Table 1, Kdock under diffusing conditions decreases from 3.7 ± 1.8 at 21 °C to 0.42 ± 0.09 at 47 °C. The equilibrium constants are in agreement for freely diffusing and immobilized constructs, confirming that the nondocking fraction is constant over this temperature range.
Thermodynamics for Tetraloop–Receptor Docking
To extract thermodynamic information from these two independent methods, we analyze both the immobilized and diffusing data sets according to the van’t Hoff equation
| (4) |
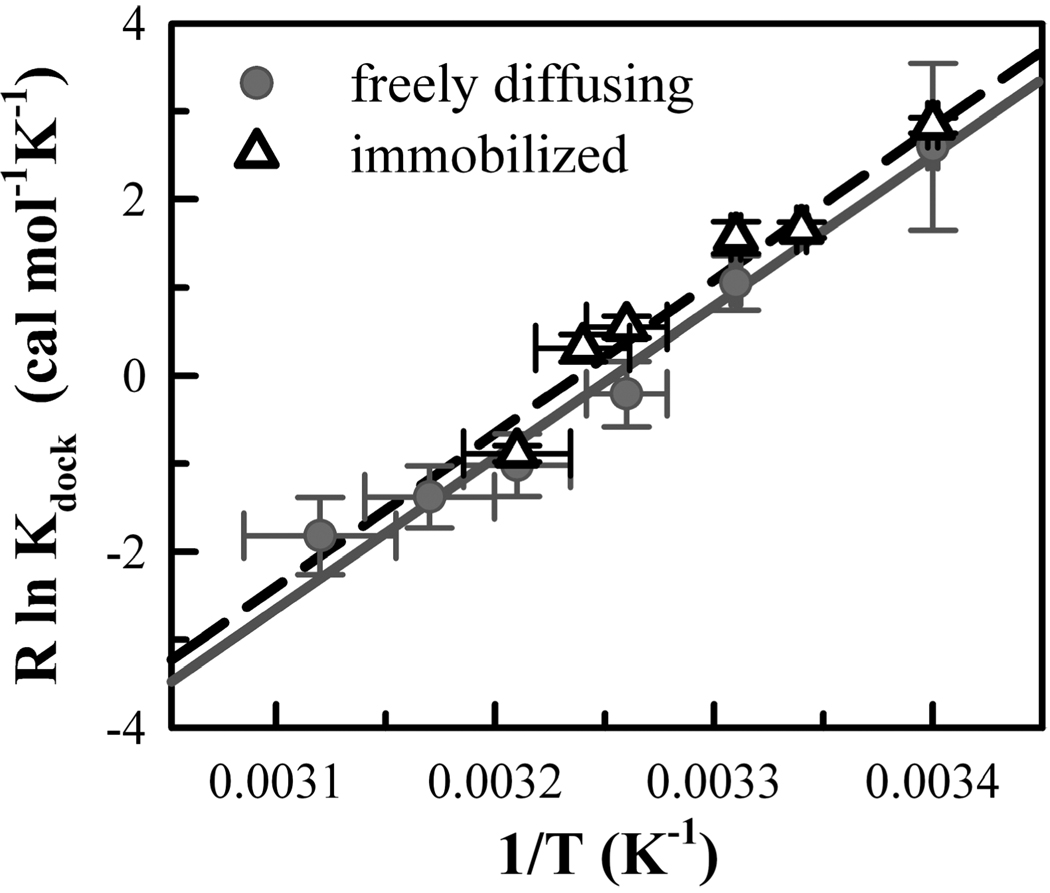
from which a plot of R ln Kdock versus 1/T yields a slope of −ΔH° and an intercept of ΔS° (see Figure 4), where R is the gas constant (1.987 cal mol−1 K−1). Both data sets yield straight line van’t Hoff plots, of which least-squares fits weighted for uncertainty of the dependent and independent variables yield the following: ΔH°immobilized = −17.4 ± 1.6 kcal/mol, ΔH°free = −17.2 ± 1.6 kcal/mol, ΔS°immobilized = −56.2 ± 5.4 cal mol−1 K−1, and ΔS°free = −55.9 ± 5.2 cal mol−1 K−1, which can be converted to standard international units with the conversion factor 4.184 J/cal. Freely diffusing and immobilized approaches for characterizing the thermodynamics of the tetraloop–receptor tertiary interaction are in excellent agreement (Figure 4). Docking of the tetraloop with the receptor results in a substantial decrease in enthalpy with an unfavorable entropy change (i.e., “enthalpy-driven folding”), where exothermicity is balanced at room temperature by a high entropic cost. More quantitatively, the standard state free energy (ΔG°) for forming the tetraloop–receptor interaction can be directly calculated as a function of temperature from the equilibrium constants (ΔG° = −RT ln Kdock), as summarized in Table 1. At 21 °C, the docked state is only marginally favored (ΔG° = −0.08 ± 0.05 kcal/ mol), with docking becoming thermodynamically unfavorable at physiological temperatures (≈37 °C).
FIGURE 4.
Thermodynamics of the tetraloop–receptor docking– undocking equilibrium from van’t Hoff plots (see eq 4). The temperature (T) dependence of the docking equilibrium constant (Kdock) is shown for freely diffusing (gray circles) and immobilized (black open triangles) molecules. Linear fits of R ln Kdock vs 1/T yield a slope of −ΔH° and an intercept of ΔS° (see Table 1) for the freely diffusing (solid gray line) and immobilized (dashed black line) data.
DISCUSSION
Accurate thermodynamic characterization of tertiary RNA folding is clearly important but has been extremely limited (15–17, 31). We demonstrate the applicability of temperature-dependent smFRET methods under both immobilized and freely diffusing conditions in deconstructing the docking thermodynamics for an isolated tetraloop–receptor tertiary motif. Freely diffusing techniques are particularly useful for monitoring RNA folding thermodynamics in the absence of surface tethering, which was demonstrated for the exothermic and entropically disfavored secondary folding of isolated RNA hairpins (33). We determine the standard state enthalpic and entropic components of tetraloop–receptor binding, revealing that the tertiary interaction like the secondary hairpin formation is enthalpy-driven, but at a large entropic cost. Extension of these methods to other isolated tertiary interactions and comparison to RNAs with multiple interactions should permit valuable insights into, for example, the thermodynamic origin of tertiary cooperativity (18, 45). Such dissections of RNA folding interactions are applicable to well-defined, modular components as in this scheme. Such a “bottom up” approach complements studies in large RNAs by avoiding potentially deleterious mutations that may disrupt the tertiary interaction through changes in global structure.
NMR and X-ray crystal structures of the docked and undocked tetraloop and receptor reveal structural contributions to the entropic cost and enthalpic favorability of tetraloop–receptor association. Specifically, tetraloop–receptor docking induces structural elements that have been generally recognized as enthalpically stabilizing and entropically unfavorable, namely, hydrogen bonding and base stacking interactions (10, 11, 46, 47). The large entropic penalty for folding may also originate from the loss of free orientational flexibility of the tetraloop in the undocked state to the specific orientation required for docking (11, 13). The proposition that the flexibility in an unfolded structure is correlated with the entropic cost of folding is supported by thermodynamic studies of the hairpin ribozyme, in which a four-way junction (4WJ) greatly decreased the entropic penalty of folding as compared to the two-way junction (2WJ). The 4WJ contributes significant rigidity and orientation to the unfolded state, leading to a decrease in the level of disorder prior to the docking event (26). Solvent effects, such as disruption of ordered water molecules, yield positive enthalpic and entropic folding contributions; this is opposite to what is observed for tetraloop–receptor folding and therefore not the dominant effect (48).
Although the measured enthalpy and entropy changes are dominated by the tetraloop–receptor interaction, we also consider thermodynamic contributions arising from the RNA construct design. Specifically, contributions could arise from the flexible A7 linker because of small differences in its base stacking in the docked and undocked forms. Additional negative entropic contributions to folding due to the reduced conformational space accessible to the linker in the docked versus undocked states are expected to be modest because this region is sufficiently long to maintain disorder. By way of confirmation, previous studies of linker length (A7 vs A14) and composition (A vs U) dependence of tetraloop–receptor docking and undocking revealed minimal changes in the equilibrium constant (15, 16).
Our results allow us to place the tetraloop–receptor interaction in thermodynamic context with global folding of the P4–P6 domain in the Tetrahymena ribozyme, which has been studied at the ensemble level by temperature gradient gel electrophoresis under similar cation conditions (see Table 2) (25). Results for the global fold indicate large net enthalpy and entropy decreases of −28 ± 3 kcal/mol (ΔH°) and −91 ± 8 cal mol−1 K−1 (ΔS°), respectively, i.e., also enthalpically driven with a magnitude only slightly larger than the tetraloop–receptor values. Although there are additional tertiary interactions and structural complexity to consider in the full P4–P6 folding problem, it would appear that the GAAA tetralop–receptor construct alone may contribute ≈60% of the ΔH° and ΔS°. The tetraloop–receptor interaction has been proposed as a thermodynamic clamp for stabilizing the global fold of large RNAs (17). This work supports such a picture by determining that thermodynamic stability may originate from the strongly favorable tetraloop–receptor docking enthalpy. Further inspection will be necessary to evaluate how the complexity of large RNAs affects the folding thermodynamics, with assessment of the role of global architecture, secondary elements, and solvent accessibility.
Table 2.
Thermodynamic Parameters for Tetraloop–Receptor Docking and Other Tertiary Folding (4.184 J/cal)
| RNA tertiary interaction | conditions | ΔH° (kcal/mol) |
ΔS° (cal mol−1 K−1) |
ΔG° at 37 °C = ΔH° – TΔS° (kcal/mol) |
|---|---|---|---|---|
| tetraloop–receptor docking | immobilized, 1 mM MgCl2, 0.1 M NaCl | −17.4 ± 1.6 | −56.2 ± 5.4 | 0.02 ± 2.3 |
| freely diffusing, 1 mM MgCl2, 0.1 M NaCl | −17.2 ± 1.6 | −55.9 ± 5.2 | 0.13 ± 2.3 | |
| P4–P6 domain folding(25) | 0.9 mM MgCl2, 10 mM NaCl | −28 ± 3 | −91 ± 8 | 0.21 ± 3.9 |
| hairpin ribozyme folding (26) | 2WJ, 1 mM MgCl2 | −9.7 ± 4.5 | −34 ± 17 | 0.84 ± 7 |
| 4WJ, 1 mM MgCl2 | −4.1 ± 2.3 | −9.6 ± 5.7 | −1.1 ± 2.9 | |
| P1 helix docking into Tetrahymena | 10 mM MgCl2 (31) | 8 ± 2 | 25 ± 8 | 0.25 ± 3.2 |
| group I intron core | 5 mM MgCl2, 135 mM NaCl (53) | 8 | 40 | −4 |
| 10 mM MgCl2 (54) | 19 ± 9 | 62 ± 30 | −0.22 ± 13 | |
| 3WJ folding of hammerhead ribozyme core (21) | 10 mM MgCl2, 0.1 M NaCl | 41 ± 1 | 120 ± 10 | 3.8 ± 3.3 |
However, the docked form of the tetraloop–receptor construct is thermodynamically disfavored (i.e., ΔG°immobilized,free = 0.022 ± 2.3 or 0.13 ± 2.3 kcal/mol) under physiologically relevant conditions (37 °C, 1 mM MgCl2, 100 mM NaCl), with similar results for the P4–P6 domain (ΔG° = 0.21 ± 3.9 kcal/ mol). This borderline stability of even strongly enthalpy-driven folding underscores that RNA folding dynamics must be addressed for a complete understanding of functionality. Folding of both P4–P6 domain and the tetraloop–receptor construct becomes favorable with increased Mg2+ concentrations (16, 18).
The thermodynamic parameters for P4–P6 and tetraloop–receptor folding are consistent with that of other RNA folding, specifically global folding of tRNA (49), the 1051–1108 rRNA fragment (50), the hairpin ribozyme (26), pseudoknot tertiary structure formation (19), ligand-induced riboswitch folding (51), and secondary loop formation (33, 52). Although differing in magnitude, each of these systems exhibits significant enthalpic gain (ΔH° < 0) and entropic penalty (ΔS° < 0) for folding. Furthermore, in a recent ITC study, Reymond et al. demonstrated through systematic mutations that various steps in the folding pathway of the hepatitis delta virus ribozyme are exothermic and entropically disfavored (24). However, these observations of enthalpy-driven RNA folding are not a universal trend, as evidenced in Table 2. The final stage of Tetrahymena ribozyme folding, i.e., the docking of the P1 duplex into the prefolded core, was investigated by smFRET of immobilized molecules and found to be enthalpically disfavored (ΔH° = 8 ± 2 kcal/ mol) and entropically favored (ΔS° = 25 ± 8 cal mol−1 K−1), in reasonable agreement with ensemble methods (53, 54). ITC studies of the three-way junction (3WJ) in the minimal hammerhead ribozyme core reveal a similar degree of endothermicity and entropic advantage (Table 2) (21). As yet a third case, smFRET investigation of the hairpin ribozyme’s 4WJ revealed no temperature dependence in the folding equilibrium constants, from which one can infer the processes to be nearly thermoneutral (ΔH° ≈ 0 kcal/mol) with only a modest decrease in disorder (ΔS° ≈ −2 ± 1 cal mol−1 K−1) between the various folded conformations (32).
As illustrated in Table 2, each tertiary association has a ΔG° near zero; thus, RNA folding can be regarded as either enthalpy- or entropy-driven. Folding events in the tetraloop–receptor, P4–P6 domain, and hairpin ribozyme constructs result in many hydrogen bonds and base stacking interactions, of which tetraloop–receptor docking forms the fewest, i.e., 10 hydrogen bonds and improved base stacking (10, 11, 55). By way of contrast, such enthalpy-driven interactions are much less prevalent in the entropy-driven P1 docking, which nets only four or five hydrogen bonds and no base stacking (56). The reduced prevalence of these secondary-like interactions (i.e., from ~11 to 4) may account for the shift from enthalpy-driven to entropy-driven (solvent-driven) folding. The net entropic drive for this folding has been proposed to arise from displacement of ordered water molecules and/or Mg2+ ions (53, 54). Furthermore, the Tetrahymena ribozyme is already highly structured prior to P1 docking, which reduces the entropic cost associated with the final folding transition.
Folding of the minimal hammerhead ribozyme is more difficult to categorize in terms of base stacking and hydrogen bonding because the structure of the core prior to coaxial stacking of the helices is not clear. However, counting hydrogen bonds and base stacking interactions that are contingent on helix alignment, i.e., not including the two noncanonical base pairs (A9-G12 and G8-A13) that likely form in the core prior to helical stacking, we identify a net of approximately seven interactions (57, 58). This is intermediate, albeit closer to the case of P1 than tetraloop–receptor docking; therefore, solvent effects, e.g., displacement of ordered water molecules, are not overcome, and entropy increases in minimal hammerhead ribozyme folding (21, 53, 54). Endothermic changes are associated with rearrangement of core residues of the hammerhead ribozyme (21), which may explain the additional enthalpic penalty for hammerhead core folding versus P1 docking. In summary, the data suggest that categorization of enthalpy- versus entropy-driven RNA folding arises from competing roles of solvent and hydrogen bonding and stacking interactions, with a net ~11 interactions providing sufficient negative ΔH° and ΔS° to overcome solvent effects. Furthermore, each of the entropy-driven RNA folds (see Table 2), as well as the thermoneutral 4WJ, was measured at significantly higher concentrations of Mg2+ than the enthalpy-driven cases [>10 mM vs ~1 mM Mg2+ (see Table 2)]. A Mg2+ environment that is more amenable to RNA folding, though less physiological, may also contribute to the observation of entropy-driven folding by increasing the magnitude of solvent effects. Further systematic study of isolated tertiary motifs with temperature-dependent smFRET methods will be invaluable in further elucidating the competing roles of solvent and hydrogen bonding and stacking interaction to categorize enthalpy- versus entropy-driven RNA folding.
As a final comment, in the course of preparing this work, we became aware of a simultaneous and independent ITC investigation of tetraloop–receptor binding by the Butcher group. Specifically, the Butcher group had cleverly designed pairs of RNA constructs, based on back-to-back copies of tetraloops (TT) and receptors (RR). These constructs can form a duplex (TRTR) through dual tetraloop–receptor docking, which permits association to be initiated by mixing of the two constructs. This work by Vander Meulen et al. published elsewhere (23) yields a ΔH° of −33.2 ± 2.0 kcal/ mol at 45 °C for double tetraloop–receptor formation under similar solvent conditions (2 mM MgCl2, 20 mM KCl). Assuming zero enthalpy cooperativity between the two tertiary interactions, this translates into a ΔH°ITC of −16.9 ± 1.0 kcal/mol for single tetraloop–receptor binding, which agrees with our single-molecule values of −17.2 ± 1.6 kcal/ mol (ΔH°free) and −17.4 ± 1.6 kcal/mol (ΔH°immobilized). This agreement between isolated and dual tetraloop–receptor docking enthalpies supports a picture in which tertiary structure formation in RNA may be largely enthalpically noncooperative. In turn, this would imply an entropic origin of tertiary cooperativity between the tetraloop–receptor and metal–core interactions observed in folding of the complete P4–P6 domain (18).
CONCLUSION
Thermodynamics of the isolated GAAA tetraloop–receptor tertiary interaction are investigated at the single-molecule level, exploiting the combination of temperature-dependent FRET and confocal microscopy methods. Results for the GAAA tetraloop–receptor motif are shown to be enthalpically driven (ΔHfree = −17.2 ± 1.6 kcal/mol, and ΔHimmobilized = −17.4 ± 1.6 kcal/mol) yet balanced by entropically unfavorable (ΔSfree = −55.9 ± 5.2 cal mol−1 K−1, and ΔSimmobilized = −56.2 ± 5.4 cal mol−1 K−1) conformational changes upon docking. These results for tetraloop–receptor folding already achieve 60% of the values obtained from ensemble studies for folding of the complete P4–P6 domain, supporting previous views that the tetraloop–receptor construct provides a dominant source of thermodynamic stabilization. However, under physiological temperature conditions, this isolated tertiary motif is found to be marginally unstable (ΔG = 0.02 and 0.13 kcal/mol from immobilized and freely diffusing, respectively), indicating that conformational change may still play a key role in RNA functionality. Further work on temperature and cation dependences of the rates, kdock and kundock, will be necessary to illuminate the transition state enthalpy and entropy changes associated with tertiary contact formation and to help develop a consensus picture for the thermodynamic origin of cation-mediated RNA folding.
Supplementary Material
ACKNOWLEDGMENT
We thank Bioptechs for their generous technical support of this study and valuable discussions. We also thank Drs. Arthur Pardi and Christopher D. Downey for their contributions to the RNA construct design and many helpful comments in the preparation of the manuscript.
Footnotes
This work was supported in part by the NSF, NIST, the W. M. Keck Foundation initiative in RNA sciences at the University of Colorado, Boulder, and PicoQuant, GmbH. J.L.F. was supported in part by Optical Science and Engineering Program NSF-IGERT and CU Biophysics Training (T32 GM-065103) grants.
Abbreviations: HPLC, high-performance liquid chromatography; HEPES, N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid; EDTA, ethylenediaminetetraacetate; ITC, isothermal titration calorimetry; NMR, nuclear magnetic resonance.
SUPPORTING INFORMATION AVAILABLE
Prediction of the effect of donor quantum yield (QD) on the observed FRET efficiency (EFRET) as a function of Cy3–Cy5 distance (R) (Figure S1) and mean cross correlations of donor and acceptor channels for the same sample containing tetraloop–receptor constructs under freely diffusing single-molecule conditions at 21 and 45 °C (Figure S2). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Tinoco I, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 2.Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 4.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JC, Gutell RR, Russell R. The UAA/GAN internal loop motif: A new RNA structural element that forms a cross-strand AAA stack and long-range tertiary interactions. J. Mol. Biol. 2006;360:978–988. doi: 10.1016/j.jmb.2006.05.066. [DOI] [PubMed] [Google Scholar]
- 6.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat. Struct. Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 8.Abramovitz DL, Pyle AM. Remarkable morphological variability of a common RNA folding motif: The GNRA tetraloop-receptor interaction. J. Mol. Biol. 1997;266:493–506. doi: 10.1006/jmbi.1996.0810. [DOI] [PubMed] [Google Scholar]
- 9.Qin PZ, Butcher SE, Feigon J, Hubbell WL. Quantitative analysis of the isolated GAAA tetraloop/receptor interaction in solution: A site-directed spin labeling study. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- 10.Butcher SE, Dieckmann T, Feigon J. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997;16:7490–7499. doi: 10.1093/emboj/16.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cate JH, Gooding AR, Podell E, Zhou KH, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 12.Jucker FM, Pardi A. GNRA tetraloops make a U-Turn. RNA. 1995;1:219–222. [PMC free article] [PubMed] [Google Scholar]
- 13.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J. Mol. Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 14.Jaeger L, Leontis NB. Tecto-RNA: One-dimensional self-assembly through tertiary interactions. Angew. Chem. Int. Ed. 2000;39:2521–2524. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 15.Downey CD, Fiore JL, Stoddard CD, Hodak JH, Nesbitt DJ, Pardi A. Metal ion dependence, thermodynamics, and kinetics for intramolecular docking of a GAAA tetraloop and receptor connected by a flexible linker. Biochemistry. 2006;45:3664–3673. doi: 10.1021/bi0520941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodak JH, Downey CD, Fiore JL, Pardi A, Nesbitt DJ. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc. Natl. Acad. Sci. U.S.A. 2005;102:10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young BT, Silverman SK. The GAAA tetraloop-receptor interaction contributes differentially to folding thermodynamics and kinetics for the P4-P6 RNA domain. Biochemistry. 2002;41:12271–12276. doi: 10.1021/bi0264869. [DOI] [PubMed] [Google Scholar]
- 18.Sattin BD, Zhao W, Travers K, Chut S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. J. Am. Chem. Soc. 2008;130:6085–6087. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nixon PL, Giedroc DP. Energetics of a strongly pH dependent RNA tertiary structure in a frameshifting pseudoknot. J. Mol. Biol. 2000;296:659–671. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- 20.Hammann C, Cooper A, Lilley DMJ. Thermodynamics of ion-induced RNA folding in the hammerhead ribozyme: An isothermal titration calorimetric study. Biochemistry. 2001;40:1423–1429. doi: 10.1021/bi002231o. [DOI] [PubMed] [Google Scholar]
- 21.Mikulecky PJ, Takach JC, Feig AL. Entropy-driven folding of an RNA helical junction: An isothermal titration calorimetric analysis of the hammerhead ribozyme. Biochemistry. 2004;43:5870–5881. doi: 10.1021/bi0360657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feig AL. Applications of isothermal titration calorimetry in RNA biochemistry and biophysics. Biopolymers. 2007;87:293–301. doi: 10.1002/bip.20816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Meulen KA, Davis JH, Foster TR, Record T, Butcher SE. Thermodynamics and folding pathway of tetraloop receptor-mediated RNA helical packing. J. Mol. Biol. 2008;384:702–717. doi: 10.1016/j.jmb.2008.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reymond C, Bisaillon M, Perreault J-P. Monitoring of an RNA multistep folding pathway by isothermal titration calorimetry. Biophys. J. 2009;96:132. doi: 10.1016/j.bpj.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szewczak AA, Podell ER, Bevilacqua PC, Cech TR. Thermodynamic stability of the P4-P6 domain RNA tertiary structure measured by temperature gradient gel electrophoresis. Biochemistry. 1998;37:11162–11170. doi: 10.1021/bi980633e. [DOI] [PubMed] [Google Scholar]
- 26.Klostermeier D, Millar DP. Helical junctions as determinants for RNA folding: Origin of tertiary structure stability of the hairpin ribozyme. Biochemistry. 2000;39:12970–12978. doi: 10.1021/bi0014103. [DOI] [PubMed] [Google Scholar]
- 27.Li PTX, Vieregg J, Tinoco I. How RNA unfolds and refolds. Annu. Rev. Biochem. 2008;77:77–100. doi: 10.1146/annurev.biochem.77.061206.174353. [DOI] [PubMed] [Google Scholar]
- 28.Bokinsky G, Zhuang XW. Single-molecule RNA folding. Acc. Chem. Res. 2005;38:566–573. doi: 10.1021/ar040142o. [DOI] [PubMed] [Google Scholar]
- 29.Zhuang XW, Kim H, Pereira MJB, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 30.Xie Z, Srividya N, Sosnick TR, Pan T, Scherer NF. Single-molecule studies highlight conformational heterogeneity in the early folding steps of a large ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2004;101:534–539. doi: 10.1073/pnas.2636333100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartley LE, Zhuang XW, Das R, Chu S, Herschlag D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J. Mol. Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- 32.Hohng S, Wilson TJ, Tan E, Clegg RM, Lilley DMJ, Ha TJ. Conformational flexibility of four-way junctions in RNA. J. Mol. Biol. 2004;336:69–79. doi: 10.1016/j.jmb.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Gell C, Sabir T, Westwood J, Rashid A, Smith DAM, Harris S-A, Stockley PG. Single-Molecule Fluorescence Resonance Energy Transfer Assays Reveal Heterogeneous Folding Ensembles in a Simple RNA Stem-Loop. J. Mol. Biol. 2008;384:264–278. doi: 10.1016/j.jmb.2008.08.088. [DOI] [PubMed] [Google Scholar]
- 34.Good NE, Winget GD, Winter W, Connolly TN, Izawa S, Singh RMM. Hydrogen ion buffers for biological research. Biochemistry. 1966;5:467–477. doi: 10.1021/bi00866a011. [DOI] [PubMed] [Google Scholar]
- 35.Ha T, Zhuang XW, Kim HD, Orr JW, Williamson JR, Chu S. Ligand-induced conformational changes observed in single RNA molecules. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9077–9082. doi: 10.1073/pnas.96.16.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rasnik I, McKinney SA, Ha T. Nonblinking and longlasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 37.Fiore JL, Hodak JH, Piestert O, Downey CD, Nesbitt DJ. Monovalent and divalent promoted GAAA-tetraloop-receptor tertiary interactions from freely diffusing single-molecule studies. Biophys. J. 2008;95:3892–3905. doi: 10.1529/biophysj.108.134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hernández B, Baumruk V, Leulliot N, Gouyette C, Huynh-Dinh T, Ghomi M. Thermodynamic and structural features of ultrastable DNA and RNA hairpins. J. Mol. Struct. 2003;651–653:67–74. [Google Scholar]
- 39.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanborn ME, Connolly BK, Gurunathan K, Levitus M. Fluorescence Properties and Photophysics of the Sulfoin-docyanine Cy3 Linked Covalently to DNA. J. Phys. Chem. B. 2007;111:11064–11074. doi: 10.1021/jp072912u. [DOI] [PubMed] [Google Scholar]
- 41.Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Phys. (Weinheim, Ger.) 1948;2:55–75. [Google Scholar]
- 42.Iqbal A, Arslan S, Okumus B, Wilson TJ, Giraud G, Norman DG, Ha T, Lilley DMJ. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11176–11181. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn871. doi: 10.1093/nar/gkn871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deniz AA, Dahan M, Grunwell JR, Ha TJ, Faulhaber AE, Chemla DS, Weiss S, Schultz PG. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: Observation of Forster distance dependence and sub-populations. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3670–3675. doi: 10.1073/pnas.96.7.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Serra MJ, Turner DH, Michael LJ, Ackers GK. Predicting thermodynamic properties of RNA. Methods Enzymol. 1995;259:242–261. doi: 10.1016/0076-6879(95)59047-1. [DOI] [PubMed] [Google Scholar]
- 47.Silverman SK, Cech TR. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry. 1999;38:8691–8702. doi: 10.1021/bi9906118. [DOI] [PubMed] [Google Scholar]
- 48.Mikulecky PJ, Feig AL. Heat capacity changes associated with nucleic acid folding. Biopolymers. 2006;82:38–58. doi: 10.1002/bip.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein A, Crothers DM. Conformational-changes of transfer-RNA: Role of magnesium(II) Biochemistry. 1976;15:160–168. doi: 10.1021/bi00646a025. [DOI] [PubMed] [Google Scholar]
- 50.Lu M, Draper DE. Bases defining an ammonium and magnesium ion-dependent tertiary structure within the large subunit ribosomal-RNA. J. Mol. Biol. 1994;244:572–585. doi: 10.1006/jmbi.1994.1753. [DOI] [PubMed] [Google Scholar]
- 51.Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Antao VP, Tinoco I. Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y, Bevilacqua PC, Mathews D, Turner DH. Thermodynamic and activation parameters for binding of a pyrene-labeled substrate by the Tetrahymena ribozyme: Docking is not diffusion-controlled and is driven by a favorable entropy. Biochemistry. 1995;34:14394–14399. doi: 10.1021/bi00044a016. [DOI] [PubMed] [Google Scholar]
- 54.Narlikar GJ, Herschlag D. Isolation of a local tertiary folding transition in the context of a globally folded RNA. Nat. Struct. Biol. 1996;3:701–710. doi: 10.1038/nsb0896-701. [DOI] [PubMed] [Google Scholar]
- 55.Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 56.Szewczak AA, Ortoleva-Donnelly L, Ryder SP, Moncoeur E, Strobel SA. A minor groove RNA triple helix within the catalytic core of a group I intron. Nat. Struct. Biol. 1998;5:1037–1042. doi: 10.1038/4146. [DOI] [PubMed] [Google Scholar]
- 57.Pley HW, Flaherty KM, McKay DB. 3-Dimen-sional structure of a hammerhead ribozyme. Nature. 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 58.Scott WG, Finch JT, Klug A. The crystal structure of an All-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.