Abstract
The steady discovery of new β-myosin mutations causing familial hypertrophic cardiomyopathy (FHCM) suggests that only a fraction of the total number of mutations has been identified so far by clinical screenings. To test this hypothesis, data derived from seven independent genetic studies were re-analyzed using statistical methods developed by ecologists for estimating the number of species in a community.
This novel approach reveals that more mutations will continue to be identified as more patients are genotyped, and highlights important differences in the distribution of β-myosin mutations across different populations. Hence, additional molecular sampling will be necessary to complete the census of the β-myosin mutations and facilitate the diagnosis and evaluation of FHCM patients.
Keywords: Cardiomyopathy, Myosin, Mutation, Community ecology
Hypertrophic cardiomyopathy (HCM) is a complex cardiac disease affecting one in 500 individuals. HCM can manifest with a wide degree of clinical variability, ranging from individuals who are largely asymptomatic to individuals who develop heart failure and sudden death. Histopathological cardiac alterations such as fibrosis are primarily associated with changes in the size of ventricular chamber(s), increased wall thickness, and atrioventricular conduction abnormalities. Missense mutations in the β-myosin heavy chain gene (β-MHC), the predominant mammalian myosin isoform in adult human heart ventricles, are among the most frequent genetic defects linked to familial hypertrophic cardiomyopathy (FHCM) [1]. Interestingly, the frequency of β-myosin mutations associated with HCM appears to vary across different populations [2]. Low-density β-myosin DNA microarrays for early FHCM genetic screening and diagnosis have been developed [3,4]. However, their potential clinical development is limited by the finding that no single β-myosin mutation appears to be predominant and, more importantly, new mutations are regularly being discovered. To estimate the number of β-myosin mutations that remain to be characterized, we applied established techniques commonly used by community ecologists to study the distribution and abundance of different organisms present in a particular environment (Fig. 1A).
Fig. 1.
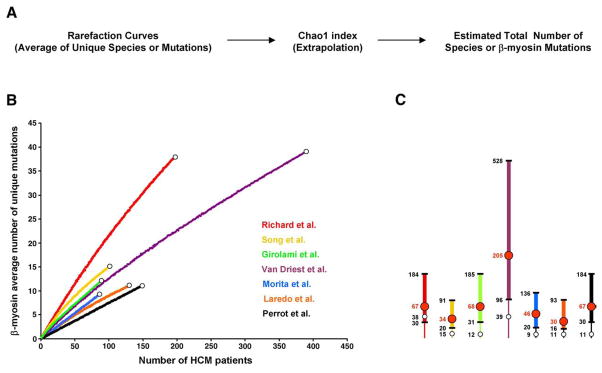
Estimation of β-myosin mutations richness. (A) Flow chart of the community ecology methods employed in this study. (B) Individual rarefaction curves derived from seven independent studies are depicted in different colors. White dots correspond to the total number of unique mutations experimentally found. (C) Graphic representation of the total number of estimated β-myosin mutations derived from each study. Intervals (95% confidence) are shown by thicker lines. Red numbers/dots correspond to the estimated mean number of mutations. White dots correspond to the total number of unique mutations experimentally found. Rarefaction curves employed to estimate the average number of unique β-myosin mutations were calculated by randomly resampling the mutation population 100 times, according to Simberloff [12]. The validity of this approach is based on the assumptions that the population is infinite, and the patients enrolled in each study represent a random sample of the total number of people with cardiomyopathy (random spatial distribution of individuals). Since the prevalence of familial hypertrophic cardiomyopathy in the global population is 0.2%, we believe that the resulting FHCM population, corresponding to about 1.3×107 individuals, is large enough to satisfy the first assumption. Since patients examined and genotyped were mainly Europeans and Americans they do not represent an equiprobable sampling. Based on this limitation, the resulting analyses should be considered approximate estimates (see text for discussion). EstimateS [http://viceroy.eeb.uconn.edu/EstimateS] was used to extrapolate the population size of β-myosin mutations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We analyzed missense mutations characterized in seven independent clinical studies that screened the whole β-myosin gene coding region [5–11]. By using rarefaction curves that allow comparisons between data sets of different sizes and analysis of small samples of individuals, we estimated the number of unique β-myosin mutations per patient in each study [12]. If a rarefaction curve, which in our case plots the number of patients with HCM versus the average number of unique mutations observed in that number of patients, rapidly levels off, we can be confident that most unique mutations have already been observed and that no additional sampling is necessary. In contrast, if the curve keeps rising, more mutations remain to be found.
The seven clinical studies analyzed a total of 1133 patients clinically diagnosed with HCM, and found 135 single β-myosin missense mutations associated with the disease (See Table 1 for mutation frequency across studies/populations). When rarefaction curves were derived from each investigation, we found that they continued to rise steeply, indicating that the experimentally observed β-myosin mutations are likely to represent only a small percentage of the total number that will be found (Fig. 1B). We next extrapolated the total number of mutations that could be found in each study population with additional sampling, by comparing the number of unique mutations to the number of mutations that occur more than once [13]. This approach is related to capture-recapture studies that estimate the size of a population by capturing, marking and releasing animals, then scoring the fraction of recaptured marked animals in the total captured population. Intuitively, if only few mutations are present in the β-myosin gene, the number of unique mutations found in every study will be the same. As shown in Fig. 1C, we obtained an estimated mean number of mutations of 67 for Richard et al. (38 unique mutations found experimentally; 57%), 34 for Song et al. (15 unique mutations found experimentally; 44%), 68 for Girolami et al. (12 unique mutations found experimentally; 18%), 205 for Van Driest et al. (39 unique mutations found experimentally; 19%), 46 for Morita et al. (9 unique mutations found experimentally in a cohort of pediatric patients; 19%), 30 for Laredo et al. (11 unique mutations found experimentally; 37%), and 67 for Perrot et al. (11 unique mutations found experimentally; 16%).
Table 1.
Study characteristics
| Author | Population | Method | Mutation frequency % | Expected mutations | % Found |
|---|---|---|---|---|---|
| Perrot | Kyn/Pol/Ger | SSCP | 7.5 | 67 | 16 |
| Laredo | Spain | SSCP | 11 | 30 | 37 |
| Morita | USA | Seq | 11 | 46 | 19 |
| Van Driest | USA | DHPLC | 15 | 205 | 19 |
| Girolami | Italy | DHPLC | 20 | 68 | 18 |
| Song | China | Seq | 23 | 34 | 44 |
| Richard | Europe | SSCP | 24 | 67 | 57 |
Kyr/Pol/Germ: Kyrgyzstan/Poland/Germany. SSCP: single-strand conformation polymorphism analysis. DHPLC: denaturing high-performance liquid chromatography analysis. Seq: complete sequencing of β-myosin exons. Mutations frequency is expressed as percentage of HCM patients carrying a β-myosin mutation. Expected mutations corresponds to the estimated mean number of mutations. % Found corresponds to the percentage of Expected mutations identified so far.
These data show that the individual studies have observed only a fraction of the total number of estimated mutations. More importantly, the finding that individual rarefaction curves are approaching saturation at different rates, suggests that i) the total number β-myosin mutations varies among different populations and ii) some mutations affect only specific populations (see Table 1).
Although our ecology-based approach suggests that additional molecular sampling will be necessary to complete the census of β-myosin mutations, our estimates are probably conservative. In fact they are based on studies that were carried out by European and American tertiary centers, which usually receive selective referrals of high-risk patients. Thus our results may only predict the total number of β-myosin mutations that can be diagnosed using the patient selection and methodologies routinely employed (see Table 1), rather than the total number of circulating mutations. Nevertheless, this study is the first attempt to quantify the number of disease-causing mutations in a human gene using an ecological model. Our findings can assist the design of future HCM patient screenings as well as the development of representative β-myosin DNA microarrys for early FHCM genetic screening and diagnosis.
Acknowledgments
We would like to thank Ada Buvoli and Robert Thompson for discussion and critical reading of the manuscript. This work was partially supported by NIH grant GM29090 to L. A. L., by the Keck RNA Bioinformatics Initiative, and by NIH training grant T32GM065103-03.
The authors of this manuscript have certified that they comply with the Principles of Ethical Publishing in the International Journal of Cardiology [14].
References
- 1.Tardiff JC. Sarcomeric proteins and familial hypertrophic cardiomyopathy: linking mutations in structural proteins to complex cardiovascular phenotypes. Heart Fail Rev. 2005;10:237–48. doi: 10.1007/s10741-005-5253-5. [DOI] [PubMed] [Google Scholar]
- 2.Navarro-Lopez F. Hypertrophic cardiomyopathy: never-ending complexity. Rev Esp Cardiol. 2006;59:994–6. doi: 10.1157/13093974. [DOI] [PubMed] [Google Scholar]
- 3.Wang D, Li Y, Zhang R, et al. Detection of known mutations in hypertrophic cardiomyopathy using oligonucleotide microarrays assisted by improved base stacking hybridization. Biotechnol Lett. 2003;25:1613–8. doi: 10.1023/a:1025616015090. [DOI] [PubMed] [Google Scholar]
- 4.Waldmuller S, Freund P, Mauch S, Toder R, Vosberg HP. Low-density DNA microarrays are versatile tools to screen for known mutations in hypertrophic cardiomyopathy. Human Mutat. 2002;19:560–9. doi: 10.1002/humu.10074. [DOI] [PubMed] [Google Scholar]
- 5.Morita H, Rehm HL, Menesses A, et al. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899–908. doi: 10.1056/NEJMoa075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laredo R, Monserrat L, Hermida-Prieto M, et al. Beta-myosin heavy-chain gene mutations in patients with hypertrophic cardiomyopathy. Rev Esp Cardiol. 2006;59:1008–18. doi: 10.1157/13093977. [DOI] [PubMed] [Google Scholar]
- 7.Girolami F, Olivotto I, Passerini I, et al. A molecular screening strategy based on beta-myosin heavy chain, cardiac myosin binding protein C and troponin T genes in Italian patients with hypertrophic cardiomyopathy. J Cardiovasc Med (Hagerstown) 2006;7:601–7. doi: 10.2459/01.JCM.0000237908.26377.d6. [DOI] [PubMed] [Google Scholar]
- 8.Perrot A, Schmidt-Traub H, Hoffmann B, et al. Prevalence of cardiac beta-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. J Mol Med. 2005;83:468–77. doi: 10.1007/s00109-005-0635-7. [DOI] [PubMed] [Google Scholar]
- 9.Song L, Zou Y, Wang J, et al. Mutations profile in Chinese patients with hypertrophic cardiomyopathy. Clin Chim Acta. 2005;351:209–16. doi: 10.1016/j.cccn.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Van Driest SL, Jaeger MA, Ommen SR, et al. Comprehensive analysis of the beta-myosin heavy chain gene in 389 unrelated patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44:602–10. doi: 10.1016/j.jacc.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 11.Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–32. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 12.Simberloff D. Properties of the rarefaction diversity measurement. Am Nat. 1972;106:414–8. [Google Scholar]
- 13.Chao A. Nonparametric estimation of the number of classes in a population. Scand J Statist. 1984;11:265–70. [Google Scholar]
- 14.Coats AJ. Ethical authorship and publishing. Int J Cardiol. 2009;131:149–50. doi: 10.1016/j.ijcard.2008.11.048. [DOI] [PubMed] [Google Scholar]