SUMMARY
Maintenance of skeletal muscle structure and function requires innervation by motor neurons, such that denervation causes muscle atrophy. We show that myogenin, an essential regulator of muscle development, controls neurogenic atrophy. Myogenin is up-regulated in skeletal muscle following denervation and regulates expression of the E3 ubiquitin ligases MuRF1 and atrogin-1, which promote muscle proteolysis and atrophy. Deletion of myogenin from adult mice diminishes expression of MuRF1 and atrogin-1 in denervated muscle and confers resistance to atrophy. Mice lacking histone deacetylases (HDACs) 4 and 5 in skeletal muscle fail to up-regulate myogenin and also preserve muscle mass following denervation. Conversely, forced expression of myogenin in skeletal muscle of HDAC mutant mice restores muscle atrophy following denervation. Thus, myogenin plays a dual role as both a regulator of muscle development and an inducer of neurogenic atrophy. These findings reveal a specific pathway for muscle wasting and potential therapeutic targets for this disorder.
INTRODUCTION
Maintenance of muscle mass depends on a balance between protein synthesis and degradation. Innervation of skeletal muscle fibers by motor neurons is essential for maintenance of muscle size, structure and function. Numerous disorders, including amyotrophic lateral sclerosis (ALS), Guillaine-Barre syndrome, polio and polyneuropathy disrupt the nerve supply to muscle, causing debilitating loss of muscle mass (referred to as neurogenic atrophy), and eventual paralysis.
Loss of the nerve supply to muscle fibers results in muscle atrophy mainly through excessive ubiquitin-mediated proteolysis via the proteasome pathway (Beehler et al., 2006). Other pathologic states and systemic disorders, including cancer, diabetes, fasting, sepsis, and disuse also cause muscle atrophy through ubiquitin-dependent proteolysis (Attaix et al., 2008; Attaix et al., 2005; Medina et al., 1995; Tawa et al., 1997). The muscle-specific E3 ubiquitin ligases MuRF1 (also called Trim63) and atrogin-1 (also called MAFbx or Fbxo32) are up-regulated during muscle atrophy and appear to represent final common mediators of this process (Bodine et al., 2001; Clarke et al., 2007; Gomes et al., 2001; Kedar et al., 2004; Lecker et al., 2004; Li et al., 2004; Li et al., 2007; Willis et al., 2009). However, the precise molecular mechanisms and signaling pathways that control the expression of these key regulators of muscle protein turnover have not been fully defined and it remains unclear whether all types of atrophic signals control these E3 ubiquitin ligase genes through the same or different mechanisms. Further understanding of the molecular pathways that regulate muscle mass is a prerequisite for the development of novel therapeutics to ameliorate muscle wasting disorders.
Myogenin is a bHLH transcription factor essential for skeletal muscle development (Hasty et al., 1993; Nabeshima et al., 1993). After birth, myogenin expression is down-regulated in skeletal muscle, but is re-induced in response to denervation (Merlie et al., 1994; Tang et al., 2008; Williams et al., 2009). Up-regulation of myogenin in denervated skeletal muscle promotes the expression of acetylcholine receptors and other components of the neuromuscular synapse (Merlie et al., 1994; Tang and Goldman, 2006; Williams et al., 2009). However, it has not been possible to address the potential involvement of myogenin in neurogenic atrophy because myogenin null mice die at birth due to failure in skeletal muscle differentiation (Hasty et al., 1993; Nabeshima et al., 1993).
Histone acetylation has been implicated in denervation-dependent changes in skeletal muscle gene expression and histone deacetylase (HDAC) inhibitors block the expression of myogenin in response to denervation (Tang and Goldman, 2006). In this regard, the class IIa HDACs, HDAC4 and HDAC5, which act as transcriptional repressors (Haberland et al., 2009; McKinsey et al., 2000; Potthoff et al., 2007), are up-regulated in skeletal muscle upon denervation and repress the expression of Dach2, a negative regulator of myogenin (Cohen et al., 2007; Tang et al., 2008).
To investigate the potential involvement of myogenin, HDAC4 and HDAC5 in neurogenic atrophy, we performed denervation experiments in mutant mice in which these transcriptional regulators were deleted in adult skeletal muscle. We show that adult mice lacking myogenin fail to up-regulate the E3 ubiquitin ligases MuRF1 and atrogin-1 following denervation and are resistant to neurogenic atrophy. We demonstrate that myogenin binds and activates the promoter regions of the MuRF1 and atrogin-1 genes, in vitro and in vivo. Similar to adult mice lacking myogenin, mice lacking Hdac4 and Hdac5 in skeletal muscle do not up-regulate myogenin following denervation and are resistant to muscle atrophy. Conversely, over-expression of myogenin in skeletal muscle is sufficient to up-regulate the expression of MuRF1 and atrogin-1 and promote neurogenic atrophy in mice lacking Hdac4 and Hdac5. These findings reveal a key role of myogenin and class IIa HDACs as mediators of neurogenic atrophy and potential therapeutic targets to treat this disorder.
RESULTS
Adult Mice Lacking Myogenin Are Resistant to Muscle Atrophy Upon Denervation
To bypass the requirement of myogenin for skeletal muscle development and investigate its functions in muscle of adult mice, we used a conditional myogenin null allele (Knapp et al., 2006), which could be deleted in adult muscle with a tamoxifen-regulated Cre recombinase transgene (Hayashi and McMahon, 2002; Knapp et al., 2006). Tamoxifen was administered to mice at 2 months of age and 89% deletion of the conditional myogenin allele occurred as measured by PCR genotyping from genomic DNA one week after tamoxifen injection (Figure S1 available online). Hereafter, we refer to these mice with deletion of myogenin during adulthood as Myog-/- mice.
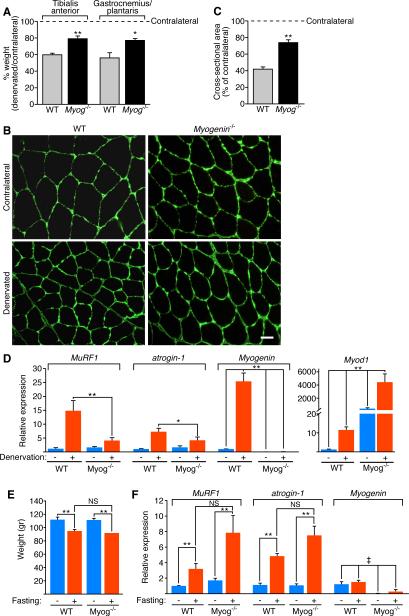
To examine the role of myogenin in denervated skeletal muscle, the sciatic nerve was severed one month following tamoxifen administration and muscle atrophy was assessed 14 days later by weighing denervated and contralateral tibialis anterior (TA) muscles. WT denervated TA showed approximately a 40% decrease in weight following denervation in comparison to the contralateral TA (Figure 1A). In contrast, denervated TA from Myog-/- mice showed a minimal decrease in muscle weight (~20%) compared to the contralateral TA (Figure 1A), suggesting that Myog-/- mice were partially resistant to muscle atrophy. Since we deleted myogenin in adult mice, muscle development and growth occurred normally prior to tamoxifen administration. As expected, the muscle weights of the non-denervated contralateral TA in Myog-/- and WT mice were similar (WT TA = 37.82 ± 0.87 mg; Myog-/- TA = 36.27 ± 0.54 mg; t test = 0.19). Comparable resistance to atrophy was observed in the gastrocnemius and plantaris (GP) weight of Myog-/- mice (Figure 1A).
Figure 1. Adult Mice Lacking Myogenin Are Resistant to Muscle Atrophy Upon Denervation.
(A) Percentage of TA or GP muscle weight of WT and Myog-/- mice 14 days after denervation, expressed relative to contralateral muscle. * p < 0.05 versus WT. ** p < 0.005 versus WT. n=4 for each sample. Data are represented as mean ± SEM.
(B) Immunostaining for laminin of contralateral and denervated TA of WT and Myog-/- mice, 14 days after denervation. Scale bar=20 microns.
(C) Morphometric analysis of contralateral and denervated TA of WT and Myog-/- mice, 14 days after denervation. Values indicate the mean of cross-sectional area of denervated TA fibers as a percentage of the contralateral fibers ± SEM. ** p < 0.005 versus WT. n=3 cross sections.
(D) Expression of MuRF1, atrogin-1, Myogenin and Myod1 in contralateral (-) and denervated (+) GP of WT and Myog-/- mice, 7 days after denervation, detected by real-time PCR. The values are normalized to WT contralateral GP. Data are represented as mean ± SEM. * p < 0.05; ** p < 0.005 versus WT. n=4 for each sample.
(E) Weight of GP muscle of WT and Myog-/- mice fed (-) or fasted (+) for 48 hours. Data are represented as mean ± SEM. ** p < 0.005 versus fed GP. NS=not significant. n=6 for each sample.
(F) Expression of MuRF1, atrogin-1 and Myogenin in fed (-) and 48 hour fasted (+) GP of WT and Myog-/- mice, detected by real-time PCR. The values are normalized to WT fed GP. Data are represented as mean ± SEM. ‡ p < 0.005 versus WT. ** p < 0.005 versus fed. NS=not significant. n=6 for each sample. See also Figure S1 and S2.
Immunostaining for laminin of TA cross-sections clearly delineated a decrease of muscle fiber size in the WT denervated TA in comparison to the contralateral muscle, indicative of muscle atrophy (Figure 1B). In contrast, the decrease in fiber size was less evident in the Myog-/- denervated TA (Figure 1B). Morphometric analysis of TA cross-sections highlighted a significant difference in myofiber size between WT and Myog-/- muscles following denervation, confirming the latter were resistant to muscle atrophy (Figure 1C).
As expected, seven days after denervation MuRF1 and atrogin-1 expression was dramatically up-regulated in the GP of denervated WT mice (Figure 1D). Remarkably, this up-regulation was significantly reduced in Myog-/- denervated GP (Figure 1D), suggesting that the lack of up-regulation of MuRF1 and atrogin-1 in denervated Myog-/- muscles was responsible for resistance to atrophy. Deletion of myogenin mRNA from adult Myog-/- muscle was confirmed by real-time PCR (Figure 1D). Of note, expression of MyoD (Myod1), another bHLH myogenic regulatory factor (Davis et al., 1987) was highly up-regulated in both the contralateral and denervated GP of the Myog-/- mice, seven days after denervation (Figure 1D). This data shows that myogenin does not regulate Myod1 expression following denervation. The dramatic up-regulation of MyoD following denervation of Myog-/- mice, which are resistant to atrophy, also argues against a major role of MyoD in promoting neurogenic atrophy. Accordingly, MyoD null mice are not resistant to muscle atrophy following denervation (Jason O'Rourke and E. Olson, unpublished data).
Denervation is known to affect skeletal myofiber composition (Herbison et al., 1979; Midrio et al., 1992; Nwoye et al., 1982; Patterson et al., 2006; Sandri et al., 2006; Sato et al., 2009). To determine whether the resistance to muscle atrophy observed in mice lacking myogenin was due to differences in fiber type composition, we performed fiber type analysis of soleus muscles two weeks after denervation. Our findings revealed no difference in fiber type composition between WT and Myog-/- mice (Figure S2). These findings suggest that myogenin, which is up-regulated following denervation, is required for maximal induction of E3 ubiquitin ligase genes and neurogenic atrophy.
We next tested whether myogenin was necessary for mediating other forms of atrophy, such as occurs in response to fasting. As shown in Figure 1E, the GP muscles of WT and Myog-/- mice displayed comparable loss in mass following a 48 hour fast. We observed the up-regulation of MuRF1 and atrogin-1 upon fasting in both WT and Myog-/- mice and validated the deletion of myogenin in Myog-/- mice (Figure 1F). These data clearly demonstrate that myogenin is not required for starvation atrophy, but rather is a specific mediator of neurogenic atrophy.
Myogenin Activates MuRF1 and Atrogin-1 Transcription
Since up-regulation of MuRF1 and atrogin-1 was impaired in Myog-/- mice, we analyzed the promoter regions of the MuRF1 and atrogin-1 genes for E-boxes (CANNTG) that might confer sensitivity to myogenin. Indeed, three E-boxes are located in the promoter of the MuRF1 gene, E1 (-143 bp), E2 (-66 bp) and E3 (-44 bp), and one conserved E-box is located 79 bp upstream of the atrogin-1 gene (Figure S3A). The E-boxes upstream of MuRF1 are contained in a genomic region near the binding site for FoxO transcription factors (Waddell et al., 2008), but several kilobases away from a region shown to be regulated by NFκB (Cai et al., 2004). The E-box upstream of atrogin-1 is embedded in a region containing multiple FoxO binding sites (Sandri et al., 2004).
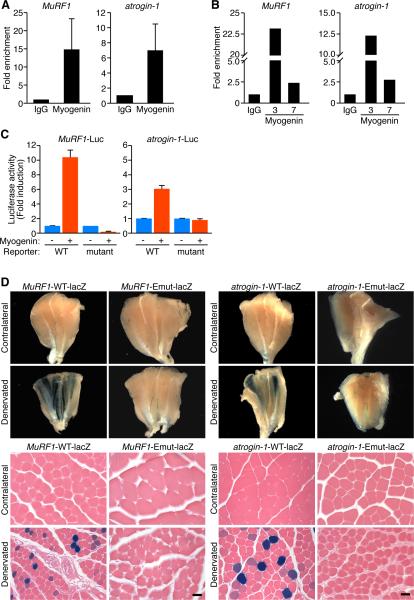
To confirm the binding of myogenin to the MuRF1 and atrogin-1 promoters, we performed chromatin immunoprecipitation (ChIP) assays using differentiated C2C12 myotubes, since myogenin expression correlates with MuRF1 and atrogin-1 expression during muscle cell differentiation (Figure S3B) (Spencer et al., 2000). After six days of differentiation, chromatin from C2C12 myotubes was immunoprecipitated with antibodies against myogenin or immunoglobulin G (IgG) as a control. Using primers flanking the E-boxes in the MuRF1 and atrogin-1 promoters, DNA was amplified by PCR (Figure 2A and Figure S3C). Clear enrichment of the corresponding promoter sequences in the DNA immunoprecipitated with antibodies against myogenin compared to IgG was indicative of myogenin binding to the endogenous MuRF1 and atrogin-1 promoters.
Figure 2. Myogenin Directly Regulates MuRF1 and Atrogin-1.
(A) ChIP assay performed in C2C12 myotubes showing myogenin binding to MuRF1 and atrogin-1 promoters. Chromatin was immunoprecipitated with antibodies against immunogloblulin G (IgG), or myogenin. Primers flanking the E-boxes on the MuRF1 and atrogin-1 promoters were used for amplifying DNA by real-time PCR. Values indicate the mean of fold enrichment over chromatin immunoprecipitated with antibodies against IgG ±SEM. n=3.
(B) ChIP assays performed using denervated TA muscle at 3 and 7 days following denervation show myogenin binding to the MuRF1 and atrogin-1 promoters. Values indicate the fold enrichment over chromatin immunoprecipitated with antibodies against IgG.
(C) Luciferase assays performed on cell extracts of C2C12 myoblasts transfected with luciferase reporter plasmids ligated to the WT (MuRF1-Luc) (atrogin-1-Luc), or the mutant constructs of MuRF1 and atrogin-1 genes, with myogenin (+) or empty (-) expression plasmid. Data are represented as mean ± SEM.
(D) β-galactosidase staining of contralateral and denervated GP muscles isolated from transgenic mice containing a lacZ transgene under the control of the WT (MuRF1-WT-lacZ) (atrogin-1-WT-lacZ) or the mutant (MuRF1-Emut-lacZ) (atrogin-1-Emut-lacZ) constructs of the MuRF1 or atrogin-1 promoters. Upper panels show whole muscles. Lower panels show H&E sections. Scale bar=20 microns. See also Figure S3.
We validated in vivo binding of myogenin to the endogenous MuRF1 and atrogin-1 promoters by performing ChIP assays using sonicated chromatin extracts from TA muscles harvested from mice at 3 days and 7 days after denervation (Figure 2B and Figure S3D). Direct binding of myogenin as a heterodimer with E12 proteins to the E-boxes E2 and E3 in the MuRF1 promoter and to the E-box in the atrogin-1 promoter was shown by gel mobility shift assays (Figure S3E).
We further tested the ability of myogenin to activate the MuRF1 and atrogin-1 promoter regions in vitro by constructing luciferase reporter plasmids containing the 600 bp genomic DNA fragment upstream of the MuRF1 gene (MuRF1-Luc) or 712 bp upstream of the atrogin-1 gene (atrogin-1-Luc) upstream of a luciferase reporter. Mutant versions of these promoter regions were generated by mutating the myogenin binding sites in the promoters. By transfecting C2C12 cells, activation of luciferase was detected in response to myogenin using the wild-type promoters (Figure 2C). This activation was blunted by mutation of the E-boxes in the promoters (Figure 2C), indicating that the MuRF1 and atrogin-1 promoter regions contain responsive myogenin binding sites. Similar results were obtained in transfected COS1 cells (Figure S3F).
To test the responsiveness of the E3 ligase gene promoters to atrophic signals in vivo, transgenic mice were generated harboring the same upstream regions of the genes ligated to a lacZ reporter (Kothary et al., 1989; Williams et al., 2009). Transgenic mice with the mutated versions of these promoter regions were also generated (MuRF1-Emut-lacZ and atrogin-1-Emut-lacZ). Seven days following denervation, β-galactosidase expression controlled by the wild-type promoters was up-regulated in denervated GP muscle fibers compared to the innervated contralateral leg muscles (Figure 2D). The expression of lacZ in only a subset of myofibers likely reflects the mosaicism of F0 transgenic mice and, perhaps, variable up-regulation of the E3 ubiquitin ligase genes in different myofibers in response to denervation (Moriscot et al., 2010). In contrast to the obvious up-regulation of the wild-type transgenes following denervation, mutation of the E-boxes in these promoters abrogated β-galactosidase expression, revealing an essential role for myogenin in denervation-dependent activation of MuRF1 and atrogin-1 in vivo (Figure 2D). These results show that the MuRF1 and atrogin-1 genes are targets of myogenin transcriptional activation in response to denervation.
Mice Null for Class II HDACs Are Resistant to Muscle Atrophy upon Denervation
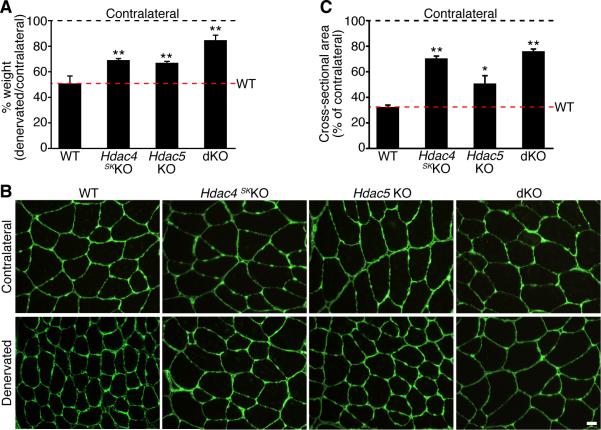
Previous studies showed that the class II HDACs, HDAC4 and HDAC5, are up-regulated in skeletal muscle in response to denervation (Bodine et al., 2001; Cohen et al., 2007; Tang et al., 2008) and are responsible for the repression of Dach2, a negative regulator of myogenin (Cohen et al., 2007; Tang et al., 2008). In light of the role of myogenin in promoting muscle atrophy, we hypothesized that mice lacking HDAC4 or HDAC5 in skeletal muscle would be resistant to atrophy following denervation owing to a block of myogenin expression via Dach2. Mice with global deletion of Hdac4 display lethal bone abnormalities (Vega et al., 2004), so we deleted Hdac4 specifically in skeletal muscle using a conditional allele and a myogenin-Cre transgene (Hdac4fl/fl; myog-Cre; hereafter referred to as Hdac4 skKO) (Potthoff et al., 2007). Deletion of HDAC4 in Hdac4 skKO GP was confirmed by Western blot analysis (Figure S4). Since mice null for Hdac5 do not display a phenotype (Chang et al., 2004), we used Hdac5-/- mice (hereafter referred to as Hdac5 KO) for these experiments. Fourteen days following denervation, WT denervated TA showed approximately a 50% decrease in weight in comparison to the contralateral TA (Figure 3A). In contrast, denervated TA muscles from Hdac4 skKO or Hdac5 KO mice showed a decrease of about 30% in muscle weight in comparison to the contralateral muscles (Figure 3A), suggesting that these mice were partially resistant to muscle atrophy. The weight of the contralateral TA was similar among the mice (data not shown).
Figure 3. HDAC4 and HDAC5 Redundantly Regulate Skeletal Muscle Atrophy.
(A) Percentage of TA muscle weight of mice of the indicated genotype 14 days after denervation, expressed relative to the contralateral muscle. Data are represented as mean ± SEM. ** p < 0.005 versus WT. n=5 for each sample.
(B) Immunostaining for laminin in contralateral and denervated TA of mice of the indicated genotype, 14 days after denervation. Scale bar=20 microns.
(C) Morphometric analysis of contralateral and denervated TA of indicated genotype, 14 days after denervation. Values indicate the mean of cross-sectional area of denervated TA fibers as a percentage of the contralateral fibers ± SEM. * p < 0.05 and ** p < 0.005 versus WT. n=3 cross sections. See also Figure S4 and S5.
HDAC4 and HDAC5 display functional redundancy in different tissues and in a variety of developmental and pathological settings (Backs et al., 2008; Haberland et al., 2009; Potthoff et al., 2007), so we generated double knockout (dKO) mice by crossing Hdac4 skKO with Hdac5 KO mice to further investigate the role of HDAC4 and HDAC5 in skeletal muscle atrophy. The dKO mice were viable and fertile and showed no obvious phenotype under normal conditions (data not shown). Strikingly, fourteen days after denervation, the TA of denervated dKO mice showed a decrease in weight of only ~10% compared to the contralateral TA (Figure 3A), revealing that the dKO mice were more resistant to muscle atrophy compared to Hdac4 skKO or Hdac5 KO mice. The weight of the contralateral TA was comparable among the mice (data not shown). Similar differences were also observed among GP muscles between WT and dKO mice (Figure S5).
Immunostaining for laminin 14 days after denervation clearly demonstrated that the denervated TA fibers from Hdac4 skKO and Hdac5 KO mice were larger than the denervated WT fibers and that the denervated TA from dKO mice had a minimal decrease in muscle fiber size compared to the contralateral dKO TA (Figure 3B). Morphometric analysis on TA sections revealed that while WT mice showed a reduction of ~70% in the myofiber cross-sectional area between denervated and contralateral TA, Hdac4 skKO denervated TA displayed ~30% reduction in myofiber cross-sectional area. Hdac5 KO denervated TA also showed a substantial reduction in myofiber area (~50%) when compared to the contralateral TA, whereas in dKO mice this reduction was only ~25% (Figure 3C). From these results, we conclude that HDAC4 and HDAC5 redundantly regulate skeletal muscle atrophy and mice lacking these HDACs in skeletal muscle are resistant to muscle atrophy upon denervation.
Aberrant Transcriptional Responses to Denervation in HDAC Mutant Mice
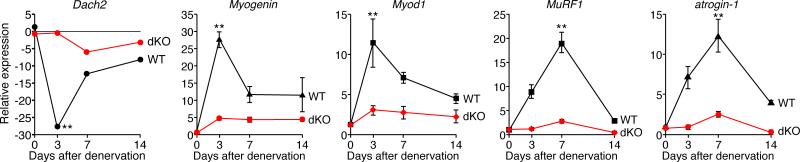
We compared the transcriptional responses to denervation in WT and dKO mice by real-time PCR analysis of denervation-responsive transcripts. As reported previously (Cohen et al., 2007; Tang et al., 2008), Dach2 expression was dramatically down-regulated upon denervation in WT mice. However, Dach2 was only modestly down-regulated in the dKO mice (Figure 4). Consistent with the repressive influence of Dach2 on myogenin expression, in WT mice myogenin and Myod1 were strongly up-regulated three days after denervation, as were MuRF1 and atrogin-1 (Figure 4). In contrast, neither myogenin nor Myod1 transcripts were up-regulated following denervation of dKO mice (Figure 4). The up-regulation of MuRF1 and atrogin-1 was also completely abolished in dKO denervated GP (Figure 4), suggesting that the lack of up-regulation of MuRF1 and atrogin-1 in denervated dKO muscles was in part responsible for resistance to atrophy.
Figure 4. dKO Mice Show Altered Gene Expression Upon Denervation.
Expression of the indicated mRNAs was detected by real-time PCR in WT and dKO denervated GP and normalized to the expression in the contralateral muscle. Data are represented as mean ± SEM. ** p < 0.005 versus dKO. n=6 for each time point.
Myogenin Over-expression in dKO Muscle Restores Neurogenic Atrophy
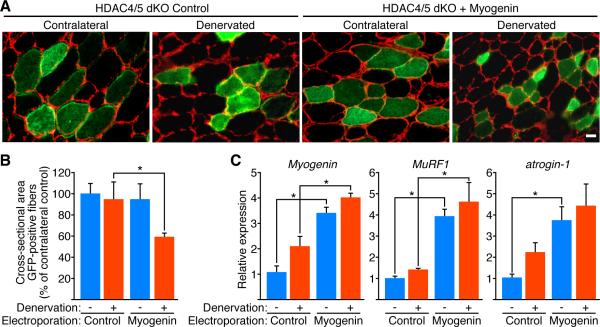
To examine whether forced expression of myogenin was sufficient to overcome the resistance of the dKO TA muscle to denervation-induced atrophy, we electroporated the TA of dKO mice with a myogenin expression plasmid or an empty expression plasmid. Gene delivery efficiency was monitored by co-electroporation with a GFP vector (Dona et al., 2003; Rana et al., 2004). Three days after electroporation, which is sufficient time for the electroporated plasmids to be expressed in skeletal muscle (Dona et al., 2003), we denervated one leg of the dKO mice by cutting the sciatic nerve; the TA muscles were harvested 10 days after denervation. As seen in Figure 5A, laminin immunostaining of dKO TA muscles clearly revealed a decrease in myofiber size in the denervated TA of dKO mice over-expressing myogenin compared to the denervated dKO TA electroporated with the control vector. Morphometric analysis performed on GFP-positive myofibers showed a significant decrease in the size of myofibers of the denervated dKO TA electroporated with myogenin versus control vector (Figure 5B). Real-time PCR analysis validated the over-expression of myogenin in electroporated TA muscle of dKO mice and showed an up-regulation of the expression of MuRF1 and atrogin-1 (Figure 5C), confirming the myogenin-dependent regulation of the E3 ubiquitin ligases.
Figure 5. Ectopic Expression of Myogenin Induces Muscle Atrophy in dKO Mice Following Denervation.
(A) Immunostaining for laminin (red) of cross-section of contralateral and denervated dKO TA electroporated with GFP expression plasmid and control plasmid (HDAC4/5 dKO Control) or GFP plasmid and myogenin (HDAC4/5 dKO + Myogenin), 10 days after denervation. Histology shows that the dKO denervated GFP-positive fibers co-electroporated with myogenin are smaller than denervated GFP-positive fibers co-electroporated with control plasmid. Scale bar=20 microns.
(B) Morphometric analysis performed on GFP-positive fibers of contralateral (-) and denervated (+) dKO TA muscles electroporated with GFP expression plasmid and control plasmid (Control) or GFP plasmid and myogenin (Myogenin), 10 days after denervation. Values indicate the mean of cross-sectional area of GFP-positive muscle fibers as a percentage of the contralateral control fibers ± SEM. * p < 0.05 versus Control. n=7 for each condition.
(C) Expression of Myogenin, MuRF1 and atrogin-1 in contralateral (-) and denervated (+) dKO TA muscles electroporated with GFP plasmid and a control plasmid (Control) or GFP plasmid and myogenin (Myogenin), 10 days after denervation. Values are normalized to the expression in the contralateral control muscles. Data are represented as mean ± SEM. * p < 0.05 versus Control. n=3 for each sample. See also Figure S6.
The potential role of myogenin in driving muscle atrophy was further investigated by over-expressing myogenin in the TA muscle of WT mice. Morphometric analysis performed on GFP-positive myofibers showed no significant size difference between myofibers electroporated with control or myogenin expression plasmid (Figure S6A and S6B). Real-time PCR analysis validated the over-expression of myogenin in electroporated TA muscle of WT mice and showed an up-regulation of the expression of MuRF1 and atrogin-1 (Figure S6C). Taken together, these findings demonstrate that over-expression of myogenin is necessary but not sufficient to induce muscle atrophy.
DISCUSSION
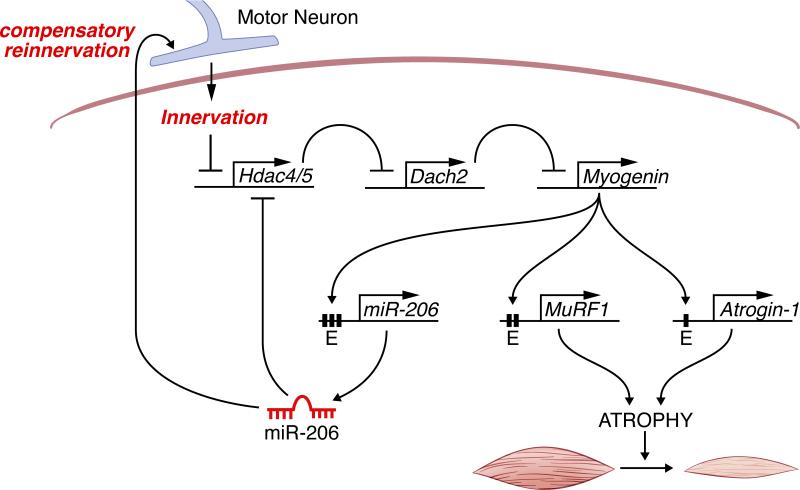
The results of this study demonstrate a key role of myogenin, well known for its function as an essential regulator of myogenesis, in controlling neurogenic atrophy. Myogenin promotes muscle atrophy upon denervation by directly activating the expression of MuRF1 and atrogin-1, which encode E3 ubiquitin ligases responsible for muscle proteolysis. Up-regulation of myogenin in response to denervation is controlled by a transcriptional pathway in which HDAC4 and 5 are initially induced and, in turn, repress the expression of Dach2 (Tang and Goldman, 2006), a negative regulator of myogenin (Figure 6).
Figure 6. Model for Neurogenic Atrophy.
Denervation of skeletal muscle results in the up-regulation of HDAC4 and HDAC5, which represses Dach2, a negative regulator of myogenin, resulting in myogenin expression. Myogenin activates the expression of MuRF1 and atrogin-1, two E3 ubiquitin ligases that participate in the proteolytic pathway resulting in muscle atrophy. Myogenin also regulates miR-206, which establishes a negative feedback loop to repress HDAC4 expression and promote reinnervation.
It is generally accepted that muscle atrophy occurs when proteolysis exceeds protein synthesis (Eley and Tisdale, 2007; Glass, 2003; Mammucari et al., 2008; Sandri et al., 2004). Up-regulation of myogenin in response to denervation has been proposed as an adaptive mechanism to prevent muscle atrophy (Hyatt et al., 2003; Ishido et al., 2004). On the contrary, we demonstrate here that myogenin directly regulates MuRF1 and atrogin-1, which promote the loss of muscle mass in response to denervation, revealing a mechanistic basis for neurogenic muscle atrophy and a previously unrecognized function for myogenin in this pathological process.
Recently, we showed that microRNA (miR) 206 is also up-regulated in denervated skeletal muscle via a series of conserved E-boxes that bind myogenin (Williams et al., 2009). miR-206, in turn, represses expression of HDAC4 and controls a retrograde signaling pathway that promotes reinnervation of denervated myofibers (Figure 6). Thus, skeletal muscle responds to denervation by activating an elaborate network of transcriptional and epigenetic pathways, involving positive and negative feedback loops, which modulate nerve-muscle interactions and muscle growth and function (Figure 6).
Dual Roles of Myogenin in Muscle Development and Atrophy
Our findings reveal the gene regulatory circuitry for muscle development is redeployed in adulthood to control aspects of muscle disease and stress responsiveness. Thus, myogenin can exert opposing effects on skeletal muscle – either promoting differentiation or degradation – depending on the developmental or pathological setting. These contrasting activities of myogenin likely reflect differential modulation by signaling pathways and cofactors that enable myogenin to regulate distinct sets of target genes.
Similar to myogenin, Dach2 is a transcription factor involved in both muscle development and muscle atrophy. Dach2 is expressed in the developing somites prior to the onset of myogenesis and has been shown to regulate myogenic specification by interacting with the Eya2 and Six1 transcription factors (Heanue et al., 1999; Kardon et al., 2002). Indeed, Dach proteins are required for activation of Six1 targets (Li et al., 2003), suggesting a possible role of Dach proteins in the Six1-mediated regulation of muscle development (Laclef et al., 2003) or fiber type specification (Grifone et al., 2004). Following denervation, Dach2 plays a role in connecting neuronal activity with myogenin expression (Cohen et al., 2007; Tang and Goldman, 2006; Tang et al., 2008).
The finding that forced expression of myogenin in HDAC4/5 mutant mice is sufficient to restore muscle atrophy following denervation indicates that myogenin is a key downstream mediator of the pro-atrophic functions of these HDACs. It is noteworthy, however, the blockade to muscle atrophy and E3 ligase expression imposed by the combined deletion of HDACs 4 and 5 is more pronounced than in Myog-/- mice. This suggests the existence of additional downstream targets of these HDACs that promote neurogenic atrophy. We also note that forced over-expression of myogenin in innervated skeletal muscle was not sufficient to induce muscle atrophy (Figure S6) (Hughes et al., 1999). These findings indicate that myogenin is necessary, but not sufficient, to regulate the genetic program for muscle atrophy and imply the existence of additional denervation-dependent signals that potentiate the ability of myogenin to promote atrophy.
MyoD, like myogenin, is up-regulated in response to denervation (Figure 4 and (Charge et al., 2008; Hyatt et al., 2003; Ishido et al., 2004). In Myog-/- mice, Myod1 expression is dramatically elevated compared to WT muscles, which express low levels of Myod1, and in response to denervation is furthermore induced (Figure 1D). The observation that MyoD null mice are not resistant to muscle atrophy following denervation (Jason O'Rourke and E. Olson, unpublished data) demonstrates a negligible role for MyoD in neurogenic atrophy and points to myogenin as the major myogenic bHLH factor involved in this process. This is consistent with the finding that although MyoD and myogenin bind the same DNA consensus sequences, they regulate distinct sets of target genes (Blais et al., 2005; Cao et al., 2006).
A Myogenin-Dependent Transcriptional Pathway for Muscle Atrophy
We show both in vivo using denervated muscles and in vitro using differentiated C2C12 cells, that myogenin binds the endogenous MuRF1 and atrogin-1 promoters. We observed a decrease in myogenin expression and binding to these E3 ubiquitin ligase promoters between days 3 and day 7 after denervation (Figure 4 and 2B), suggesting an especially important role of myogenin in triggering the transcriptional cascade leading to atrophy. Consistent with our finding that myogenin regulates MuRF1 and atrogin-1 expression, these E3 ubiquitin ligases are up-regulated upon C2C12 differentiation (Figure S3B; (Spencer et al., 2000)), a process known to be regulated by myogenin. While it is well established that MuRF1 and atrogin-1 function in driving skeletal muscle atrophy (Bodine et al., 2001; Clarke et al., 2007; Gomes et al., 2001; Kedar et al., 2004; Lecker et al., 2004; Li et al., 2004; Li et al., 2007; Willis et al., 2009), their potential roles in myogenesis have not been explored. Considering the important role of ubiquitination in regulating proteolysis, endocytosis, signal transduction (Hicke, 2001) and transcription (Salghetti et al., 2001), it will be interesting to investigate the potential involvement of MuRF1 and atrogin-1 in muscle development and regeneration.
Therapeutic Implications
Numerous disorders, including motor neuron disease, fasting, cancer cachexia, and sarcopenia cause muscle atrophy and the E3 ubiquitin ligase genes are thought to function as final common mediator of different atrophic stimuli. Myogenin is up-regulated upon denervation and spinal cord isolation (Hyatt et al., 2003), but is not induced in response to other forms of atrophy such as fasting, cancer cachexia or diabetes (Lecker et al., 2004; Sacheck et al., 2007). In this regard, we have found that Myog-/- mice display a normal loss of skeletal muscle mass in response to fasting, further demonstrating that myogenin is dedicated to neurogenic atrophy and sensing the state of motor innervation. The fact that MuRF1 and atrogin-1 are up-regulated in other atrophy conditions in the absence of myogenin up-regulation (Lecker et al., 2004; Sacheck et al., 2007) strongly suggests that other transcription factors known to regulate the expression of these ubiquitin ligases, such as the FoxO family or NFκB (Bodine et al., 2001; Sandri et al., 2004; Waddell et al., 2008), play a role in driving muscle atrophy in a myogenin-independent manner.
Our finding that myogenin, in addition to HDAC4 and HDAC5, acts as a regulator of neurogenic muscle atrophy through the activation of E3 ubiquitin ligases provides a new perspective on potential therapies for muscle wasting disorders. Class II HDACs are regulated by a variety of calcium-dependent signaling pathways that control their nuclear export through signal-dependent phosphorylation (Backs et al., 2008; McKinsey et al., 2000). In a pathological condition such as muscle denervation, HDAC4 and HDAC5 are up-regulated, shuttle into the myonuclei adjacent to neuromuscular junctions (Cohen et al., 2007) and are critical regulators of muscle atrophy. Modulation of the activity of class II HDACs, through pharmacologic inhibition compatible with the maintenance of steady state transcription of genes regulated by class II HDACs, may represent a new strategy for ameliorating muscle atrophy following denervation.
EXPERIMENTAL PROCEDURES
Mouse Lines
Mice used in this study are described in Supplemental Information.
Denervation
In anaesthetized adult mice, the sciatic nerve of the left leg was cut and a 3 mm piece was excised. The right leg remained innervated and was used as control. Mice were sacrificed after 3, 7, 10 or 14 days.
DNA Delivery by Electroporation
For gene delivery by electroporation, adult dKO mice were anesthetized; TA muscles exposed, injected with 30 μg of DNA in a solution of 5% mannitol and immediately subjected to electroporation. Electroporation was performed by delivering ten electric pulses of 20 V each (five with one polarity followed by five with inverted polarity). A pair of 3 × 5 mm Genepaddle electrodes (BTX, San Diego, CA) placed on opposite sides of the muscle was used to deliver the electric pulses. pCMV-Snap25-GFP (provided by Tullio Pozzan, University of Padua, Padua, Italy) was used in a 1:1 ratio with pcDNA3.1 (Invitrogen) or EMSV-myogenin plasmid (Rana et al., 2004).
Immunohistochemistry
Cryosections of TA or soleus were fixed in 4% paraformaldehyde in PBS for 10 minutes at 4°C and washed in PBS. After incubating 30 minutes with 0.1% Triton X100 in PBS, the samples were fixed for 1h in 15% goat serum in PBS supplemented with M.O.M. Mouse IgG blocking reagent (Vector Laboratories) (BB) at room temperature. Primary antibodies were incubated overnight at 4°C (1:100 dilution of rabbit polyclonal anti-laminin antibody; 1:16000 anti-type I myosin heavy chain (MHC) (Sigma). Primary antibodies were detected by Alexa Fluor-488 or -555 goat anti-rabbit antibody (Invitrogen) diluted 1:800 in BB. DAB staining (Vector Laboratories) was used on soleus muscle for detecting type I MHC. Soleus muscles were used for metachromatic ATPase staining as previously described (Ogilvie and Feeback, 1990). Staining of transgenic lines positive for β-galactosidase was performed on GP muscles, as previously described (Williams et al., 2009).
Morphometric Analysis
Myofiber area was assessed on TA cryosections using ImageJ software (http://rsb.info.nih.gov/ij/) (NIH). Three H&E stained cross sections from three different mice for each genotype were analyzed. Between 100 to 350 GFP-positive fibers were analyzed for each electroporated TA muscle. The values are calculated as the percentage of the average of the cross-sectional area of each TA over the average cross-sectional area of the contralateral TA fibers.
RNA Isolation and RT-PCR
Total RNA was isolated from GP muscles using Trizol reagent (Invitrogen) following the manufacturer's instructions. Three micrograms of RNA was converted to cDNA using random primers and Superscript III reverse transcriptase (Invitrogen). Gene expression was assessed using real-time PCR with the ABI PRISM 7000 sequence detection system and TaqMan or with SYBR green Master Mix reagents (Applied Biosystems). Real-time PCR values were normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH). A list of Taqman probes and Sybr Green primers are available in the Supplemental Information.
Plasmid Constructs
A list of the plasmids used in this study is available in the Supplemental Information.
Cell Culture
COS cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). C2C12 myoblasts were grown in DMEM supplemented with 20% FBS and antibiotics and differentiated in DMEM supplemented with 2% horse serum and antibiotics.
Chromatin Immunoprecipitation Assay
ChIP assays were performed using C2C12 myotubes at day six of differentiation or using TA muscles three and seven days after denervation with the ChIP assay kit (Upstate) following the manufacturer's instructions. Chromatin was immunoprecipitated with antibodies against immunogloblulin G (Sigma) or myogenin (M-225; Santa Cruz). The sequences of the ChIP primers are available in the Supplemental Information.
Luciferase assay
C2C12 transfections were performed using Lipofectamine 2000 (Invitrogen) as previously described (Mercer et al., 2005). COS cells were plated and transfected 12 hours later using FuGENE (Roche Applied Science) following the manufacturer's instructions. The MuRF1 and atrogin-1 reporter plasmid cloning strategy is described in the Supplemental Information. Luciferase assays were performed with the Luciferase Assay kit (Promega) according to the manufacturer's instructions.
Site-Directed Mutagenesis
Mutations were introduced into E-boxes E2 and E3 of the MuRF1 promoter region and in the E-box of the atrogin-1 promoter by using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene). The same E-box mutations as those used in electrophoretic mobility shift assays were introduced within each E-box site in the promoters.
Statistical Analysis
Data are presented as mean ± SEM. Statistical significance was determined using two-tailed t-test with a significance level minor of 0.05.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marco Sandri for scientific input; Cheryl Nolen and Svetlana Bezprozvannaya for technical assistance; Jose Cabrera for graphics and Jennifer Brown for editorial assistance. Work in the laboratory of E.N.O. was supported by grants from NIH and the Robert A. Welch Foundation. W.H.K. was supported by a grant from the Muscular Dystrophy Association and Robert A. Welch Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- Myogenin, a key regulator of myogenesis controls muscle atrophy upon denervation
- Adult mice lacking myogenin are resistant to muscle atrophy upon denervation
- Myogenin transcriptionally activates E3 ubiquitin ligases which promote atrophy
- Mice lacking histone deacetylases 4 and 5 mimic mice lacking myogenin
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and supplemental experimental procedures can be found with this article online.
REFERENCES
- Attaix D, Combaret L, Bechet D, Taillandier D. Role of the ubiquitin-proteasome pathway in muscle atrophy in cachexia. Curr. Opin. Support Palliat. Care. 2008;2:262–266. doi: 10.1097/spc.0b013e3283196ac2. [DOI] [PubMed] [Google Scholar]
- Attaix D, Ventadour S, Codran A, Bechet D, Taillandier D, Combaret L. The ubiquitin-proteasome system and skeletal muscle wasting. Essays Biochem. 2005;41:173–186. doi: 10.1042/EB0410173. [DOI] [PubMed] [Google Scholar]
- Backs J, Backs T, Bezprozvannaya S, McKinsey TA, Olson EN. Histone deacetylase 5 acquires calcium/calmodulin-dependent kinase II responsiveness by oligomerization with histone deacetylase 4. Mol. Cell. Biol. 2008;28:3437–3445. doi: 10.1128/MCB.01611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beehler BC, Sleph PG, Benmassaoud L, Grover GJ. Reduction of skeletal muscle atrophy by a proteasome inhibitor in a rat model of denervation. Exp. Biol. Med. 2006;231:335–341. doi: 10.1177/153537020623100315. [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Kluger Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19:553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Cai D, Frantz JD, Tawa NE, Jr., Melendez PA, Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ, et al. IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 2004;24:8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charge SB, Brack AS, Bayol SA, Hughes SM. MyoD- and nerve-dependent maintenance of MyoD expression in mature muscle fibres acts through the DRR/PRR element. BMC Dev. Biol. 2008;8:5–18. doi: 10.1186/1471-213X-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, et al. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6:376–385. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- Cohen TJ, Waddell DS, Barrientos T, Lu Z, Feng G, Cox GA, Bodine SC, Yao TP. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J. Biol. Chem. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Dona M, Sandri M, Rossini K, Dell'Aica I, Podhorska-Okolow M, Carraro U. Functional in vivo gene transfer into the myofibers of adult skeletal muscle. Biochem. Biophys. Res. Commun. 2003;312:1132–1138. doi: 10.1016/j.bbrc.2003.11.032. [DOI] [PubMed] [Google Scholar]
- Eley HL, Tisdale MJ. Skeletal muscle atrophy, a link between depression of protein synthesis and increase in degradation. J. Biol. Chem. 2007;282:7087–7097. doi: 10.1074/jbc.M610378200. [DOI] [PubMed] [Google Scholar]
- Glass DJ. Molecular mechanisms modulating muscle mass. Trends Mol. Med. 2003;9:344–350. doi: 10.1016/s1471-4914(03)00138-2. [DOI] [PubMed] [Google Scholar]
- Gomes MD, Lecker SH, Jagoe RT, Navon A, Goldberg AL. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R, Laclef C, Spitz F, Lopez S, Demignon J, Guidotti JE, Kawakami K, Xu PX, Kelly R, Petrof BJ, et al. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol. Cell. Biol. 2004;24:6253–6267. doi: 10.1128/MCB.24.14.6253-6267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Heanue TA, Reshef R, Davis RJ, Mardon G, Oliver G, Tomarev S, Lassar AB, Tabin CJ. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13:3231–3243. doi: 10.1101/gad.13.24.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbison GJ, Jaweed MM, Ditunno JF. Muscle atrophy in rats following denervation, casting, inflammation, and tenotomy. Arch. Phys. Med. Rehabil. 1979;60:401–404. [PubMed] [Google Scholar]
- Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106:527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Chi MM, Lowry OH, Gundersen K. Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. J. Cell Biol. 1999;145:633–642. doi: 10.1083/jcb.145.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt JP, Roy RR, Baldwin KM, Edgerton VR. Nerve activity-independent regulation of skeletal muscle atrophy: role of MyoD and myogenin in satellite cells and myonuclei. Am. J. Physiol. Cell. Physiol. 2003;285:C1161–1173. doi: 10.1152/ajpcell.00128.2003. [DOI] [PubMed] [Google Scholar]
- Ishido M, Kami K, Masuhara M. In vivo expression patterns of MyoD, p21, and Rb proteins in myonuclei and satellite cells of denervated rat skeletal muscle. Am. J. Physiol. Cell. Physiol. 2004;287:C484–493. doi: 10.1152/ajpcell.00080.2004. [DOI] [PubMed] [Google Scholar]
- Kardon G, Heanue TA, Tabin CJ. Pax3 and Dach2 positive regulation in the developing somite. Dev. Dyn. 2002;224:350–355. doi: 10.1002/dvdy.10107. [DOI] [PubMed] [Google Scholar]
- Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc. Natl. Acad. Sci. USA. 2004;101:18135–18140. doi: 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp JR, Davie JK, Myer A, Meadows E, Olson EN, Klein WH. Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development. 2006;133:601–610. doi: 10.1242/dev.02249. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Laclef C, Hamard G, Demignon J, Souil E, Houbron C, Maire P. Altered myogenesis in Six1-deficient mice. Development. 2003;130:2239–2252. doi: 10.1242/dev.00440. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, Patterson C. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Invest. 2004;114:1058–1071. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, Patterson C. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J. Clin. Invest. 2007;117:3211–3223. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Schiaffino S, Sandri M. Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy. 2008;4:524–526. doi: 10.4161/auto.5905. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Wing SS, Goldberg AL. Increase in levels of polyubiquitin and proteasome mRNA in skeletal muscle during starvation and denervation atrophy. Biochem. J. 1995;307(Pt 3):631–637. doi: 10.1042/bj3070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer SE, Ewton DZ, Deng X, Lim S, Mazur TR, Friedman E. Mirk/Dyrk1B mediates survival during the differentiation of C2C12 myoblasts. J. Biol. Chem. 2005;280:25788–25801. doi: 10.1074/jbc.M413594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlie JP, Mudd J, Cheng TC, Olson EN. Myogenin and acetylcholine receptor alpha gene promoters mediate transcriptional regulation in response to motor innervation. J. Biol. Chem. 1994;269:2461–2467. [PubMed] [Google Scholar]
- Midrio M, Danieli-Betto D, Megighian A, Velussi C, Catani C, Carraro U. Slow-to-fast transformation of denervated soleus muscle of the rat, in the presence of an antifibrillatory drug. Pflugers Arch. 1992;420:446–450. doi: 10.1007/BF00374618. [DOI] [PubMed] [Google Scholar]
- Moriscot AS, Baptista IL, Bogomolovas J, Witt C, Hirner S, Granzier H, Labeit S. MuRF1 is a muscle fiber-type II associated factor and together with MuRF2 regulates type-II fiber trophicity and maintenance. J. Struct. Biol. 2010;170:344–353. doi: 10.1016/j.jsb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Nwoye L, Mommaerts WF, Simpson DR, Seraydarian K, Marusich M. Evidence for a direct action of thyroid hormone in specifying muscle properties. Am. J. Physiol. 1982;242:R401–408. doi: 10.1152/ajpregu.1982.242.3.R401. [DOI] [PubMed] [Google Scholar]
- Ogilvie RW, Feeback DL. A metachromatic dye-ATPase method for the simultaneous identification of skeletal muscle fiber types I, IIA, IIB and IIC. Stain Technol. 1990;65:231–241. doi: 10.3109/10520299009105613. [DOI] [PubMed] [Google Scholar]
- Patterson MF, Stephenson GM, Stephenson DG. Denervation produces different single fiber phenotypes in fast- and slow-twitch hindlimb muscles of the rat. Am. J. Physiol. Cell. Physiol. 2006;291:C518–528. doi: 10.1152/ajpcell.00013.2006. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Wu H, Arnold MA, Shelton JM, Backs J, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana ZA, Ekmark M, Gundersen K. Coexpression after electroporation of plasmid mixtures into muscle in vivo. Acta Physiol. Scand. 2004;181:233–238. doi: 10.1111/j.1365-201X.2004.01282.x. [DOI] [PubMed] [Google Scholar]
- Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, Lecker SH, Goldberg AL. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- Salghetti SE, Caudy AA, Chenoweth JG, Tansey WP. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc. Natl. Acad. Sci. USA. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Shimizu M, Mizunoya W, Wariishi H, Tatsumi R, Buchman VL, Ikeuchi Y. Differential expression of sarcoplasmic and myofibrillar proteins of rat soleus muscle during denervation atrophy. Biosci. Biotechnol. Biochem. 2009;73:1748–1756. doi: 10.1271/bbb.90085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JA, Eliazer S, Ilaria RL, Jr., Richardson JA, Olson EN. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J. Cell Biol. 2000;150:771–784. doi: 10.1083/jcb.150.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Goldman D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc. Natl. Acad. Sci. USA. 2006;103:16977–16982. doi: 10.1073/pnas.0601565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Macpherson P, Marvin M, Meadows E, Klein WH, Yang XJ, Goldman D. A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol. Biol. Cell. 2008;20:1120–1131. doi: 10.1091/mbc.E08-07-0759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawa NE, Jr., Odessey R, Goldberg AL. Inhibitors of the proteasome reduce the accelerated proteolysis in atrophying rat skeletal muscles. J. Clin. Invest. 1997;100:197–203. doi: 10.1172/JCI119513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega RB, Matsuda K, Oh J, Barbosa AC, Yang X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Waddell DS, Baehr LM, van den Brandt J, Johnsen SA, Reichardt HM, Furlow JD, Bodine SC. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am. J. Physiol. Endocrinol. Metab. 2008;295:E785–797. doi: 10.1152/ajpendo.00646.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AH, Valdez G, Moresi V, Qi X, McAnally J, Elliott JL, Bassel-Duby R, Sanes JR, Olson EN. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Rojas M, Li L, Selzman CH, Tang RH, Stansfield WE, Rodriguez JE, Glass DJ, Patterson C. Muscle ring finger 1 mediates cardiac atrophy in vivo. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H997–H1006. doi: 10.1152/ajpheart.00660.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.