Abstract
Filopodia are finger-like protrusive structures, containing actin bundles. By incubating frog egg extracts with supported lipid bilayers containing phosphatidylinositol(4,5)bisphosphate, we have reconstituted the assembly of filopodia-like structures (FLSs). The actin assembles into parallel bundles and known filopodial components localize to the tip and shaft. The filopodia tip complexes self-organize—they are not templated by preexisting membrane microdomains. The F-BAR domain protein toca-1 recruits N-WASP, followed by the Arp2/3 complex and actin. Elongation proteins, Diaphanous-related formin, VASP and fascin are recruited subsequently. Although the Arp2/3 complex is required for FLS initiation, it is not essential for elongation, which involves formins. We propose that filopodia form via clustering of Arp 2/3 complex activators, self-assembly of filopodial tip complexes on the membrane, and outgrowth of actin bundles.
Actin assembly is largely responsible for cell shape and movement in eukaryotic cells (1). Actin filaments underlie a range of morphological structures, among these, filopodia, which contain closely apposed bundled parallel actin filaments, are of particular interest (2, 3). They have numerous biological roles but their mechanism of formation is poorly understood. The persistence of actin growth in a parallel bundle in filopodia suggests roles for proteins with anti-capping and/or processive elongation activity (e.g. VASP and formins) (4–8). But, formins are also important in lamellipodia (9). The role of the N-WASP/Arp2/3 complex pathway in filopodia formation is also contentious (8, 10–14). Although there are in vitro models of actin bundling (15–17), no current experimental system recapitulates two physiological aspects of filopodia: (1) spontaneous formation of actin bundles in the presence of capping activity and (2) the assembly of a membrane-localized tip complex.
To investigate actin polymerization at membranes and to exploit advanced microscopic techniques like TIRF microscopy, we have replaced liposomes as the source of lipids with supported lipid bilayers (18). Purified Cdc42.GTPγS, N-WASP-WIP, toca-1, Arp2/3 complex, and actin comprise a minimal set of proteins for stimulation of actin nucleation by PI(4,5)P2-containing liposomes in vitro (19). When these proteins are supplied to the supported bilayer, a thin actin layer is formed at the membrane surface (Fig. 1A and B). When we substituted concentrated frog egg extracts for the purified proteins, unusual dense, focal and long actin structures, with a diameter of 0.3–1.5 µm, rose from the surface of the bilayer (Fig. 1C–E, fig. S1A, Supporting Videos 1–4). We term these filopodia-like structures (FLSs).
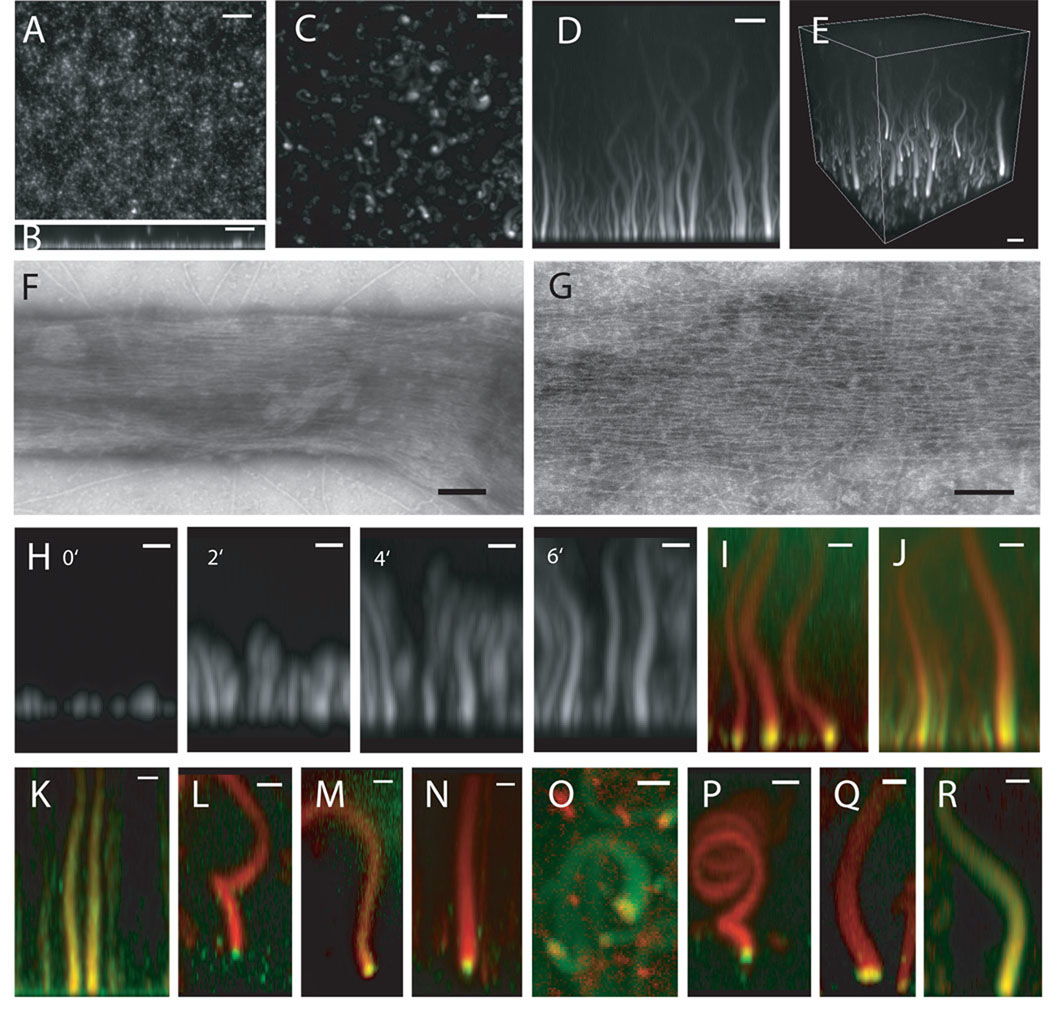
Figure 1. Reconstitution of filopodia-like structures on supported lipid bilayers containing 45% PC (Phosphatidylcholine), 45% PI (Phosphatidylinositol), 10% PI(4,5)P2.
(A) Confocal microscopy of supported bilayers with Cdc42, N-WASP-WIP, toca-1, Arp2/3 complex, and actin generates uniform, short polymerized actin. (B) Z-stack reconstruction of (A) showing growth of actin in the z-axis. (C) Confocal microscopy of supported lipid bilayers with Xenopus egg extracts and Alexa647-actin (seen in x–y) shows growth of focal actin structures from the bilayer in the z-axis. (D) Z-stack reconstruction of x–z shows the height of the actin structures. (E) 3D reconstruction. Bars: 5 µm. (F–G) Negative-stain electron microscopy of the phalloidin-stabilized actin structures shows that these are made of bundled, unbranched actin filaments Bars: 100 nm (H) Time-lapse sequence of FLS formation seen. Bars: 2 µm. (I and J) Pulse-chase experiments show that actin polymerization occurs at membrane. The reaction is started with Alexa 647-actin (red) and chased by Alexa 488-actin (green). Bars: 2 µm. (I) 1 min timepoint. (J) 2 min 40 sec timepoint. (K) Immunostained FLSs from the side, green: fascin immunostain, red: Alexa568-phalloidin (L) 60 degree tilt from x–y, green: Drf1 immunostain, red: Alexa568-phalloidin (M) 60 degree tilt from x–y, green: GFP-VASP, red: Alexa 647-actin (N) 60 degree tilt from x–y, green: profilin immunostain, red: Alexa568-phalloidin (O) top view, red: mCherry-Cdc42, green: Alexa 488-actin (P) 60 degree tilt from x–y, green: N-WASP immunostain, red: Alexa568-phalloidin (Q) tilted view, green: GFP-toca-1, red: Alexa 647-actin (R) side view, red: Alexa 568-Arp2/3 complex, green: Alexa 647-actin. Bars: 2 µm.
By electron microscopy using negative stain, we observe bundled actin filaments in the FLSs, which are characteristic of filopodia and distinct from the dendritic networks made by Listeria or ActA beads (Fig. 1F and G). The parallel alignment of actin filaments is revealed, when smaller FLSs spread out two dimensionally (Fig. 1G). We observe actin bundles of the complete size range seen by light microscopy (fig. S1B). Prefixing the FLSs produces similar actin bundles (fig. S1C).
If FLSs recapitulate filopodia, we would predict that new actin monomers would be added at the membrane-localized tip with dynamics similar to filopodial growth in vivo (20, 21). Time-lapse confocal imaging and z-stack reconstructions show that the typical initial rate of FLS growth is 2.5 µm/min, within the range of filopodia (Fig. 1H and Supporting Videos 1–4). A pulse-chase experiment starting actin growth with Alexa 647-actin and adding Alexa 488-actin after 20 min shows that actin polymerization occurs at the membrane-localized tip (Fig. 1I and J, fig. S1D–G). Actin monomers are added at 2.8 µm/min (fig. S1H). Growth of the bundles into the membrane is prevented by the underlying glass support; hence assembly is constrained to occur away from the membrane. With this difference, the assembly of bundled and parallel actin filaments at the membrane recapitulates filopodial formation and growth.
For filopodia in vivo a distinct collection of proteins is observed by electron microscopy and is termed the tip complex (5, 20). To determine if such a structure is present in the FLSs, we have immunostained for known filopodial components or added fluorescently tagged proteins. Like filopodia, FLSs immunostain for fascin along the length of the structure (22) (Fig. 1K and antibody specificity is shown in fig. S1I). The tip of the FLSs contains proteins that localize to the tips of filopodia - diaphanous related formins, VASP, profilin, N-WASP (5, 8, 23–26) (Fig. 1L, M, N, P). Cdc42 and toca-1 also localize to the tip (Fig. 1O and Q). Alexa-568 labeled Arp2/3 complex is enriched at the tip and also decorates the actin shaft (Fig. 1R). In some filopodia Arp2/3 complex is excluded from the shaft, in others it is present (5, 27).
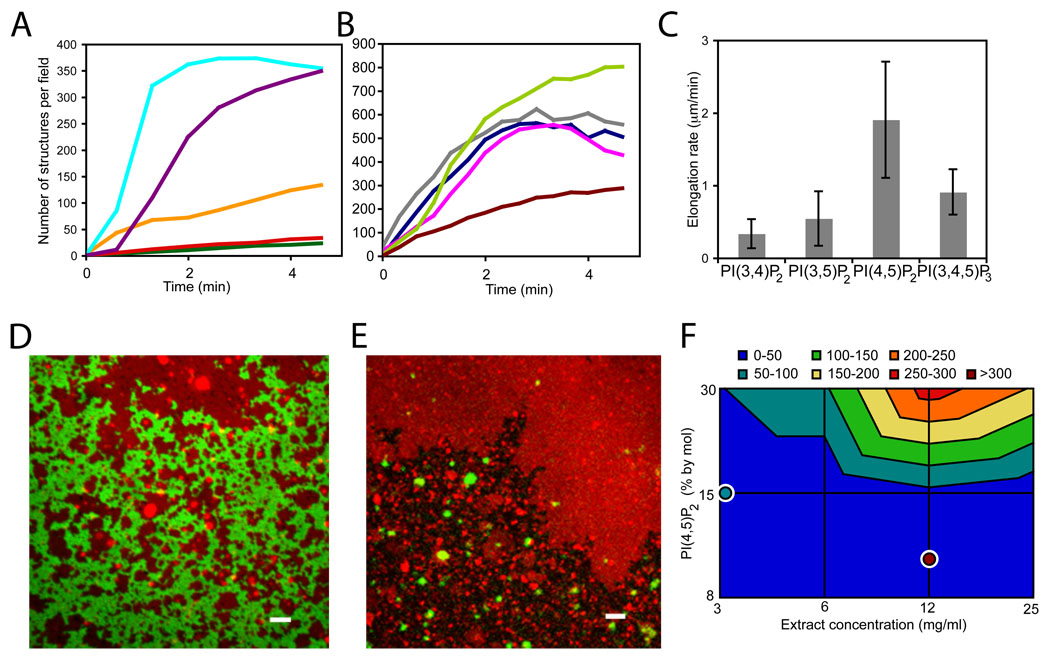
Tip complex assembly requires negatively-charged membranes as no FLSs form on glass or phosphatidylcholine (PC) bilayers alone. A direct comparision of PC/phosphatidylserine (PS) and PC/PS/PI(4,5)P2 membranes of equivalent net charge shows that there is specificity for PI(4,5)P2, which supports a 2.5 fold higher density of FLSs at steady state (20 min); their initial rate of appearance is also 5-fold faster than with just PS (shown in cyan and orange, Fig. 2A). Substitution of PS for PI (phosphatidylinositol) leads to a 2-fold decrease in the rate of appearance, which may reflect the role of PS in binding some F-BAR proteins (Fig. 2A, 28). All phosphoinositides (PIPs) nucleate actin spots on the membrane (bis/tris-PIPs are shown in Fig. 2B and monophosphorylated PIPs in fig. S2A) though few of the actin spots nucleated by monophosphorylated PIPs elongate. Among bisphosphorylated PIPs and PI(3,4,5)P3, PI(4,5)P2 gives the highest number and elongation rate (Fig. 2B and C), consistent with filopodia forming on the plasma membrane. One caveat in comparing the different PIPs is that the extracts may convert one form into another.
Figure 2. Membrane requirements for FLS formation.
(A) Time course of FLS appearance shows negatively-charged lipids are essential for FLS formation and there is specificity for PI(4,5)P2 and PS. Cyan: 60% PC/30% PS/10% PI(4,5)P2, purple: 60% PC/30% PI/10% PI(4,5)P2, orange:40% PC/60% PS, red: 55% PC/45% PS, green: 70% PC/30% PS. (B) Time course of FLS appearance. All PIPs can nucleate FLSs but there is preference for PI(4,5)P2. Compositions: 60% PC/30% PS/10% PIP, where lime:PI(4,5)P2, gray:PI(3,4)P2 magenta:PI(3,5)P2 navy:PI(3,4,5)P3, brown:PI. Data are the mean of 3 timecourses normalized to the average number of structures from the 3 experiments from 5 or more pictures over each supported bilayer. (C) Rate of actin addition (using the pulse-chase approach) for the different PIPs shows PI(4,5)P2 specificity, error bars are s.d., n = 18, 19, 16, 20. (4-way ANOVA, p<0.001). (D) Confocal microscopy of supported bilayer surface. GFP-PH domain (green) addition to supported bilayers including rhodamine-PE (red) shows membrane domains. Bar: 2 µm. The lipid composition was 45% PC, 45% PI, 10% PI(4,5)P2, similar data was obtained with 60% PC, 30% PS, 10% PI(4,5)P2. (E) FLSs grow preferentially from rhodamine-PE depleted (PH domain binding) regions. Alexa-647 actin is in green, rhodamine-PE in red. Bar: 2 µm (F) Contour plot of the number of FLSs per field of view at steady state (20 min) from fluid membranes in response to PI(4,5)P2 and extract concentrations. The lipid composition was 45% PC, 45% PI, 10% PI(4,5)P2. For comparison, overlaid single points show the number of FLSs formed from the gel phase. Data is plotted logarithmically. Example pictures and control data are in Fig. S4.
The tendency of lipid mixtures to segregate into domains suggests two different models for tip assembly and FLS nucleation. In the first, the FLSs would be templated by small domains enriched in PI(4,5)P2. In this model the size of the tips and the girth of the FLSs should reflect the size of the domains. In the second, tip complexes would selfassemble on the lipid bilayer in relatively homogeneous lipid domains. We find no evidence for preformed domains that template FLS formation, as GFP-PLCδ PH domain, which has specificity for PI(4,5)P2, binds evenly over large interconnected domains and there is no enrichment of GFP-PLCδ PH domains at sites of FLS growth (Fig. 2D and fig. S2B–D). Furthermore, the kinetics of PH domain binding to the bilayer does not reveal hotspots of PI(4,5)P2 (fig. S2E and F). Rhodamine-PE, which labels the liquid-disordered phase of membranes (29), has the inverse distribution to GFP-PH domain, confirming that lipid is present in regions depleted of the GFP-PLCδ PH domain. This also indicates that either PI(4,5)P2 partitions differently from rhodamine-PE, or that the PH domain cannot bind PI(4,5)P2 within these domains (Fig. 2D). Domains of rhodamine-PE enrichment vary in size with 53% less than 10 µm2, 38% 10–100 µm2, 7% 100–1000 µm2 and 2% >1000 µm2. Rhodamine-PE enriched regions nucleate 20-fold fewer FLSs than the rhodamine-PE depleted regions (Fig. 2E). DiI, another fluorescent membrane marker that labels the disordered phase, also localizes to areas with fewer FLSs (fig. S3A). Fluorescence recovery after photobleaching (FRAP) experiments confirm that the rhodamine-PE positive regions are fluid (fig. S3B and C). The irregular boundaries between rhodamine-PE regions and GFP-PH domain regions also suggest the coexistence of the liquid-disordered (rhodamine-PE) and gel phases (GFP-PH domain) (30). FRAP of GFP-PH domain confirms the low fluidity of these regions (fig. S3D and E). Thus FLS formation occurs preferentially but not exclusively in the gel phase. The tendency of FLS not to form from the liquid-disordered phase can be overcome by increasing the mole fraction of PI(4,5)P2 or by increasing the extract concentration (Fig. 2F and fig. S4) and therefore the gel phase is not an absolute prerequisite for FLS formation.
Thus, the assembly of FLSs does not occur by direct templating at preformed domains in the membrane, but instead occurs by a process of self-assembly driven by proteins at a permissive membrane surface. Several mechanisms could contribute to FLS formation: (1) Clustering of oligomeric activation proteins (e.g. BAR domain superfamily proteins) (31); (2) Positive feedback on small Arp2/3 complex-catalyzed clusters of polymerized actin through recruitment of more nucleation factors (32), as occurs during symmetry breaking by N-WASP on lipid coated glass beads; (3) Lattices generated by cooperative protein-protein interactions between signaling molecules; (4) Cooperative association of proteins with PI(4,5)P2, as has previously been shown for N-WASP (33). Any small local fluctuations in PI(4,5)P2 could be magnified in such a mechanism.
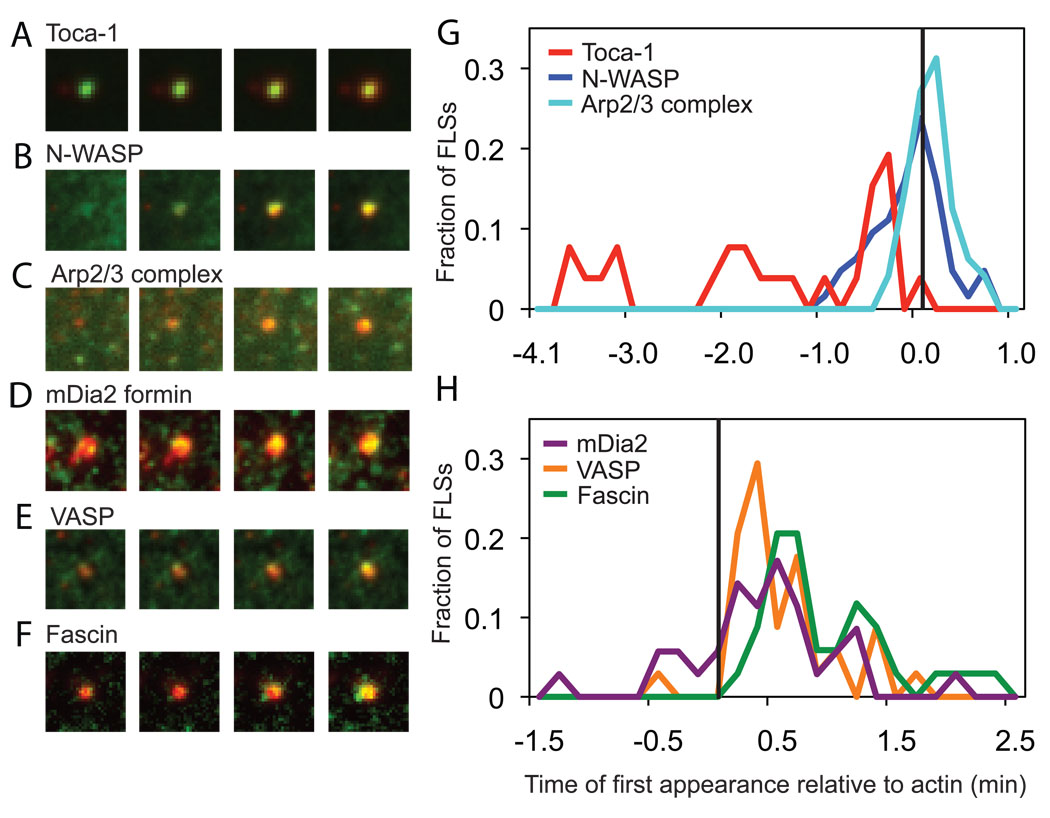
To investigate the kinetics of the self-assembly process of the FLSs, we added fluorescently labeled filopodial proteins to the extracts, and followed their recruitment to sites of FLS formation by total internal reflection fluorescence microscopy (TIRFM) (Fig. 3 and fig. S5). The recruitment times are normalized to the recruitment time of labeled actin, with the time differences plotted as a histogram (Fig. 3). The membrane-binding, F-BAR domain protein toca-1 is recruited earliest, at sites that later go on to form FLSs (Fig. 3A and G). After a variable time, N-WASP is recruited, again before actin (Fig. 3B and G). The Arp2/3 complex is recruited concomitantly with actin (Fig. 3C and G). VASP and mDia2 proteins, which are implicated in the formation of long, unbranched actin filaments, are recruited to the tip complex after the first appearance of actin (Fig. 3D, E and H). The bundling protein fascin is recruited last (Fig. 3F and H).
Figure 3. Kinetics of signaling protein recruitment to filopodia-like structures – Arp2/3 complex signaling proteins are recruited before actin and formin, VASP and fascin are recruited later.
(A–F) Total internal reflection fluorescence images of FLS tips with fluorescently labeled proteins (GFP-toca-1, GFP-N-WASP, Alexa568-Arp2/3 complex, GFP-mDia2, GFP-VASP, GFP-fascin) in green and Alexa-647 actin in red. 20 s time intervals are shown at the time of first recruitment of the signaling protein or actin. 5 nM labeled proteins were added to the extracts. (G) Histogram showing the relative time of first recruitment of Arp2/3 complex (n=48), toca-1 (n=26), and N-WASP (n=34) compared to the first appearance of actin. (H) Histogram showing the relative time of first recruitment for filopodia elongation and bundling proteins, VASP (n=34), mDia2 (n=35) and fascin (n=34) compared to actin. Using the KS-test, p=0.000 for toca-1-N-WASP, N-WASP-Arp2/3 complex, Arp2/3 complex-VASP; p=0.005 for Arp2/3 complex-mDia2; p=0.003 for mDia2-fascin; p=0.004 for VASP-fascin and no significant difference for mDia2-VASP. The lipid composition was 60% PC, 30% PS, 10% PI(4,5)P2.
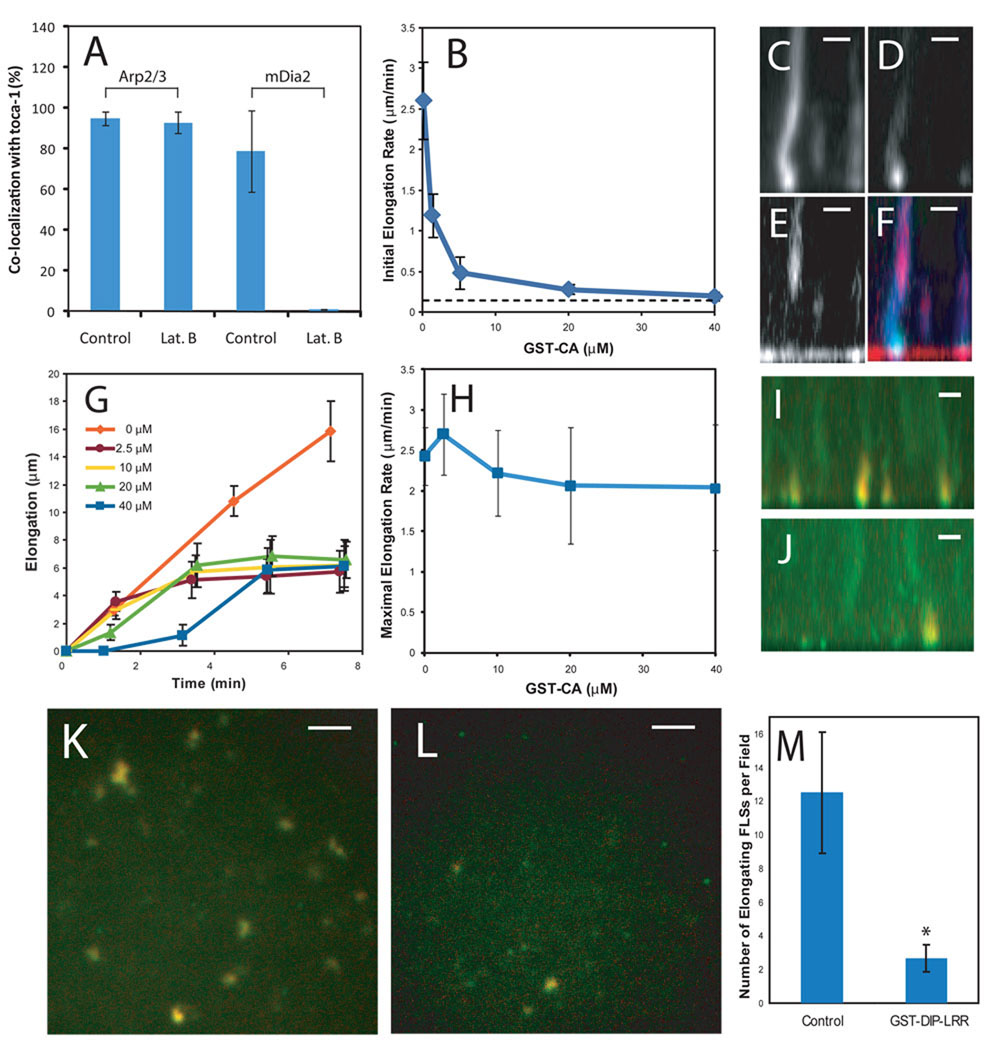
These data suggest that an Arp2/3 complex-driven initiation step nucleates an initial branched actin structure in a small patch, which stimulates the recruitment of filament elongation and bundling factors. To test this mechanism further we looked at the recruitment of Arp2/3 complex and mDia2 to toca-1 spots in the presence of actin monomer sequestering drug, latrunculin B. The number of toca-1 spots that recruit Arp2/3 complex are unaffected by latrunculin (Fig. 4A), whereas it completely blocks toca-1-mDia2 colocalization (Fig. 4A).
Figure 4. Initiation and elongation of FLSs occur by separable molecular mechanisms.
(A) Latrunculin B does not affect the co-localization of Arp2/3 complex with toca-1, but completely inhibits mDia2 recruitment to toca-1 sites (n=4). (B) Dose-response of FLS initial elongation rate with GST-CA preincubated in the extract before addition to the supported bilayer (n=20). The dotted-line indicates the minimum elongation rate (0.1 µm/min) due to the axial resolution limit of confocal microscope (~0.7 µm) (C–F) Pulse-chase experiment starting with Alexa-647 actin and Alexa 568-labeled Arp2/3 complex in the extract, with later addition of 40 µM GST-CA and Alexa-488 actin. Panels show Alexa 647-actin (C, blue); Alexa 488-actin (D, green); Alexa-568-Arp2/3 complex (E, red) and three color overlay (F). Addition of new actin monomers continues in the absence of the Arp2/3 complex recruitment into the FLS. Bars: 2 µm. (G) The time course of FLS elongation (measured by the second color of actin) at increasing GST-CA concentrations (n=20). (H) Dependence of rate of FLS elongation on the concentration of GST-CA. Maximal FLS elongation rate is unchanged by addition of GST-CA. (I–M) Similar pulse-chase experiment explained in (C–H), with the additional use of GST-LRR to inhibit formin activity. (I and K) Control addition of GST-CA plus the second color of actin. Alexa-647 (first, red) and Alexa-488 (second, green). (I) Side view. (K) Actin on the bilayer. (J and L) Inclusion of 5 µM GST-LRR in the extract then addition of GST-CA and Alexa-488 actin. GST-LRR leads to the detachment of the FLSs so fewer punctae are present at the membrane surface. (J) Side view. (L) Actin on bilayer. Bars: 2 µm (I and J); 5 µm (K and L) (M) Quantification of GST-DIP-LRR addition (n=6). *p<0.001. All error bars are s.d. The lipid composition was 45% PC, 45% PI, 10% PI(4,5)P2.
To probe the role of the Arp2/3 complex in FLS formation, we add GST-CA domain, which inhibits N-WASP activation of the Arp2/3 complex; this reduces the number of nucleation sites and abolishes FLS elongation (Fig. 4B, fig. S6A–B). Immunodepletion of N-WASP significantly decreases but does not completely inhibit FLS formation and elongation (fig. S6D, H–I), consistent with studies in cultured cells suggesting the involvement of other Arp2/3 complex nucleation-promoting factors (11, 12). Immunodepletion of toca-1 has only a minor effect on elongation (fig. S6E, H–I), however there are more than a dozen candidate BAR domain superfamily proteins that could compensate for loss of toca-1. We conclude that signaling through the Arp2/3 complex plays a vital role in the initiation of FLS formation, although these proteins alone are not sufficient to generate FLSs (Fig. 1A and B).
The usual product of Arp2/3 complex activation is an array of disorganized or branched actin structures, rather than organized parallel actin bundles. The kinetics of protein recruitment to the nascent FLSs suggests that a formin-driven elongation process occurs after the formation of the first actin nucleus. To test whether the elongation phase is dependent on the Arp2/3 complex, we start the reaction with Alexa-647 actin and Alexa-568 Arp2/3 complex and after 20 min add GST-CA to block further Arp2/3 complex function and Alexa-488 actin to record any further actin polymerization (Fig. 4C–H). We find that new actin monomers are still added at the FLS tip after Arp2/3 complex inhibition (Fig. 4C–F). Significantly, in the presence of GST-CA, there is no further incorporation of Arp2/3 complex into the FLS. The region of newly assembled actin in the shaft lacks Arp2/3 complexes (Fig. 4E and F, fig. S7A). After ~5 min addition of new actin to the FLS slows, indicating a requirement for Arp2/3 complex to maintain FLS growth over the long term (Fig. 4G). At high concentrations of GST-CA, there is a noticeable lag in the incorporation of new actin monomers (Fig. 4G), likely because the occupancy of GST-CA on N-WASP binding sites of the Arp2/3 complex stimulates a reorganization of the tip complex. Even at high doses of GST-CA, an Arp2/3 complex-independent component of elongation is maintained (Fig. 4H).
Diaphanous-related formins are the obvious engine for filopodial growth in the absence of the Arp2/3 complex, as formins are thought to drive filopodial elongation in different cell types. As there are many formin proteins, we employed a dominant-negative approach known to inhibit filopodia formation (34, 35). The leucine-rich region (LRR) of diaphanous interacting protein (DIP) binds and inhibits Drf1 and Drf3 (35). When GST-DIP-LRR is added to extracts, FLS formation still occurs with no significant reduction in elongation rate or the number of structures. However, when Arp2/3 complex function is inhibited by GST-CA in the presence of GST-DIP-LRR, the number of structures that incorporates new actin monomers is significantly reduced (Fig. 4I–M). This is accompanied by the detachment of many FLSs from the lipid bilayer, suggesting that the tip complex undergoes conformational changes, and that continuing reorganization of the tip complex cannot occur in the presence of GST-LRR (Fig. 4M). Furthermore, as diaphanous-related formins are activated by RhoA, we also tested dominant negative RhoA, GST-RhoA-N19. This produced similar results to GST-DIP-LRR (fig. S7B–F). Thus there are important functions contributed by both the Arp2/3 complex and formins. In the presence of capping activities, the Arp2/3 complex may be required for generating new actin nuclei (7, 36).
Currently there are two principal models for filopodial assembly - convergent elongation (5) and de novo nucleation (37). In the former Arp2/3 complex driven actin filaments continually coalesce into parallel arrays, by the continuous action of bundling proteins like fascin. In the latter, filopodia are formed through the establishment of a tip complex of formins, which nucleates long actin filaments which are then bundled. In the model we propose (fig. S8), signaling by a negatively charged membrane leads first to the recruitment and clustering of BAR domain superfamily proteins and N-WASP or other nucleation promoting factors at the membrane, leading to subsequent recruitment of the Arp2/3 complex and formation of a small patch of short actin filaments. This early clustering step represents the key difference from the convergent elongation model, as symmetry breaking occurs by focal recruitment of activators of the Arp2/3 complex rather than by a coalescence of preexisting actin barbed ends. In the proposed clustering-outgrowth model, local assembly of actin initiated by the Arp2/3 complex is converted into a filopodial tip complex by the recruitment of formins and VASP. This recruitment enables linear outgrowth of the filopodium, with short actin filaments elongated by formins and/or VASP and bundled by fascin (15, 16). Actin filaments generated by the Arp2/3 complex appear to be continually required to feed the elongation process, though other actin nucleators could fulfill a similar role. The observation of short actin filaments at the tips of filopodia in Dictyostelium by electron tomography is consistent with this model (38). The clustering of actin assembly proteins and outgrowth by elongation factors may be served by different components in different circumstances. In this view we should expect that, while the overall mechanism would be conserved but there could be flexibility in filopodial composition (39).
Supplementary Material
References and Notes
- 1.Pollard TD, Cooper JA. Science. 2009;326:1208. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davenport RW, Dou P, Rehder V, Kater SB. Nature. 1993;361:721. doi: 10.1038/361721a0. [DOI] [PubMed] [Google Scholar]
- 3.Lendvai B, Stern EA, Chen B, Svoboda K. Nature. 2000;404:876. doi: 10.1038/35009107. [DOI] [PubMed] [Google Scholar]
- 4.Krugmann S, et al. Curr Biol. 2001;11:1645. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- 5.Svitkina TM, et al. J Cell Biol. 2003;160:409. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lebrand C, et al. Neuron. 2004;42:37. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- 7.Romero S, et al. Cell. 2004;119:419. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 8.Steffen A, et al. Mol Biol Cell. 2006;17:2581. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C, et al. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miki H, Sasaki T, Takai Y, Takenawa T. Nature. 1998;391:93. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 11.Snapper SB, et al. Nat Cell Biol. 2001;3:897. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- 12.Lommel S, et al. EMBO Rep. 2001;2:850. doi: 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson-Dykstra SM, Higgs HN. Cell Motil Cytoskeleton. 2008;65:904. doi: 10.1002/cm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korobova F, Svitkina T. Mol Biol Cell. 2008;19:1561. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vignjevic D, et al. J Cell Biol. 2003;160:951. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brieher WM, Coughlin M, Mitchison TJ. J Cell Biol. 2004;165:233. doi: 10.1083/jcb.200311040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu AP, et al. Nat Phys. 2008;4:789. doi: 10.1038/nphys1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan YH, Boxer SG. Curr Opin Chem Biol. 2007;11:581. doi: 10.1016/j.cbpa.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho HY, et al. Cell. 2004;118:203. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 20.Mooseker MS, Pollard TD, Wharton KA. J Cell Biol. 1982;95:223. doi: 10.1083/jcb.95.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallavarapu A, Mitchison T. J Cell Biol. 1999;146:1097. doi: 10.1083/jcb.146.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vignjevic D, et al. J Cell Biol. 2006;174:863. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pellegrin S, Mellor H. Curr Biol. 2005;15:129. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Ho HY, Rohatgi R, Ma L, Kirschner MW. Proc Natl Acad Sci U S A. 2001;98:11306. doi: 10.1073/pnas.211420498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arasada R, et al. Biochim Biophys Acta. 2007;1773:631. doi: 10.1016/j.bbamcr.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Sarmiento C, et al. J Cell Biol. 2008;180:1245. doi: 10.1083/jcb.200708123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnston SA, Bramble JP, Yeung CL, Mendes PM, Machesky LM. BMC Cell Biol. 2008;9:65. doi: 10.1186/1471-2121-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh T, et al. Dev Cell. 2005;9:791. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Crane JM, Tamm LK. Biophys J. 2004;86:2965. doi: 10.1016/S0006-3495(04)74347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwille P, Kahya N, Bacia K. In: Protein-Lipid Interactions: From Membrane Domains to Cellular Networks. Tamm L, editor. Weinheim: Wiley-VCH; 2005. pp. 337–365. [Google Scholar]
- 31.Padrick SB, et al. Mol Cell. 2008;32:426. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Co C, Wong DT, Gierke S, Chang V, Taunton J. Cell. 2007;128:901. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papayannopoulos V, et al. Mol Cell. 2005;17:181. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 34.Higgs HN, Peterson KJ. Mol Biol Cell. 2005;16:1. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenmann KM, et al. Curr Biol. 2007;17:579. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 36.Mejillano MR, et al. Cell. 2004;118:363. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 37.Faix J, Breitsprecher D, Stradal TE, Rottner K. Int J Biochem Cell Biol. 2009;41:1656. doi: 10.1016/j.biocel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 38.Medalia O, et al. Curr Biol. 2007;17:79. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 39.Gupton SL, Gertler FB. Sci STKE. 2007;2007:re5. doi: 10.1126/stke.4002007re5. [DOI] [PubMed] [Google Scholar]
- 40.K.L. was the recipient of KOSEF Scholarship and Harvard-MIT HST Fellowship. J.L.G was the recipient of an EMBO Fellowship. This work was funded by NIH grant number GM26875. We thank Gary Bokoch for the gift of the GST-RhoA-N19 plasmid, Seth Field for the gift of the pEGFP GFP-PLCδ PH domain plasmid, Orion Weiner for the RhoGDI plasmid, Rogen Tsien for the mCherry plasmid, Arthur Alberts for the mDia2 plasmid, Danijella Vignjevic for the fascin plasmid. We thank Andres Lebensohn for helping extract preparation and valuable discussions and Margaret Coughlin, Tom Walz and Debbie Kelly for help with electron microscopy. We thank Jennifer Waters and Nikon Imaging Center at Harvard Medical School.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.