Abstract
Epistasis or gene-gene interaction is a fundamental component of the genetic architecture of complex traits such as disease susceptibility. Multifactor dimensionality reduction (MDR) was developed as a nonparametric and model-free method to detect epistasis when there are no significant marginal genetic effects. However, in many studies of complex disease, other covariates like age of onset and smoking status could have a strong main effect and may potentially interfere with MDR's ability to achieve its goal. In this paper, we present a simple and computationally efficient sampling method to adjust for covariate effects in MDR. We use simulation to show that after adjustment, MDR has sufficient power to detect true gene-gene interactions. We also compare our method with the state-of-art technique in covariate adjustment. The results suggest that our proposed method performs similarly, but is more computationally efficient. We then apply this new method to an analysis of a population-based bladder cancer study in New Hampshire.
Key Words: Covariate adjustment, Multifactor dimensionality reduction, Epistasis
Introduction
The major burden of ill-health in western society, and to a growing extent in developing societies, is due to common chronic diseases such as coronary heart disease, stroke, cancer, and diabetes. Although heritability estimates suggest that genetic factors play an important role in chronic diseases, susceptibility cannot be predicted by individual DNA sequence variations. One explanation is that a significant proportion of heritability is due to complexities in the genotype-phenotype mapping relationship resulting from gene-gene interactions (i.e. epistasis), and other phenomena such as gene-environment interactions, epigenetics and locus heterogeneity. Before these phenomena can be fully explored, we must develop the statistical and computational methods that are powered to detect and characterize them.
Multifactor dimensionality reduction (MDR) was developed as a nonparametric and model-free data mining method for detecting, characterizing, and interpreting epistasis in the absence of significant marginal effects in genetic and epidemiologic studies of complex traits such as disease susceptibility [Ritchie et al., 2001, 2003; Hahn et al., 2003; Hahn and Moore, 2004; Moore, 2004; Moore et al., 2006; Moore, 2007]. The goal of MDR is to change the representation of the data using a constructive induction algorithm to make non-additive interactions easier to detect using any classification method such as naïve Bayes or logistic regression [Moore et al., 2006; Moore, 2007]. One limitation of MDR has been the lack of a covariate adjustment mechanism to remove the effects of some well-known risk factors, such as age of onset and smoking status. Lou et al. [2007] recently proposed a generalized version of MDR (GMDR) that can account for covariant effects in MDR. The GMDR approach works under the generalized linear model framework and uses score-based statistics to adjust for any covariate effect. In short, GMDR first fits an appropriate generalized linear model with outcome and covariates and then uses the residuals to replace the outcome and run MDR. Since the residuals have no covariate effects, none of the covariates are expected to be chosen as the best model.
While GMDR has been shown to be very effective for adjusting for covariates [Lou et al., 2007], it does suffer from the drawback of being computationally inefficient and complex. GMDR redefines MDR's balanced accuracy to remove the covariate contribution. This step makes it an attractive method for detecting the true model in the presence of a covariate effect, but it also introduces greater computation burden for every genotype combination considered. The goal of the present study was to develop and evaluate a simple, effective and computationally efficient method for adjusting for covariate effects. We introduce here a simple sampling method for covariate adjustment. Using data simulated from 70 different epistasis models, we compared the power of our proposed method with that of GMDR, and found that our method is simpler and more computationally efficient while retaining equivalent power. We then present an application of the new method to an analysis of gene-gene interactions and bladder cancer susceptibility. The results of this study will play an important role in detecting epistasis using MDR when computational efficiency is critically important. We provide this new sampling method to adjust for covariate effects in the latest version of the freely available and open-source MDR software.
Methods
Overview of Multifactor Dimensionality Reduction
The goal of MDR is to change the representation space of the data using constructive induction to make interactions easier to detect. This is done by combining two or more variables into a single attribute that can be modeled using a discrete data classifier. The general process of defining a new attribute as a function of two or more other attributes is referred to as constructive induction or attribute construction and was first described by [Michalski, 1983]. Constructive induction using MDR was accomplished in this study as described below. Given a threshold T, a multilocus genotype combination, for example, is considered ‘high-risk’ if the cases:controls ratio exceeds T; otherwise it is considered low-risk. Utilizing this model, genotypes were labeled as either ‘high-risk’ or ‘low-risk’, and a new binary attribute was created using the two levels. Here, we set T to the cases:controls ratio in the dataset being analyzed as recommended by Velez et al. [2007]. Figure 1 illustrates this process for a dataset of 200 cases and 200 controls that was simulated using the penetrance function in table 1.
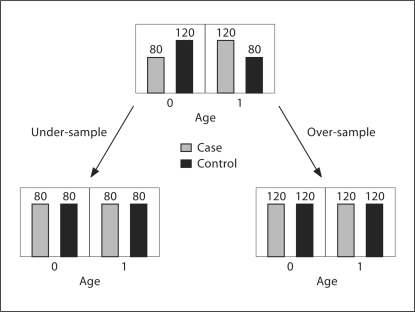
Fig. 1.
Demonstration of the over-sample and under-sample method for covariate adjustment. Assuming that age is the covariate of interest and it only has 2 levels: in under-sample remove the age effect by deletion of 40 samples from the controls in age 0 group and another 40 samples from the cases in age 1 group. In over-sample use 40 more cases from the cases in age 0 group and use another 40 samples from the controls in age 1 group.
Table 1.
Power comparison on 70 epistasis models
| Allele frequency | Heritability | MDR | Over-sample | Under-sample | GMDR |
|---|---|---|---|---|---|
| 0.8 | 0.400 | 0.738 (0.711,0.766) | 0.995 (0.991,0.999) | 0.987 (0.979, 0.994) | 0.997 (0.993,1) |
| 0.6 | 0.400 | 0.935 (0.919,0.95) | 0.998 (0.995, 1) | 0.998 (0.995, 1) | 0.999 (0.997, 1) |
| 0.8 | 0.300 | 0.615 (0.584,0.645) | 0.959 (0.947, 0.972) | 0.938 (0.923,0.953) | 0.975 (0.965,0.985) |
| 0.6 | 0.300 | 0.903 (0.885,0.922) | 0.994 (0.989,0.999) | 0.990 (0.984, 0.996) | 0.998 (0.995, 1) |
| 0.8 | 0.200 | 0.588 (0.558,0.619) | 0.937 (0.922,0.952) | 0.907 (0.889,0.925) | 0.953 (0.94,0.966) |
| 0.6 | 0.200 | 0.457 (0.426, 0.488) | 0.809 (0.784, 0.833) | 0.751 (0.724, 0.778) | 0.849 (0.827,0.871) |
| 0.8 | 0.100 | 0.309 (0.28,0.337) | 0.659 (0.63,0.688) | 0.609 (0.579,0.639) | 0.712 (0.684,0.74) |
| 0.6 | 0.100 | 0.638 (0.609,0.668) | 0.804 (0.779, 0.829) | 0.765 (0.739, 0.792) | 0.832 (0.808,0.855) |
| 0.8 | 0.050 | 0.087 (0.069,0.104) | 0.243 (0.216,0.27) | 0.216 (0.19,0.241) | 0.265 (0.237,0.292) |
| 0.6 | 0.050 | 0.716 (0.688, 0.744) | 0.818 (0.794,0.842) | 0.792 (0.767,0.818) | 0.838 (0.815,0.861) |
| 0.8 | 0.025 | 0.179 (0.155,0.203) | 0.308 (0.279,0.336) | 0.287 (0.259,0.315) | 0.325 (0.296,0.354) |
| 0.6 | 0.025 | 0.552 (0.521,0.583) | 0.640 (0.611,0.67) | 0.616 (0.586,0.646) | 0.649 (0.619,0.679) |
| 0.8 | 0.010 | 0.022 (0.013,0.031) | 0.058 (0.043, 0.072) | 0.051 (0.037,0.064) | 0.059 (0.044, 0.073) |
| 0.6 | 0.010 | 0.18 (0.157,0.204) | 0.235 (0.209,0.261) | 0.215 (0.19,0.24) | 0.26 (0.232,0.287) |
The numbers in parentheses represent the 95% confidence interval for the power estimate.
We used a simple probabilistic classifier that is similar to naïve Bayes [Hahn and Moore, 2004] to model the relationship between variables constructed using MDR and case-control status. Naïve Bayes classifiers were assessed using balanced accuracy as recommended by Velez et al. [2007]. Balanced accuracy is defined as the arithmetic mean of sensitivity and specificity:
where TP are true positives, TN are true negatives, FP are false positives, and FN are false negatives. For each dataset we evaluated all possible MDR attributes that are functions of two SNPs. Ten-fold cross-validation was used to select an MDR model with maximum testing accuracy (i.e. most likely to generalize) and maximum cross-validation consistency (CVC) as described previously [Ritchie et al., 2001; Hahn et al., 2003; Ritchie et al., 2003; Moore, 2004]. An open-source MDR software package is freely available at www.epistasis.org.
Sampling-Based Approaches for Covariate Adjustment
We assume that the covariate of interest is a discrete variable with K levels. When it is continuous, we can use a median or quantile cutoff to generate a discrete variable. Let T be the cases:controls ratio for the whole dataset; at each level in the discrete variable, we count the number of cases and controls, nk and mk, k = 1, …, K. If the covariate has a marginal effect, it would indicate that there exists an index k satisfying nk ≠ T·mk. We propose two sampling methods, over-sampling and under-sampling, to remove the covariate effect (fig. 1). At each level, when nk > T·mk, the over-sampling method selects nk – T·mk samples from the controls in the k-thlevel, and the under-sampling method randomly deletes nk – T·mk samples from the cases in the k-th level. This approach works in the same fashion when nk < T·mk. Note here that for some k, if nk = 0 or mk = 0, we delete all samples in level k. The goal of both the over-sampling and under-sampling methods is to make nk = T·mk for all k via sampling so that the main effect of the covariate is removed.
It is important to note that if there are multiple covariates that need to be adjusted, we can generate a single discrete variable using an interaction term. For example, if we need to adjust for age and smoking status, we will assume that age and smoking status are both discrete variables with two levels each. We will then create their interaction term as a four level, discrete variable, and follow the procedure described above to remove its effects. After the covariate effect is removed, we can run MDR on the reduced or over-sampled dataset to identify the top genotype interaction models.
The above sampling methods are derived from the assumption that covariate effects and genotype effects are independent of each other. The sampling methods are based on the covariate effect and thus should not affect our ability to detect the genotype effect.
Data Simulation
We simulated datasets of 400 samples with balanced numbers of cases and controls using a total of 70 previously published two-locus epistasis models [Velez et al., 2007]. These purely epistatic models were distributed evenly across seven broad-sense heritabilities (0.01, 0.025, 0.05, 0.1, 0.2, 0.3, and 0.4) and two different minor allele frequencies (0.2 and 0.4). Five models for each of the 14 heritability-allele frequency combinations were generated for a total of 70 models. We simulated 1,000 datasets for each model to estimate power. Each pair of functional polymorphisms was embedded within a set of 20 independent SNPs.
For any given model, we first generated a continuous risk factor, age, from a normal distribution of mean 60 and standard deviation 10. Then we generated the case-control status based on the corresponding penetrance function and age (assuming SNP 1 and 2 are the two functional polymorphisms):
Here fij is the element from the i-th row and j-th column of the penetrance function. In this model, age has a continuous effect on the phenotype. When age = 80, the probability of being a case is the same as the one given in penetrance function, fij. When age = 60, this probability will drop to fij/2. With age increases of one year, there is an approximate 3.5% increase in risk of being a case.
We used a median cutoff to dichotomize age so that it could be treated as a covariate in the proposed sampling methods. We used over-sampling and under-sampling to remove the age effect and then ran MDR to select the best model from all one-, two-, and three-way models. We also run GMDR on the same datasets with true age as a continuous variable.
The best model was defined as the one with highest balanced testing accuracy. If a tie occurred, CVC was used to break the tie. If there was still a tie, we chose the most parsimonious model. Power or success rate was defined as the number of times the best model contained the two functional polymorphisms in the absence of age out of each set of 1,000 datasets. We then averaged these results over the five models with the heritability-allele frequency combination. We also estimated the 95% confidence interval for the power or success rate estimation based on binomial distribution with normal approximation.
Application to Bladder Cancer
We demonstrated the use of the new covariate adjustment method with real data by applying it to a genetic epidemiology study that examined the relationship between DNA repair gene SNPs, smoking, and bladder cancer susceptibility that was previously analyzed using MDR and a 1,000-fold permutation test [Andrew et al., 2006]. The study analyzed 355 bladder cancer cases and 559 controls ascertained from the state of New Hampshire and focused specifically on genes that play an important role in the repair of DNA sequences that have been damaged by chemical compounds (e.g. carcinogens). Seven SNPs were measured, including two from the X-ray repair cross-complementing group 1 (XRCC1) gene, one from the XRCC3 gene, two from the Xeroderma pigmentosum group D (XPD) gene, one from the nucleotide excision repair (XPC) gene, and one from the AP endonuclease 1 (APE1) gene. Each of these genes plays an important role in DNA repair. Smoking is a known risk factor for bladder cancer and was included in the analysis along with gender and age for a total of 10 attributes. Age was discretized to > or ≤50 years.
A parametric linear statistical analysis of each attribute individually revealed a significant independent marginal effect of smoking as expected (p < 0.05). However, none of the measured SNPs were significant predictors of bladder cancer individually (p > 0.05). Andrew et al. [2006] used MDR to exhaustively evaluate all possible two-, three-, and four-way interactions among the genetic and environmental attributes. For each combination of attributes, a single constructed attribute was evaluated using a naïve Bayes classifier. Training and testing accuracy were estimated using 10-fold cross-validation. A best model was selected that maximized the testing accuracy. The best model had a testing accuracy of approximately 0.63 and included two SNPs from the XPD gene and smoking. They statistically evaluated this model with a 1,000-fold permutation test and determined these results to be highly significant (p < 0.001). Post hoc analysis of the MDR model using entropy-based measures of interaction information revealed that the two XPD polymorphisms had evidence of nonlinear interaction or synergy in the near complete absence of marginal effects. Interestingly, the joint effect of the two XPD SNPs was larger than the independent effect of smoking. As such, these data provide an ideal test case for the proposed covariate adjustment methods. Is the nonlinear interaction between the two XPD SNPs statistically significant after adjusting for the effects of smoking and other covariates?
Results
Simulation Results
In table 1, we can see that over-sampling, under-sampling and GMDR all have ample power compared to unadjusted MDR. Over-sampling yields slightly higher power than under-sampling, which one would expect given the increased sample size. When heritability is high (e.g. 0.1–0.4), over-sampling and GMDR both have a high power or success rate. When heritability is low (e.g. 0.01–0.05), there is virtually no difference between the power of GMDR and the power of over-sampling given their confidence intervals. Interestingly, this simulation is designed to favor GMDR, which can take full advantage of the continuous covariate.
When applying the over-sampling and under-sampling methods, we dichotomized the covariate variable first, which could potentially lead to the loss of some information. Intuitively, one would predict that GMDR should significantly outperform our methods. The major reason it did not is due to the true nonparametric nature of the two sampling methods. In GMDR, score statistics need to be derived based on a specific generalized linear model. For dichotomous outcomes, the default model for GMDR is a logit model, which assumes that the logit of the probability of being a case has a linear relationship with the covariate variable. In our simulation, we allowed the log of the probability of being a case to linearly correlate with the covariate variable. As we can see from the results, GMDR is no better than the sampling methods, even when the true underlying relationship between the outcome and the covariate is not linear. On the contrary, our proposed sampling methods have no assumption about the true relationship between outcome and covariate. Thus, it is expected to be more robust for real data that are very often not linear.
It is important to note that our over-sampling and under-sampling methods are much more computationally efficient than GMDR. Run time for over-sampling or under-sampling was less than 2 min for the analysis of 100 datasets exhaustively exploring all possible one-, two-, and three-way interactions. In comparison, GMDR took more than 14 min to achieve the same task on the same server. Thus, our sampling methods for covariate adjustment were more than seven times faster than GMDR.
Real Data Analysis
We applied our methods to a previously published population-based bladder cancer study in New Hampshire [Andrew et al., 2006]. The dataset included seven DNA repair SNPs and age, pack-years of smoking and gender as covariates. The previous analysis using MDR showed pack-years of smoking (smoke) to be the strongest predictor in the one-way model and XPD751 and XPD 312 to be the best two-way model (table 2). After adjusting for the covariate effect of pack-years of smoking (table 2), we can see that both the under-sampling and over-sampling approaches successfully removed smoking from the top models while choosing the same two-way SNP model and keeping its balanced accuracy almost the same as the unadjusted one. The results also show that both methods picked XPD.751 XPD.312 and XRCC1.194 as the best three-way model with good testing balanced accuracy. In the unadjusted case, MDR picked XPD.751 XPD.312 and smoking as the best three-way model. Since smoking has strong marginal effect, it can interfere with MDR's ability to identify the ‘true interaction model’. Via under-sampling or over-sampling, we can correct this and identify the interaction models that consist of markers with relatively small marginal effect.
Table 2.
Top models selected by MDR and covariate adjustment
| Model | Training bal. ace. | Testing bal. ace. | CVC | p value |
|---|---|---|---|---|
| Top models selected by MDR | ||||
| Smoke | 0.6139 | 0.6140 | 10/10 | <0.001 |
| xpd.751 xpd.312 | 0.6377 | 0.6295 | 10/10 | <0.001 |
| xpd.751 xpd.312 smoke | 0.6640 | 0.6296 | 10/10 | <0.001 |
| Adjusted by smoking using under-sample | ||||
| Gender | 0.5582 | 0.5579 | 10/10 | 0.026 |
| xpd.751 xpd.312 | 0.6377 | 0.6324 | 10/10 | <0.001 |
| xpd.751 xpd.312 xl.194 | 0.6549 | 0.6296 | 9/10 | <0.001 |
| Adjusted by smoking using over-sample | ||||
| Gender | 0.5552 | 0.5550 | 10/10 | 0.004 |
| xpd.751 xpd.312 | 0.6366 | 0.6366 | 10/10 | <0.001 |
| xpd.751 xpd.312 xl.194 | 0.6545 | 0.6429 | 10/10 | <0.001 |
| Adjusted by all covariates using under-sample | ||||
| xpd.312 | 0.5357 | 0.4744 | 6/10 | 0.97 |
| xpd.751 xpd.312 | 0.6374 | 0.6254 | 10/10 | <0.001 |
| xpd.751 xpd.312 xl.194 | 0.6578 | 0.6206 | 10/10 | <0.001 |
| Adjusted by all covariates using over-sample | ||||
| apel | 0.5384 | 0.5142 | 7/10 | 0.4 |
| xpd.751 xpd.312 | 0.6372 | 0.6374 | 10/10 | <0.001 |
| xpd.751 xpd.312 xl.194 | 0.6491 | 0.6136 | 4/10 | <0.001 |
bal. ace. = Balanced accuracy; CVC = cross-validation consistency.
In addition to smoking, we also adjusted for all covariates (table 2), resulting in only genetic makers in the top models. Since the best two- and three-way models from table 2 do not consist of any covariates, we adjusted for all covariates and found that those models still remain on the top with almost the same balanced accuracy. This verifies our assumption that when covariate effects are independent of genetic effects, under-sampling and over-sampling both can successfully remove the covariate effect while keeping the genetic effect intact.
Discussion
The goal of this study was to develop a simple, effective and computationally efficient approach for adjusting for covariates within the context of an MDR analysis of gene-gene interactions. We introduced above the over-sampling and under-sampling approaches to covariate adjustment. We used extensive simulation studies to demonstrate that our proposed methods can effectively filter out the covariate effect to identify the true gene-gene interaction model. Finally, we applied these methods to a previously published bladder cancer study to show that the adjustment does not interfere with the ability to identify gene-gene interaction models.
A clear advantage of our proposed method is its simplicity and the resulting computation efficiency. The total time and cost for any of the three methods is only slightly higher than the original MDR method. On the other hand, GMDR is more than seven times slower that the original MDR software. This computational efficiency will be very important as MDR is applied to high-order interactions and/or genome-wide data. In addition, we have implemented these new covariate adjustment methods in the open-source MDR software package available at www.epistasis.org. The simplicity of this approach makes it easy and intuitive to implement from within the MDR software framework. It is important to note that the simplicity and computational efficiency does not come at the price of power. Our proposed methods maintain the same power as the model-based GMDR approach but without the significant computational overhead and complexity of fitting a generalized linear model. Another advantage is that our sampling approach is truly nonparametric and uses only counts of cases and controls to over-sample or under-sample. This will be important when the assumptions of linearity are violated, as is likely to be the case in real data.
One potential drawback is that our proposed method might not work well if we adjust for too many covariates at the same time. For example, if we have 10 covariates with two levels each, then their interaction is a 1,024-level variable. Adjusting for these 10 covariates simultaneously would require enormous sample sizes due to the number of levels in the interaction term. As a result, the sampled data may risk losing a significant amount of sample due to zero count for case or controls. One way to overcome this limitation is to run the covariate adjustment sequentially. However, this assumes that there is no synergistic effect among all the covariates. We also found that our method is not highly sensitive to the number of levels used in the covariate variable. We used quantiles to create a 4-level discrete variable from age and re-ran the simulation and only observed a 5% loss in power compared to the original simulation.
We recommend several future studies with the proposed covariate adjustment methods. First, it will be important to apply this approach to other real datasets where covariate effects are likely to be important. Availability of the methods as part of the open-source MDR software package will make this possible. Second, it will be interesting to compare the covariate adjustment methods to the new explicit test of epistasis that holds marginal effects constant during permutation testing for MDR [Greene et al., 2010]. Can covariate adjustment methods be used in conjunction with these special permutation test methods? Additionally, does adjusting for covariates violate any of the assumptions of using an extreme value distribution (as proposed by Pattin et al. [2008]) to reduce the total number of permutations that need to be performed to assess the statistical significance of MDR models? These MDR-based methods and others will play an important role as genetic epidemiology transitions from the testing of single SNPs to combinations of SNPs as part of a research strategy that embraces the complexity of the genotype-phenotype mapping relationship.
Acknowledgements
This work was supported by NIH grants R01 LM009012, LM010098, AI59694, CA57494 and ES007373. We greatly appreciate the time and effort of the anonymous reviewers for their assistance in improving the manuscript.
References
- Andrew AS, Nelson HN, Kelsey KT, Moore JH, Meng A, Casella DP, Tosterson TD, Schned AR, Karagas MR. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking, and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–1037. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- Greene CS, Himmelstein DS, Kelsey KT, Williams SM, Andrew AS, Karagas MR, Moore JH. Enabling personal genomics with an explicit test of epistasis. Pac Symp Biocomput. 2010:327–336. doi: 10.1142/9789814295291_0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- Hahn LW, Moore JH. Ideal discrimination of discrete clinical endpoints using multilocus genotypes. In Silico Biol. 2004;4:183–194. [PubMed] [Google Scholar]
- Lou X, Chen G, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A generalized combinatorial approach for detecting gene by gene and gene by environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80:1125–1137. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski RS. A theory and methodology of inductive learning. Artif Intel. 1983;20:111–161. [Google Scholar]
- Moore JH, Williams SM. New strategies for identifying gene-gene interactions in hypertension. Ann Med. 2002;34:88–95. doi: 10.1080/07853890252953473. [DOI] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Moore JH. Computational analysis of gene-gene interactions in common human diseases using multifactor dimensionality reduction. Expert Rev Mol Diagn. 2004;4:795–803. doi: 10.1586/14737159.4.6.795. [DOI] [PubMed] [Google Scholar]
- Moore JH. Global view of epistasis. Nat Genet. 2005;37:13–14. doi: 10.1038/ng0105-13. [DOI] [PubMed] [Google Scholar]
- Moore JH, Williams SM. Traversing the conceptual divide between biological and statistical epistasis: Systems biology and a more modern synthesis. BioEssays. 2005;27:637–646. doi: 10.1002/bies.20236. [DOI] [PubMed] [Google Scholar]
- Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden W, Barney N, White BC. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol. 2006;241:252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Moore JH. Genome-wide analysis of epistasis using multifactor dimensionality reduction: feature selection and construction in the domain of human genetics. In: Zhu X, Davidson I, editors. Knowledge Discovery and Data Mining: Challenges and Realities with Real World Data. Hershey: IGI Press; 2007. pp. 17–30. [Google Scholar]
- Pattin KA, White BC, Barney N, Gui J, Nelson HH, Kelsey KR, Andrew AS, Karagas MR, Moore JH. A computationally efficient hypothesis testing method for epistasis analysis using multifactor dimensionality reduction. Genet Epidemiol. 2008;33:87–94. doi: 10.1002/gepi.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor dimensionality reduction reveals high-order interactions among estrogen metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- Velez DR, White BC, Motsinger AA, Bush WS, Ritchie MD, Williams SM, Moore JH. A balanced accuracy metric for epistasis modeling in imbalanced datasets using multifactor dimensionality reduction. Genet Epidemiol. 2007;31:306–315. doi: 10.1002/gepi.20211. [DOI] [PubMed] [Google Scholar]