Abstract
Transcription factor Stat5a/b is critical for prostate cancer cell survival and for prostate xenograft tumor growth. In addition, the Stat5a/b signaling pathway may contribute to progression of organ-confined prostate cancer to castration-resistant and/or metastatic disease. Expression of nuclear Stat5a/b is clustered to high grade human prostate cancers, and nuclear Stat5a/b in primary prostate cancer predicts early disease recurrence after initial treatment. Here, we show by Western blotting and electromobility shift assay that Stat5a/b protein in human prostate cancer is N-terminally truncated. This short form of Stat5a/b is generated post-translationally in vivo in prostate cancer cells and is the predominant form of Stat5a/b that binds to DNA. We further demonstrate by mutagenesis and co-immunoprecipitations that the N-domain of Stat5a/b is required for binding to PIAS3, and that PIAS3 inhibits transcriptional activity of Stat5a/b in breast cancer cells but not in prostate cancer cells. Thus, the proteolytic cleavage of the N-terminus of Stat5a/b may be a mechanism by which Stat5 evades the transcriptional repression by PIAS3 in prostate cancer cells, and results in increased Stat5-driven gene expression and prostate cancer progression.
Keywords: prostate cancer, Stat5a/b, N-terminal truncation, PIAS3
1. Introduction
Transcription factor Stat5a/b critically regulates viability and growth of human prostate cancer cells (Liao et al.,; Tan and Nevalainen, 2008). Inhibition of Stat5a/b induces massive apoptotic death of human prostate cancer cells in vitro (Ahonen et al., 2003; Dagvadorj et al., 2008) and blocks prostate cancer subcutaneous and orthotopic xenograft tumor growth in nude mice (Dagvadorj et al., 2008; Gu et al., 2010b). Stat5 is one of the seven members of Stat gene family of transcription factors (Darnell, 1997; Hennighausen and Robinson, 2008; Ihle, 2001; Tan and Nevalainen, 2008). Two highly homologous isoforms of Stat5, the 94-kDa Stat5a and the 92-kDa Stat5b, are encoded by separate genes and are latent cytoplasmic proteins that act as both cytoplasmic signaling proteins and nuclear transcription factors (Darnell, 1997; Hennighausen and Robinson, 2008; Ihle, 2001; Tan and Nevalainen, 2008). Phosphorylation of a specific tyrosine residue in the COOH-terminal domain by a tyrosine kinase, typically of the Janus-activated kinase protein family (Rui et al., 1992; Rui et al., 1994), activates Stat5a/b. After phosphorylation, Stat5a and Stat5b homodimerize or heterodimerize and translocate to the nucleus where they bind to specific Stat5a/b response elements of target gene promoters. Stat5 proteins are divided into five structurally and functionally conserved domains. These include the NH2-terminal domain (N-domain) which is involved in stabilizing interactions between two Stat5 dimers to form tetramers (John et al., 1999), the N-domain and the coiled-coil (CC) domain that mediate protein-protein interactions (Becker et al., 1998; Chen et al., 1998), the DNA-binding domain, the SH2 domain critical for dimerization and the C-terminal transactivation domain which binds to critical coactivators/corepressors (Kisseleva et al., 2002; Levy and Darnell, 2002).
Stat5a/b is constantly active in human prostate cancer but not in normal human prostate epithelium (Ahonen et al., 2003; Li et al., 2004). In addition, Stat5a/b activation in prostate cancer is associated with high histological grade (Li et al., 2004), and activation of Stat5a/b in primary prostate cancer predicted early prostate cancer recurrence (Li et al., 2005). Several different molecular mechanisms regulate the duration and magnitude of Stat5a/b activation in the cytoplasmic and nuclear compartments. These mechanisms involve: (1) local expression of autocrine growth factors that activate Stat proteins, (2) activating mutations in the tyrosine kinases responsible for Stat activation, (3) protein inhibitors of activated Stat proteins (PIAS) (4) cytoplasmic and nuclear protein tyrosine phosphatases (PTP); and (5) suppressors of cytokine signaling (SOCS) proteins (Darnell, 1997; Hennighausen and Robinson, 2008; Ihle, 2001; James et al., 2005; Kralovics et al., 2005; Scott et al., 2007; Tan and Nevalainen, 2008).
The PIAS family of proteins are localized within the nucleus and function as constitutive repressors of STAT activity by direct association (Schmidt and Muller, 2003; Shuai, 2006). The PIAS family members include PIAS1, PIAS3, PIASx, PIASy, and alternative splicing variants of PIASx (Palvimo, 2007). PIAS3 is the only member of the PIAS family that has been shown to directly interact with Stat5a/b and repress Stat5-mediated transcription in CHO and lymphoid Nb2 cells (Rycyzyn and Clevenger, 2002). PIAS3 is expressed in prostate cancer tissues and cell lines (Wang and Banerjee, 2004) but little is known about its function in prostate cancer. Furthermore, PIAS1 expression is 33% higher in primary prostate cancers compared to normal prostates (Li et al., 2002). In addition, PIAS1 expression has been shown to be significantly lower in hormone-refractory prostate cancer than in untreated prostate tumors (Linja et al., 2004).
In this work, we demonstrate that Stat5a/b protein is cleaved in human prostate cancer cells and clinical prostate cancers but not in COS-7 cells or breast cancer cells. This cleaved short form of Stat5a/b is N-terminally truncated and constitutes the predominant form of Stat5a/b that binds to DNA in human prostate cancer cells. We further demonstrate that the N-domain of Stat5a/b binds to PIAS3, and that PIAS3 inhibits transcriptional activity of Stat5a/b in breast cancer cells but not in prostate cancer cells. Thus, the short form of Stat5 in prostate cancer cells evades the transcriptional inhibition by PIAS3 by proteolytical cleavage of its N-terminus. This may represent a molecular mechanism underlying increased Stat5-driven gene transcription that contributes to prostate cancer growth and progression.
2. Materials and methods
2.1. Cell lines and clinical human prostate cancer specimens
CWR22Rv1, DU145, PC-3 and LNCaP prostate cancer cells or T47D or MCF-7 breast cancer cells (American Type Culture Collection (ATCC), Manassas, VA) were grown in RPMI-1640 medium (Mediatech, Manassas, VA). The basal media contained 10% fetal bovine serum (FBS) (Atlanta Biologicals, Norcross, GA), 2 mM L-glutamine (Mediatech), 5 mM HEPES (Mediatech), pH 7.3, and penicillin-streptomycin (Mediatech) (50 IU/ml and 50 μg/ml, respectively), at 37°C with 5% CO2. LNCaP cells were cultured in the presence of 0.5 nM dihydrotestosterone (DHT) (5α-androstan-17β-ol-3one; Sigma, St. Louis, MO). Construct expression and functional tests were performed by transient transfection of COS-7 cells (ATCC) which were grown in RPMI 1640 medium (Mediatech) containing 10% fetal bovine serum, 2 mM L-glutamine, and penicillin-streptomycin (50 IU/ml and 50 μg/ml, respectively) at 37 °C with 5% CO2. Sub-confluent COS-7 cells in 6-well plates were transfected using FuGENE6 (Roche) according to the manufacturer's protocol and were serum starved in RPMI 1640 with 0.5% fetal bovine serum for 12 h followed by stimulation with 10 nM prolactin (Prl) for 30 min.
Paraffin-embedded prostate cancers were obtained from the Turku University Hospital (n=20) (approved by the Thomas Jefferson University Institutional Review Board). The fresh prostate cancer specimens were obtained from 3 patients undergoing radical prostatectomy at the Turku University Hospital by the urologist (M.N). The prostate tissues were obtained after informed consent of the patients and the approval of the Ethical Committee of the Turku University Hospital. Within 1–3 hours of the surgery, a board-certified pathologist (K.A. or T.M.) made a selection of the tissue slices of prostate cancer nodules that were available for the analysis. The selection of the area was assisted by the clinical information of the localization of the cancer based on the location of the needle biopsy taken at the time of diagnosis and by frozen sections.
2.2. Solubilization of proteins, immunoprecipitation, and immunoblotting
Cells were lysed and fresh human prostate cancer specimens were homogenized in lysis buffer [10 mM Tris-HCl (pH 7.5), 5 mM EDTA, 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM sodium fluoride, 1 mM sodium orthovanadate, 1% Triton X-100, 1 mM phenylmethylsulphonylfluoride, 5 μg/ml aprotinin, 1 μg/ml pepstatin A, and 2 μg/ml leupeptin], and the protein concentrations of the whole cell lysates were determined by the Bradford method (BioRad Laboratories Inc., Hercules, CA). In Figure 2B: 1) Frozen cell pellets were pulverized in liquid nitrogen and solubilized directly with SDS sample buffer by heating for 10 min at 99°C. 2) After harvest and centrifugation, 10 vol of SDS sample buffer was added to the cell pellets and the cells were lysed by heating for 10 min at 99°C. 3) Cell pellets were lysed in the cell lysis buffer [10 mM Tris-HCl (pH 7.5), 5 mM EDTA, 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM sodium fluoride, 1 mM sodium orthovanadate, 1% Triton X-100, 1 mM phenylmethylsulphonylfluoride, 5 μg/ml aprotinin, 1 μg/ml pepstatin A, and 2 μg/ml leupeptin] and heated with the SDS sample buffer for 10 min at 99°C. In the indicated the experiments, cell lysates were immunoprecipitated for 2 h at 4°C with anti-Stat5a or anti-Stat5b pAbs (each 1.2 μg/ml; Advantex Bioreagents, Conroe, TX). Antibodies were captured by incubation with protein A-Sepharose beads (GE Healthcare, Piscataway, NJ) for 60 min. The filters were blotted with anti-phosphotyrosine-Stat5a/b (Y694/Y699) mAb (1 μg/ml, Advantex Bioreagents), anti-panStat5ab mAb (1:250) (Transduction Laboratories, Inc.), anti-C-terminus Stat5a/b mAb (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-C-terminus Stat5a mAb (Santa Cruz Biotechnology), anti-C-terminus Stat5b mAb (Santa Cruz Biotechnology), anti-N-terminus-Stat5a/b pAb (Santa Cruz Biotechnology), anti-actin pAb (1:4000) (Invitrogen), anti-HDAC (1:500) (Invitrogen) and anti-tubulin (1:1000) (Millipore). The immunoreaction was detected by horseradish peroxidase-conjugated secondary antibodies in conjunction with enhanced chemiluminescence substrate mixture (GE Healthcare), and exposed to film.
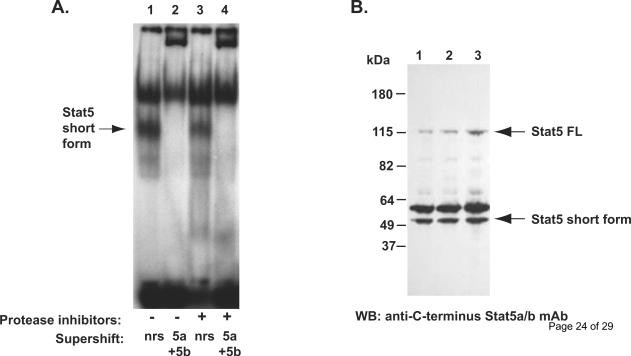
Figure 2. Short form STAT5 occurs naturally in prostate cancer cells.
A, Electromobility shift assay of nuclear extract of DU145 cells prepared from flash frozen cell pellets in the absence (lane 1 and 2) or presence (lane 3 and 4) of protease inhibitors (phenylmethylsulfonylfluoride, aprotinin, leupeptin and pepstatin-A). The short form Stat5a/b (arrow) was detected in DU145 cells in the presence or in the absence of protease inhibitors, and the short form Stat5a/b was super-shifted by anti-Stat5a and anti-Stat5b antibodies. B, The short form Stat5a/b is not generated in vitro after disruption of the cell membranes. DU145 cell pellets were flash frozen in liquid nitrogen and directly lysed in boiling SDS loading buffer (lane 1), or the cell pellets were lysed in cell lysis buffer in the presence (lane 2) or in the absence of proteases inhibitors (lane 3) and boiled in SDS loading buffer and analyzed by Western blotting using anti-C-terminus Stat5a/b mAb.
2.3. Co-IPs of FLAG-tagged Stat5a and HA-tagged PIAS3
Full length Stat5a was amplified by PCR and subcloned to pCMV-3FLAG vector (Agilent Biotechnologies, La Jolla, CA) with an EcoR1 and a SalI sites. Plasmid CMV3FLAG-Stat5a, pCMV-HAPIAS3 (GeneCopeia, Maryland) and pPrlR were co-transfected using FuGENE6 (Roche, Indianapolis, IN) into COS-7 cells seeded 24 h earlier (2 μg of each plasmid per 1 × 107 cells) (Fig. 4). The cells were starved for 20 h, then stimulated with hPrl (10 nM) in RPMI 1640 without serum for 16 h. The cell lysates were immunoprecipitated with 25 μl anti-FLAG M2 polyclonal affinity gel (2 μg/ml, Sigma), anti-HA pAb (Bethyl Laboratories, Montgomery, TX) or IgG (Sigma). The primary antibodies were used in the immunoblottings at the following concentrations: anti-FLAG mAb (1:1000; Agilent) and anti-HA mAb (1:1000; Bethyl Laboratories) detected by horseradish peroxidase-conjugated secondary antibodies.
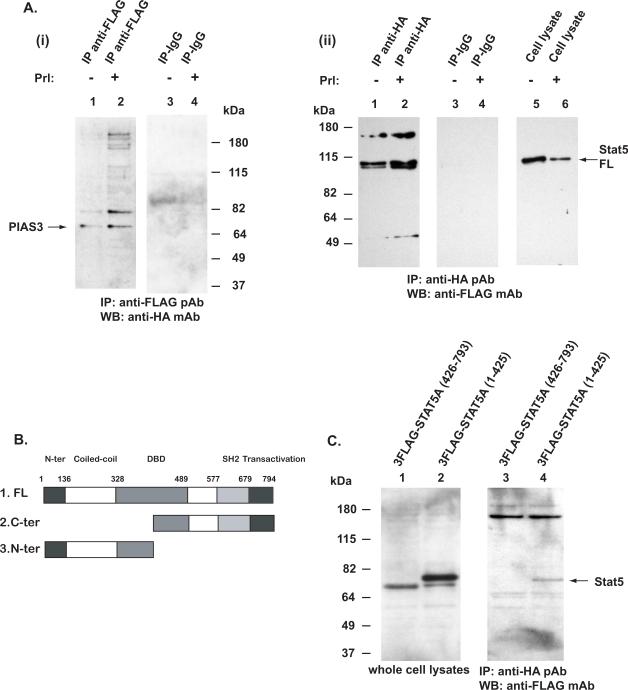
Figure 4. PIAS physically interacts with the N-terminus of STAT5a.
A, COS-7 cells were co-transfected with pFlag-Stat5a, pHA-PIAS3 and pPrlR, serum-starved for 16 h, then stimulated with 10 nM human Prl (Pr) for 30 min with unstimulated cells as control. Stat5a was immunoprecipitated with anti-FLAG pAb and blotted with anti-HA mAb (i). Stat5a formed a complex with PIAS3 (lanes 1 and 2). In the converse experiments, PIAS3 was immunoprecipitated with anti-HA pAb and the immunoprecipitations were blotted with anti-FLAG mAb (ii). B, Schematic presentation of the deletion constructs of Stat5a that were either C-terminally truncated or N-terminally truncated and tagged with the FLAG-epitopes. C, COS-7 cells were co-transfected with pFlag-Stat5a (426–793), pFlag-Stat5a (1–415) or pHA-PIAS3. PIAS3 was immunoprecipitated with anti-HA pAb and immunoblotted with anti-FLAG mAb recognizing the Flag-tagged N-terminally or C-terminally truncated Stat5a. The C-terminally truncated Stat5a (1–425) formed complexes with PIAS3 (lane 4) while the N-terminally truncated Stat5a (426–793) failed to co-immunoprecipitate with PIAS3 (lane 3).
2.4. Generation of adenoviruses for gene delivery of wild-type (WT) Stat5a
We cloned pcDNA-CMV-WTStat5a (AdWTStat5a) into adenoviral vector using BD Adeno-X™ Expression System 2 (BD Biosciences Clontech, San Jose, CA) according to the manufacturer's protocol as described previously (Abdulghani et al., 2008). Viral stocks were expanded in large-scale cultures, purified by double cesium chloride gradient centrifugation, and titered side-by-side by a standard plaque assay method in QBI-293A cells as per the manufacturer's instructions (MP biomedicals, OH).
2.5. Immunohistochemistry
Sections of formalin-fixed tissues were deparaffinized by two 15-min washes in xylene, followed by rehydration in graded alcohol. Slides containing deparaffinized tissue sections or paraformaldehyde-fixed cells were microwave-treated in a pressure cooker with antigen-retrieval solution AxAR1 (Advantex BioReagents) for use with the anti-pTyrStat5a/b mAb. After the antigen retrieval procedure, endogenous peroxidase activity was blocked by incubating slides in 0.3% hydrogen peroxide for 10 min at room temperature, and nonspecific binding of immunoglobulins was minimized by preincubation in normal goat serum for 2 h at RT. The anti-pTyrStat5 mAb was diluted in 1% BSA in PBS at a final concentration of 0.6 μg/ml. Antigen-antibody complexes were detected using biotinylated goat anti-mouse IgG secondary antibody (BioGenex Laboratories, Inc., San Ramon, CA) followed by streptavidin-horseradish-peroxidase complex. As a chromogen, 3,3'-diaminobenzidine (DAB) was used, and Mayer hematoxylin was used as a counterstain. For controls, subtype-specific mouse IgG was used as appropriate.
2.6. Electrophoretic mobility shift assay (EMSA)
Cells were starved in serum-free medium for 16 h, then treated with or without 10 nM hPrl as indicated, pelleted by centrifugation and immediately solubilized in EMSA lysis buffer [20 mM HEPES (pH 7.0), 10 mM KCl, 1 mM MgCl2, 20% glycerol, 0.2% NP-40, 1 mM orthovanadate, 25 mM NaF, 200 μM phenylmethylsulfonylfluoride, 5 μg/ml aprotinin, 1 μg/ml pepstatin A, and 2 μg/ml leupeptin]. The cell lysates were pelleted by centrifugation at 800 × g for 10 min at 4 C, and the pellets were solubilized in EMSA lysis buffer containing 300 mM NaCl. Lysates were incubated on ice for 10 min, then clarified by centrifugation at 20,000 × g for 10 min at 4 C. [32P]-labeled oligonucleotide probe corresponding to the Prl response element (5′agatttctaggaattcaaatc 3′) of the rat β-casein gene was used. The samples were pre-incubated with either normal rabbit serum (NRS), anti-C-terminus-Stat5a pAb, anti-C-terminus-Stat5b pAb or anti-N-terminus-Stat5a/b pAb (all from Santa Cruz Biotechnology). In Fig. 1C, nuclear extracts from COS-7 cells transfected with full-length pWtStat5a were mixed at indicated proportions with nuclear extracts from CWR22Rv1 cells before running on the EMSA gels. In Fig. 1D, CWR22Rv1 cells were infected at increasing MOIs (multiplicity of infection) with AdWtStat5a 24 h before the cells were harvested for EMSA. In Fig. 5B, MCF-7 cells and LNCaP cells (1.5 × 106 cells/dish) were transiently co-transfected with genomic pStat5a (2.0 μg) and pPrl-receptor (PrlR) (2.0 μg). The cells were serum-starved for 20 h and treated with (+) or without (−) 10 nM human hPrl for 30 min. The nuclear extracts were prepared for EMSA assay. Polyacrylamide gels (4%) containing 5% glycerol and 0.25 × TBE were prerun in 0.25 × TBE buffer at 4–10 °C for 1.5 h at 300 V. After loading of samples, the gels were run at room temperature for 3 h at 250 V. Gels were dried by heating under vacuum and exposed to X-ray film.
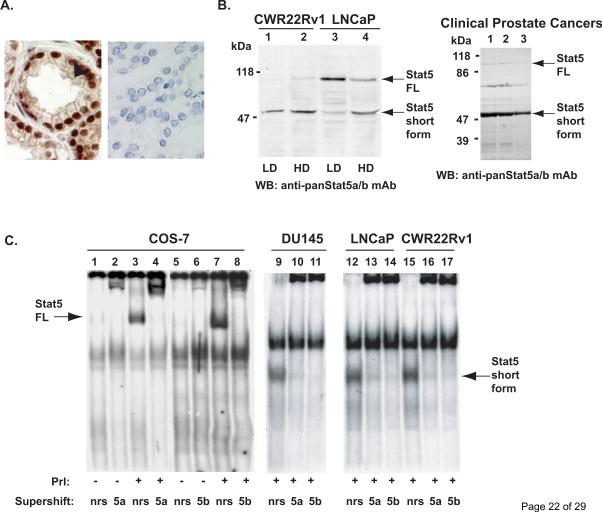
Figure 1. Stat5a/b is cleaved to a truncated protein in human prostate cancer cells.
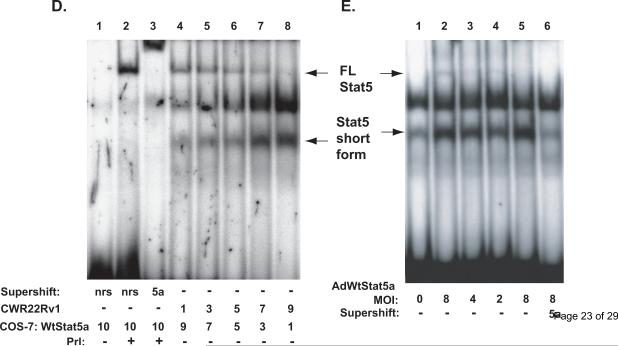
A, Immunohistochemical detection of Stat5a/b in human prostate cancer using a monoclonal anti-pYStat5 antibody (left panel) with isotype-specific IgG as control (right panel). B, Western blotting of whole cell extracts of CWR22Rv1 and LNCaP cells grown at low density (LD) or high density (HD) and tissue extracts of three clinical human prostate cancers with anti-Stat5a/b mAb showing the full length (FL) and a truncated Stat5a/b (Stat5 short form) of approximately 50 kDa in size (arrows). C, EMSA of nuclear extracts of COS-7 cells transfected with Stat5a and stimulated with human prolactin (hPrl) using the Prl-response element of the beta–casein gene promoter as the probe (lanes 3, 4, 7 and 8). Stat5a and Stat5b complexes were supershifted with anti-Stat5a (lane 4) and anti-Stat5b (lane 8) pAbs indicating the specificity of the DNA-binding complexes. EMSA of nuclear extracts of human prostate cancer cell lines DU145 (lanes 9–11), LNCaP (lanes 12–14) and CWR22Rv1 (lanes 15–17) showing the presence of a smaller truncated form of Stat5a/b (Stat5 short form) which was supershifted by anti-Stat5a and anti-Stat5b pAbs. D, Nuclear extracts of COS-7 cells transfected with Stat5a, stimulated with hPrl for 30 min and supershifted with anti-Stat5a pAb (lanes 1–3) were mixed in various ratios with nuclear extracts from CWR22Rv1 cells and analyzed by EMSA (lanes 4–8). E, Full-length (FL) wild-type (Wt) Stat5a was introduced at decreasing doses (MOI 8, 4 and 2) to CWR22Rv1 cells using adenovirus as the expression vector (AdWtStat5a). EMSA shows the conversion of the full-length WtStat5a to the short form of Stat5a (the lower arrows on the left) (lanes 2–5). The short form Stat5a was supershifted by anti-Stat5a pAb (lane 6).
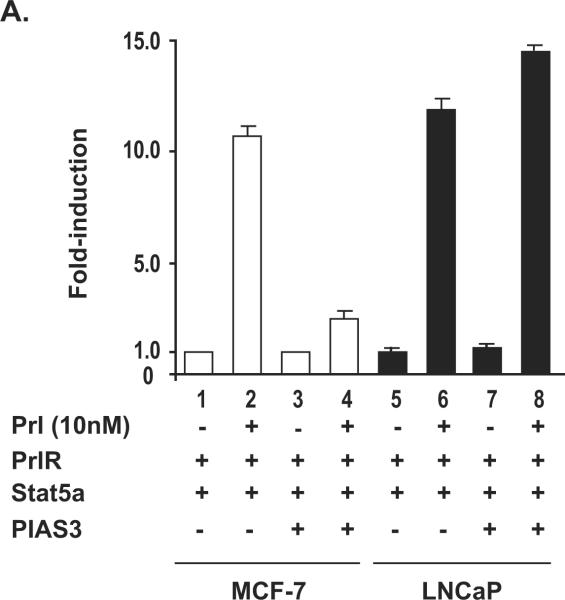
Figure 5. Transcriptional activity of ligand-induced Stat5a in beta-Casein promoter-luciferase assay is repressed by PIAS3 in human breast cancer cells but not in human prostate cancer cells.
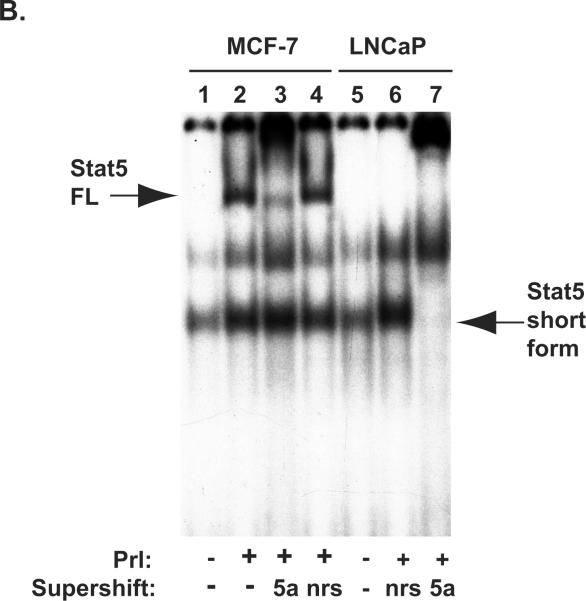
A, MCF-7 cells and LNCaP cells (0.25 × 106 cells/well) were transiently co-transfected with genomic beta-Casein-promoter-luciferase plasmid (0.5 μg), pRL-TK (0.025 μg), pPrl-receptor (PrlR) (0.25 μg), pStat5a (0.25 μg), and/or pPIAS3 (0.5 μg), as indicated. The total amount of plasmid DNA transfected was normalized to 1.275μg/well by addition of the empty vector (pMod-DNR). The cells were serum-starved for 20 h and treated for 16 h with (+) or without (−) 10 nM human hPrl as indicated. The mean values of three independent experiments performed in triplicates are presented, and S.E. values are indicated by bars. B, MCF-7 cells and LNCaP cells (1.5 × 106 cells/dish) were transiently co-transfected with genomic pStat5a (2.0 μg) and pPrl-receptor (PrlR) (2.0 μg). The cells were serum-starved for 20 hr and treated with (+) or without (−) 10 nM human hPrl for 30 min. The nuclear extracts were prepared for EMSA assay. EMSA shows the presence of short form Stat5a in LNCaP cells but not in MCF-7 cells.
2.7. Northern Blot Analysis
Total RNA was isolated from T47D and CWR22Rv1 cells by using RNeasy kit (Qiagen, Valecia, CA). Thirty micrograms of total RNA was denatured in 50% formamide at 60°C, electrophoresed on 1% agarose with 6% formaldehyde gel, and blotted to Hybond-N (GE Healthcare). The filter was incubated at 42°C in 50% formamide, 3 × Denhardt's reagent (0.06% polyvinylpyrrolidone, 0.06% bovine serum albumin, 0.06% Ficoll), 5 × SSC (standard saline citrate; 1 × SSC=0.15 M NaCl, 0.015 M trisodium citrate), 1% SDS, 200 g/ml of denatured salmon sperm DNA, and randomly primed (Agilent Technologies) 32P-labeled cDNA probe. After hybridization, the filter was washed in 0.1 × SSC, 0.1% SDS at room temperature, and autoradiographed. Reprobing was carried out after washing the filter in a stripping buffer of 10 mM Tris–HCl pH 7.5, 1 mM EDTA, 0.1% SDS and 0.3 Denhardt's reagent at 90°C for 20 min. The probes used were the fragment of the mouse Stat5a cDNA containing the N-terminus and the coiled-coiled domain.
2.8. Generation of the Stat5a deletion constructs
The Stat5a truncation constructs were first amplified by PCR from pcDNAStat5a using an upper primer that includes an EcoRI site and a lower primer with a SalI site. The amplified sequences were then subcloned into the pCRII-TOPO vector (Invitrogen, Carlsbad, CA). Stat5a fragments in pCRII-TOPO were then excised with the restriction enzymes EcoRI and SalI and then subcloned into similarly digested pCMV-3Tag1A (Agilent) plasmid. The plasmids express the truncated STAT5a tagged with N-terminal 3xFLAG epitope. STAT5a-N-ter (1M-425K) was amplified with the upper primer #1, GAATTCATGGCGGGCTGGATTCAGGCCCAGCAGCTT, and lower primer #5, GTCGACCTTGATTCTTTTCAGTGACATGTTTCTGAA. STAT5 C-ter (426R-794S) was amplified with the upper primer #6, GAATTCGATCGCGCCGACAGGCGTGGTGCAGAGT, and lower primer #2, GTCGACTCAGGACAGGGAGCTTCTAGCGGAGGTGAA.
2.9. Luciferase reporter gene assays
LNCaP cells and MCF-7 breast cancer cells were transiently transfected using FuGENE6 (Roche) with 0.25 μg of each pStat5a or pPIAS3, pPrlR and either 0.5 μg of pβ-casein–luciferase and 0.025 μg of pRL-TK (Renilla luciferase) as an internal control. The total amount of plasmid DNA per well was normalized by pMod-DNR to 1.275 μg per well. The cells were starved in serum-free medium for 20 h and stimulated with 10 nM hPrl in the starvation medium for 16 h. The lysates were assayed for firefly and Renilla luciferase activities using the Dual-Luciferase reporter assay system (Promega, Madison, WI). Three independent experiments were carried out in triplicate using at least two sets of plasmids that were prepared separately. The firefly luciferase activity was normalized to the Renilla luciferase activity of the same sample, and the mean was calculated from the parallels experiments. From the mean values of each independent run, the overall mean and its SE were calculated.
3. Results
3.1. Stat5a/b binds to DNA as a truncated protein in human prostate cancer cell
First, we demonstrated active Stat5a/b expression in clinical human prostate cancer by immunohistochemical detection using anti-pTyrStat5a/b mAb (Fig. 1A, left panel) with IgG as control (Fig. 1A, right panel). Western blotting of whole cell lysates of CWR22Rv1 and LNCaP and clinical prostate cancer specimens indicate the full-length Stat5a/b band of approximately 92 kDa in size, as expected (Fig. 1B). Cells were harvested at low density (LD) or high density (HD) for Western blot analysis. In addition to the full-length Stat5a/b, all prostate cancer cell lines and clinical prostate cancer specimens show a predominant shorter band of about 50 kDa in Western blotting (Fig. 1B). Minor bands visible in the Western blot analyses are likely due to unspecific binding of the antibodies to the filters because whole cell lysates were analyzed instead of immunoprecipitated Stat5a/b.
To examine the truncated Stat5a/b by another methodological approach, we performed electrophoretic mobility shift assay (EMSA) using the Stat5a/b-response element of beta–casein gene (Ahonen et al., 2002; Nevalainen et al., 2002). In nuclear extracts of COS-7 cells transfected with Stat5a and stimulated with human Prolactin (hPrl), only full-length of Stat5a and Stat5b were detected (Fig. 1C, lanes 3 and 7). These complexes were supershifted with polyclonal anti-Stat5a (Fig. 1C, lane 4) and anti-Stat5b (Fig. 1C, lane 8) antibodies indicating the specificity of the bands in EMSA. In contrast, nuclear extracts of prostate cancer cell lines DU145 (lanes 9–11), LNCaP (lanes 12–14), and CWR22Rv1 (lanes 15–17) showed the presence of a smaller truncated form of Stat5a/b in EMSA. Supershift with anti-Stat5a and anti-Stat5b antibodies (Fig. 1C, lanes 9–17) demonstrated that the shorter band was indeed the cleaved form of Stat5a/b.
In the next set of experiments, we transfected COS-7 cells with full-length Stat5a and stimulated the cells with hPrl for 30 min. Nuclear extracts of the full-length Stat5a expressing cells were mixed with nuclear extracts from CWR22Rv1 cells at various ratios and analyzed by EMSA. As demonstrated in Figure 1D, the full length Stat5 and the short form Stat5 bind to DNA according to the ratio of the nuclear extracts. To further examine whether the full-length Stat5 is cleaved to the short form Stat5a/b in prostate cancer cells, we expressed full-length wild-type (Wt) Stat5 in a dose-dependent manner using adenovirus as the expression vector (AdWtStat5a) in CWR22Rv1 human prostate cancer cells. As demonstrated in Figure 1E, full-length WtStat5a was efficiently converted to the truncated Stat5a, and the amount of the short form Stat5a was decreased according to the amount of full-length WtStat5a introduced to the cells (MOI 8, 4 and 2). This indicated that the full-length Stat5a/b was converted to the truncated form according to the amount of full-length Stat5a/b introduced to the cells. Importantly, the band representing the truncated Stat5a complex was super-shifted by anti-Stat5a antibody (lane 6) demonstrating that the shorter band was Stat5a. Based on these results, we concluded that prostate cancer cells express a short form of Stat5a/b, and the short Stat5a/b is capable of binding to DNA (Stat5-response element).
3.2. The short form of Stat5a/b in human prostate cancer cells occurs naturally
To determine whether the short forms of Stat5a/b in prostate cancer cells are generated in vitro during the preparation of the cell extracts, we first carried out EMSA of nuclear extracts prepared from DU145 prostate cancer cells in the presence or in the absence of protease inhibitors (phenylmethylsulfonylfluoride, aprotinin, leupeptin and pepstatin-A) in the lysis buffer. The short form Stat5a/b complexes were formed regardless of the presence (Fig. 2A, lanes 1 and 2) or absence (Fig. 2A, lanes 3 and 4) of protease inhibitors, respectively, and the short form Stat5a/b was supershifted by anti-Stat5a and anti-Stat5b antibodies (Fig. 2A, lanes 2 and 4). This indicated that the short form Stat5a/b is present in the prostate cancer cells prior to the sample preparation. A further possibility for the generation of the truncated Stat5a/b forms would be by proteases that are either resistant to the action of the protease inhibitors present during the lysis and harvest of the cells or proteases that are not immediately inactivated by the inhibitors. Proteolysis could then occur during the extraction procedure after disruption of the cellular structures. To exclude this possibility, we prepared DU145 cell pellets that were flash frozen in liquid nitrogen and directly lysed in boiling SDS loading buffer (Fig. 2B, lane 1). In addition, cells were lysed in cell lysis buffer in the presence or (Fig. 2B, lane 2) in the absence of proteases inhibitors (Fig. 2B, lane 3) and boiled in SDS loading buffer. The short form 50 kDa Stat5a/b was present in human prostate cancer cells following each of the three different sample preparations indicating that the short form Stat5a/b is generated in vivo and not in vitro.
3.3. The short form of Stat5a/b in human prostate cancer cells lacks the N-terminus
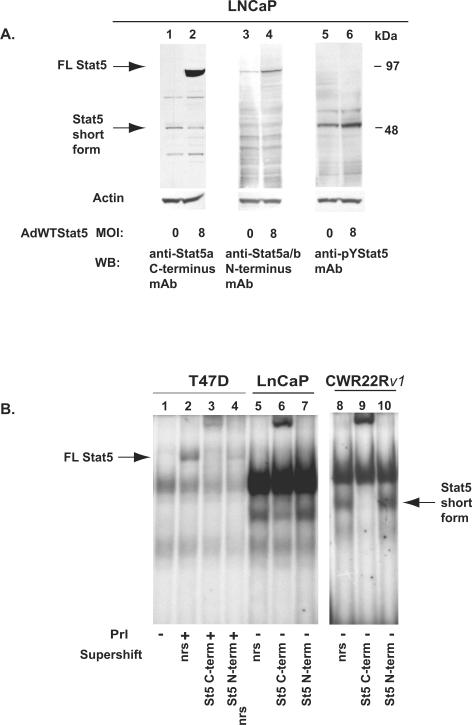
To determine whether the short form Stat5a/b is truncated at its N- or C-terminus, we blotted whole cell lysates of LNCaP cells with antibodies recognizing the C-terminus of Stat5a/b (Fig. 3A; lanes 1 and 2), N-terminus of Stat5a/b (lanes 3 and 4) or activated, C-terminally tyrosine phosphorylated Stat5a/b (lanes 5 and 6). LNCaP cells were infected with AdWTStat5 at the MOI of 8 (lanes 2, 4 and 6) or mock-infected (lanes 1, 3 and 5), and equal amounts of cell lysates were loaded in each lane. The anti-Stat5a/b mAb raised against C-terminal epitopes recognized both the long forms and short forms of Stat5a/b (lanes 1 and 2; left arrows). In contrast, immunoblotting with an anti-Stat5a/b antibody raised against epitopes in the N-terminus failed to recognize the short form of Stat5a/b in the parallel samples (lanes 3 and 4). Immunoblotting with the anti-pYStat5a/b antibody showed that the majority of the N-terminally truncated Stat5a/b was phosphorylated (lanes 5 and 6). These results suggested that the short form Stat5a/b in human prostate cancer cells is N-terminally truncated.
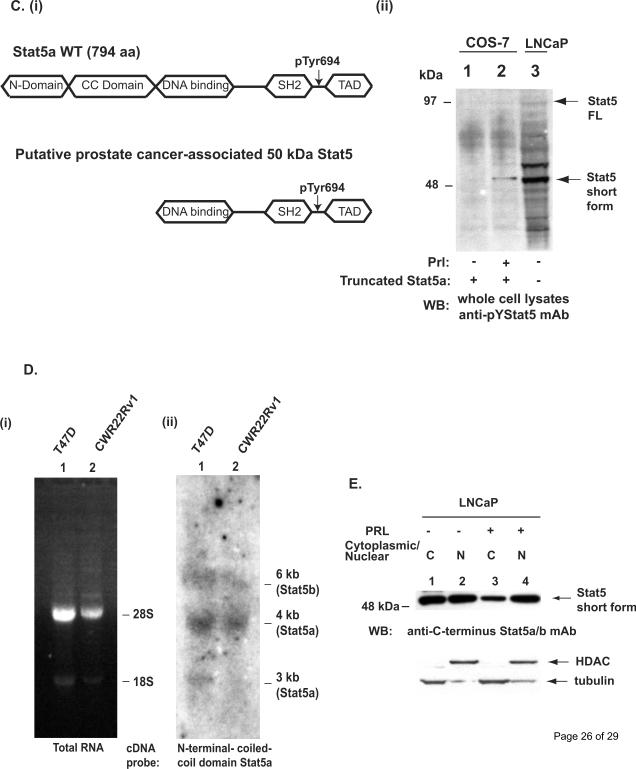
Figure 3. The short form Stat5a/b expressed in human prostate cancer cells is N-terminally truncated and generated at posttranslational level.
A, LNCaP cells were mock-infected or infected with AdWtStat5a at MOI of 8. Equal amounts of proteins were loaded in each lane, separated and immunoblotted with anti-Stat5a/b (lanes 1–2) mAbs raised against the C-terminal epitopes. The full-length (FL) and short Stat5a/b forms are indicated by arrows (left). Immunoblotting with anti-Stat5a/b mAb against epitopes in the N-terminus of Stat5a/b failed to recognize the short form of Stat5a/b in the parallel samples (lanes 3–4). Immunoblotting with the anti-pYStat5a/b mAb of parallel samples indicated that the majority of the N-terminally truncated short form Stat5a/b was phosphorylated. Actin immunoblotting demonstrates equal amounts of proteins loaded. B, EMSA of T47D human breast cancer cells stimulated with hPrl for 30 min with unstimulated cells as control show the full-length Stat5a/b expression in human breast cancer cells (arrow on left) (lanes 1–4) while not in human prostate cancer cells LNCaP (lanes 5–7) or CWR22Rv1 cells (lanes 8–10). The full-length Stat5a/b in T47D breast cancer cells was supershifted with the anti-Stat5a/b antibodies against the N-terminus or C-terminus of Stat5a/b (lanes 3 and 4), while the short form Stat5a/b in LNCaP and CWR22Rv1 cells (arrow on right) was not supershifted with only the antibody against the N-terminus of Stat5a/b (lanes 7 and 10). C, Schematic presentation of the truncated Stat5a generated by cloning (i). The truncated Stat5a lacking the N-terminal and coiled-coil (CC)-domain was transfected to COS-7 cells (ii). COS-7 cells were starved and stimulated with Prl (30 min) with unstimulated cells as control. Whole cell lysates of COS-7 cells were immunoblotted side-by-side with lysates of LNCaP cells with anti-pYStat5a/b mAb. D, Ethidium bromide staining of total RNA extracted from T47D human breast cancer cells or CWR22Rv1 prostate cancer cells and separated on 1% agarose gel (i). Hybridization of the Northern blot with a cDNA probe of the N-terminal and CC-domain of Stat5a demonstrates equal sizes of the mRNA transscripts in breast cancer cells expressing the full-length Stat5a protein and prostate cancer cells expressing the short form Stat5a/b protein (ii). E, Western blotting of cytoplasmic and nuclear (N) extracts of LNCaP cells with anti-C-terminus Stat5a/b mAb. Before preparation of the cytoplasmic and nuclear extracts, the cells had been starved for 16 h and stimulated with 10 nM human prolactin (Prl) for 30 min. Anti-HDAC and anti-tubulin immunoblotiing verify the quality of the nuclear and cytoplasmic extracts (lower panel).
To establish by another methodological approach that the short form Stat5a/b in prostate cancer cells lacks the N-terminus, we supershifted Stat5a/b in nuclear extracts of T47D human breast cancer cells, LNCaP and CWR22Rv1 human prostate cancer cells using antibodies raised against either the C-terminus or the N-terminus of Stat5a/b (Fig. 3B). In T47D cells, only the full length Stat5a/b was detected in prolactin-stimulated cells, which was supershifted by the antibodies against the C-terminus or N-terminus of Stat5a/b (lanes 1–4). In LNCaP and CWR22Rv1 cells, only the short form Stat5a/b was detected and the short form Stat5a/b was supershifted only by the antibody against the C-terminus but not by the antibody against the N-terminus (lanes 5–10).
We next generated a Stat5a construct lacking both the N-terminal domain and the coiled-coiled (CC) domain (Fig. 3C; (i)) and expressed the construct in COS-7 cells (Fig.3C; (ii)). The size of the transfected Stat5a construct lacking the N-terminus and CC-domain (Fig. 3C; (ii) lane 2) corresponded to the size of the naturally occurring short form Stat5a/b in LNCap cells (Fig. 3C; (ii) lane 3).
Given that Stat5a/b protein expressed in human prostate cancer cell lines is truncated, we next aimed to establish whether the short form Stat5a/b is generated at mRNA or protein level. Total RNA extracted from T47D breast cancer cells and CWR22Rv1 prostate cancer cells (Fig. 3D; (i)) was hybridized with a cDNA probe corresponding to the N-terminal and CC-domain of Stat5a (Fig. 3D; (ii)). The Northern blotting demonstrates that the sizes of the Stat5 transcripts in T47D and CWR22Rv1 cells are essentially the same indicating that the short form Stat5a/b is not generated at mRNA level resulting from an alternatively spliced mRNA but rather generated at post-translational level.
To examine whether the N-terminally truncated short form Stat5a/b is generated in the nucleus or cytoplasm of human prostate cancer cells, cytoplasmic and nuclear extracts were prepared from LNCaP cells unstimulated or stimulated with Prl for 30 min. Immunoblotting of the cytoplasmic and nuclear extracts suggest that the short form Stat5a/b is present both in the cytoplasm and the nucleus in unstimulated prostate cancer cells while the majority of the short form in prolactin-stimulated cells is present in the nucleus (Fig. 3E). Collectively, these results shown in Figure 3 indicated that the short form of Stat5a/b expressed in human prostate cancer cells is N-terminally truncated at post-translational level predominantly in the nucleus.
3.4. PIAS3 physically interacts with the N-terminus of Stat5
It is well established that PIAS3 interacts with Stat5 and mediates transcriptional repression of Stat5a/b (Gross et al., 2001; Rycyzyn and Clevenger, 2002). To identify the domain of Stat5a/b mediating the physical interaction with PIAS3, we first co-immunoprecipitated FLAG-tagged full-length Stat5a with HA-tagged PIAS3 transfected to COS-7 cells (Fig. 4A). The cells were starved, stimulated with Prl with unstimulated cells as control, and Stat5 was immunoprecipitated with anti-FLAG pAb and immunoblotted with anti-HA mAb (Fig. 4A(i), left panel). PIAS3 (68 kDa) co-immunoprecipitation with Stat5 was slightly increased in Prl-stimulated cells and immunoprecipitation with IgG shows that the second major band of approximately 85 kDa is a result of unspecific binding of the anti-HA antibody. Conversely, IP of the cell lysates with anti-HA pAb yielded a 90–110 kDa protein corresponding to the FLAG-tagged STAT5a, as shown by immunoblotting with anti-FLAG mAb (Fig. 4A (ii), lane1–2 and 5–6). The IP with IgG was loaded in lanes 3 and 4.
In the third set of experiments, we generated deletion constructs of Stat5a that were either C-terminally truncated or N-terminally truncated (Fig. 4B) and tagged these constructs with the FLAG-epitopes (Fig. 4C, lanes 1 and 2). COS-7 cells were transfected with the deletion constructs, HA-tagged PIAS3 was immunoprecipitated with the anti-HA pAb and blotted with anti-FLAG mAb. IP of PIAS3 pulled down the C-terminally-deleted Stat5a construct of approximately 70 kDa (Fig. 4C, lane 4) but not the N-terminally deleted Stat5a indicating that PIAS3 interacts with the N-terminus of Stat5a/b. In summary, these results suggest that Stat5 physically interacts with PIAS3 and this interaction is mediated by the N-terminus of Stat5.
3.5. PIAS3 down-regulates the transcriptional activity of Stat5 in breast cancer cells but not in prostate cancer cells
PIAS3 is expressed in prostate cancer tissues and cell lines (Assikis et al., 2004). Our finding showing that Stat5a/b and PIAS3 physically interact in prostate cancer cells through the N-terminus of Stat5a/b led us to test whether PIAS3 is able to inhibit transcriptional activity of Stat5a/b in human prostate cancer cells. MCF-7 human breast cancer cells and LNCaP human prostate cancer cells were cotransfected with beta-Casein-luciferase reporter gene, Prl receptor (PrlR), Stat5a and PIAS3, as indicated, serum-starved for 20 h, and stimulated with human Prl (hPrl) (10 nM) with non-stimulated cells as control (Fig. 5). In both MCF-7 and LNCaP cells not transfected with PIAS3 plasmid, Prl induced transcriptional activity of Stat5a by approximately 10-fold. In MCF-cells, co-expression of PIAS3 blocked the Prl-induced increase of Stat5a transcriptional activity. In contrast, PIAS3 co-expression did not affect Stat5a transcriptional activity in LNCaP cells. EMSA shows the absence of the full-length form in LNCaP cells due to the conversion of the full-length Stat5 to the short form Stat5 (Fig. 5B). These results suggest that PIAS3 fails to repress the transcriptional activity of Stat5 in human prostate cancer cells.
4. Discussion
The Jak2-Stat5 signaling pathway is of significant interest in the search for new molecular targets for therapy development for prostate cancer. Stat5a/b is critical for prostate cancer cell viability and tumor growth in vivo (Abdulghani et al., 2008; Ahonen et al., 2003; Dagvadorj et al., 2007), nuclear active Stat5a/b expression is increased in high-grade prostate cancers (Li et al., 2004; Li et al., 2005), and nuclear active Stat5a/b predicts poor clinical outcome of prostate cancer (Li et al., 2005). In addition, Stat5a/b is activated in the majority of distant prostate cancer metastases to lymph nodes and bone, and active Stat5a/b promotes metastatic behavior of human prostate cancer cells in vitro and in vivo (Gu et al., 2010a). The mechanisms underlying increased nuclear Stat5a/b in prostate cancer cells are unclear. In this work, we demonstrated that the N-terminally cleaved form of Stat5a/b is the predominant form binding to DNA in prostate cancer cells. This truncated form of Stat5a/b is generated in prostate cancer cells in vivo, not in vitro during the sample preparation. We further demonstrate that the N-domain of Stat5a/b binds to PIAS3 and PIAS3 inhibits transcriptional activity of Stat5a/b in breast cancer cells but not in prostate cancer cells.
The key finding of this work was the demonstration that N-terminally truncated Stat5a/b is the predominant form that binds to DNA in human prostate cancer cells. This was shown by electromobility shift assay, and Western blotting verified the short form Stat5a/b protein expression both in human prostate cancer cell lines and clinical prostate cancers. We have recently identified various Stat5a/b-induced genes in human prostate cancer cells including Cyclin-D1 (Dagvadorj et al., 2008), Bcl-Xl (Dagvadorj et al., 2008), Bcl-2 (Gu et al., 2010a; Gu et al., 2010b), KLF-4 (Gu et al., 2010a; Gu et al., 2010b) and PDCD4 (Gu et al., 2010a; Gu et al., 2010b). Future work will determine in human prostate cancer cells whether N-terminally truncated Stat5a/b binds to the promoters of these genes in vivo by chromatin immunoprecipitation assays.
This cleaved short form Stat5a/b is generated post-translationally in vivo in prostate cancer cells, not during the preparation of the cell extracts. This concept is supported by our findings showing that the short form Stat5a/b was present in the prostate cancer cell nuclear extracts in EMSA prepared regardless whether protease inhibitors were added in the extraction buffer suggesting the presence of the short form Stat5a/b in the cells prior to sample preparation. Alternatively, proteolysis of Stat5a/b could occur by the action of a protease that gains access to Stat5 after the disintegration of the cellular structures. However, the truncated Stat5a/b was present in Western blot analysis of human prostate cancer cells after direct lysis of the frozen cell pellets in boiling SDS loading buffer. Previously, C-terminally truncated and transcriptionally inactive Statt5a/b forms have been described in hematopoietic cells (Azam et al., 1997; Garimorth et al., 1999; Schuster et al., 2007). However, C-terminally truncated Stat5a/b isoforms have been reported to be likely generated in vivo during the sample preparation (Azam et al., 1997; Garimorth et al., 1999; Schuster et al., 2007).
The results of our work presented here also indicate that the short form of Stat5a/b is generated at the protein level and not at the transcriptional level. Given that PIAS3 binds to the N-terminus of Stat5a/b to repress Stat5a/b-driven transcription, N-terminal cleavage of Stat5a/b may represent a molecular mechanism by which Stat5a/b is able to evade transcriptional repression induced by PIAS3 in human prostate cancer cells. Since increased Stat5a/b-driven gene transcription promotes prostate cancer growth and progression, it will be important to purify and amino acid-sequence the N-terminally truncated Stat5a/b to identify the type of protease(s) responsible for proteolytic cleavage of Stat5a/b in prostate cancer. These enzymes may provide molecular targets for prostate cancer therapy by restoring the PIAS inhibition of Stat5a/b-driven gene expression in human prostate cancer cells.
In conclusion, this study provides the first evidence of N-terminal cleavage of Stat5a/b in human prostate cancer cells. The protease(s) responsible for generation of the short form Stat5a/b in prostate cancer may present new therapeutic target proteins.
Acknowledgements
Shared Resources of KCC are partially supported by NIH Grant CA56036-08 (Cancer Center Support Grant, to KCC). This work was supported by ACS (RSG-04-196-01-MGO), DOD (W81XWH-05-01-0062 and W81XWH-07-1-0411) and NIH NCI (1RO1CA113580-01A1) grants.
Abbreviations
- PIAS3
Protein inhibitor of activated Stat3
- DHT
dihydrotestosterone
- Prl
prolactin
- PSA
prostate specific antigen
- Stat5
signal transducer and activator of transcription 5
- EMSA
Electromobility Shift Assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdulghani J, Gu L, Dagvadorj A, Lutz J, Leiby B, Bonuccelli G, et al. Stat3 promotes metastatic progression of prostate cancer. Am J Pathol. 2008;172:1717–28. doi: 10.2353/ajpath.2008.071054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahonen TJ, Harkonen PL, Rui H, Nevalainen MT. PRL signal transduction in the epithelial compartment of rat prostate maintained as long-term organ cultures in vitro. Endocrinology. 2002;143:228–38. doi: 10.1210/endo.143.1.8576. [DOI] [PubMed] [Google Scholar]
- Ahonen TJ, Xie J, LeBaron MJ, Zhu J, Nurmi M, Alanen K, et al. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. J Biol Chem. 2003;278:27287–92. doi: 10.1074/jbc.M304307200. [DOI] [PubMed] [Google Scholar]
- Assikis VJ, Do KA, Wen S, Wang X, Cho-Vega JH, Brisbay S, et al. Clinical and biomarker correlates of androgen-independent, locally aggressive prostate cancer with limited metastatic potential. Clin Cancer Res. 2004;10:6770–8. doi: 10.1158/1078-0432.CCR-04-0275. [DOI] [PubMed] [Google Scholar]
- Azam M, Lee C, Strehlow I, Schindler C. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997;6:691–701. doi: 10.1016/s1074-7613(00)80445-8. [DOI] [PubMed] [Google Scholar]
- Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–51. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr., Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–39. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- Dagvadorj A, Collins S, Jomain JB, Abdulghani J, Karras J, Zellweger T, et al. Autocrine prolactin promotes prostate cancer cell growth via Janus kinase-2-signal transducer and activator of transcription-5a/b signaling pathway. Endocrinology. 2007;148:3089–101. doi: 10.1210/en.2006-1761. [DOI] [PubMed] [Google Scholar]
- Dagvadorj A, Kirken RA, Leiby B, Karras J, Nevalainen MT. Transcription factor signal transducer and activator of transcription 5 promotes growth of human prostate cancer cells in vivo. Clin Cancer Res. 2008;14:1317–24. doi: 10.1158/1078-0432.CCR-07-2024. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr. STATs and gene regulation. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Garimorth K, Welte T, Doppler W. Generation of carboxy-terminally deleted forms of STAT5 during preparation of cell extracts. Exp Cell Res. 1999;246:148–51. doi: 10.1006/excr.1998.4289. [DOI] [PubMed] [Google Scholar]
- Gross M, Liu B, Tan J, French FS, Carey M, Shuai K. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene. 2001;20:3880–3887. doi: 10.1038/sj.onc.1204489. [DOI] [PubMed] [Google Scholar]
- Gu L, Dagvadorj A, Lutz J, Leiby B, Bonuccelli G, Lisanti MP, et al. Transcription factor Stat3 stimulates metastatic behavior of human prostate cancer cells in vivo, whereas Stat5b has a preferential role in the promotion of prostate cancer cell viability and tumor growth. Am J Pathol. 2010a;176:1959–72. doi: 10.2353/ajpath.2010.090653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Vogiatzi P, Puhr M, Dagvadorj A, Lutz J, Ryder A, et al. Stat5 promotes metastatic behavior of human prostate cancer cells in vitro and in vivo. Endocr Relat Cancer. 2010b;17:481–93. doi: 10.1677/ERC-09-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Interpretation of cytokine signaling through the transcription factors STAT5A and STAT5B. Genes Dev. 2008;22:711–21. doi: 10.1101/gad.1643908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. 2001;13:211–7. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–8. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- John S, Vinkemeier U, Soldaini E, Darnell JE, Jr., Leonard WJ. The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol. 1999;19:1910–8. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–90. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Li H, Ahonen TJ, Alanen K, Xie J, LeBaron MJ, Pretlow TG, et al. Activation of signal transducer and activator of transcription 5 in human prostate cancer is associated with high histological grade. Cancer Res. 2004;64:4774–82. doi: 10.1158/0008-5472.CAN-03-3499. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang Y, Glass A, Zellweger T, Gehan E, Bubendorf L, et al. Activation of signal transducer and activator of transcription-5 in prostate cancer predicts early recurrence. Clin Cancer Res. 2005;11:5863–8. doi: 10.1158/1078-0432.CCR-05-0562. [DOI] [PubMed] [Google Scholar]
- Li P, Yu X, Ge K, Melamed J, Roeder RG, Wang Z. Heterogenous expression and functions of androgen receptor co-factors in primary prostate cancer. Am J Pathol. 2002;161:1467–1474. doi: 10.1016/S0002-9440(10)64422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Z, Lutz J, Nevalainen MT. Transcription factor Stat5a/b as a therapeutic target protein for prostate cancer. Int J Biochem Cell Biol. 42:186–92. doi: 10.1016/j.biocel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linja MJ, Porkka KP, Kang Z, Savinainen KJ, Janne OA, Tammela TL, et al. Expression of androgen receptor coregulators in prostate cancer. Clin Cancer Res. 2004;10:1032–40. doi: 10.1158/1078-0432.ccr-0990-3. [DOI] [PubMed] [Google Scholar]
- Nevalainen MT, Xie J, Bubendorf L, Wagner KU, Rui H. Basal activation of transcription factor signal transducer and activator of transcription (Stat5) in nonpregnant mouse and human breast epithelium. Mol Endocrinol. 2002;16:1108–24. doi: 10.1210/mend.16.5.0839. [DOI] [PubMed] [Google Scholar]
- Palvimo JJ. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem Soc Trans. 2007;35:1405–8. doi: 10.1042/BST0351405. [DOI] [PubMed] [Google Scholar]
- Rui H, Djeu JY, Evans GA, Kelly PA, Farrar WL. Prolactin receptor triggering. Evidence for rapid tyrosine kinase activation. J Biol Chem. 1992;267:24076–81. [PubMed] [Google Scholar]
- Rui H, Lebrun JJ, Kirken RA, Kelly PA, Farrar WL. JAK2 activation and cell proliferation induced by antibody-mediated prolactin receptor dimerization. Endocrinology. 1994;135:1299–306. doi: 10.1210/endo.135.4.7925093. [DOI] [PubMed] [Google Scholar]
- Rycyzyn MA, Clevenger CV. The intranuclear prolactin/cyclophilin B complex as a transcriptional inducer. Proc Natl Acad Sci U S A. 2002;99:6790–5. doi: 10.1073/pnas.092160699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci. 2003;60:2561–74. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster B, Hendry L, Byers H, Lynham SF, Ward MA, John S. Purification and identification of the STAT5 protease in myeloid cells. Biochem J. 2007;404:81–7. doi: 10.1042/BJ20061877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–68. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006;16:196–202. doi: 10.1038/sj.cr.7310027. [DOI] [PubMed] [Google Scholar]
- Tan SH, Nevalainen MT. Signal transducer and activator of transcription 5A/B in prostate and breast cancers. Endocr Relat Cancer. 2008;15:367–90. doi: 10.1677/ERC-08-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Banerjee S. Differential PIAS3 expression in human malignancy. Oncol Rep. 2004;11:1319–24. [PubMed] [Google Scholar]