Abstract
The emergence of serious infections due to community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) has fueled interest in the contributions of specific staphylococcal virulence factors to clinical disease. To assess the contributions of agr-dependent factors to the fate of organisms in polymorphonuclear neutrophils (PMN), we examined the consequences for organism and host cells of feeding PMN with wild-type CA-MRSA (LAC) or CA-MRSA (LAC agr KO) at different multiplicities of infection (MOIs). Phagocytosed organisms rapidly increased the transcription of RNAIII in a time- and MOI-dependent fashion; extracellular USA300 (LAC) did not increase RNAIII expression despite having the capacity to respond to autoinducing peptide-enriched culture medium. HOCl-mediated damage and intracellular survival were the same in the wild-type and USA300 (LAC agr KO). PMN lysis by ingested USA300 (LAC) was time- and MOI-dependent and, at MOIs >1, required α-hemolysin (hla) as USA300 (LAC agr KO) and USA300 (LAC hla KO) promoted PMN lysis only at high MOIs. Taken together, these data demonstrate activation of the agr operon in human PMN with the subsequent production of α-hemolysin and PMN lysis. The extent to which these events in the phagosomes of human PMN contribute to the increased morbidity and mortality of infections with USA300 (LAC) merits further study.
Key Words: Accessory gene regulator, α-Hemolysin, Polymorphonuclear neutrophil, Quorum sensing, Staphylococcus aureus, community-associated, methicillin-resistant
Introduction
An estimated 32% of the US population is colonized with Staphylococcus aureus, with the highest prevalence among children [1]. The proportion of strains that is methicillin resistant is increasing, with 64% of screened isolates from hospital intensive care units testing positive for methicillin resistance [2]. In recent years, more virulent community-associated methicillin-resistant strains of S. aureus (CA-MRSA) have emerged and their increasing prevalence in otherwise healthy individuals is a matter of great clinical concern [3,4]. Of the CA-MRSA strains, pulse field gel electrophoresis type USA300 has become the predominant cause of these infections [4].
Optimal host response to infection with S. aureus relies on the efficient ingestion and killing of microorganisms by polymorphonuclear neutrophils (PMN) [5]. However, phagocytosed bacteria react within minutes of exposure to antimicrobial agents present in PMN phagosomes, mounting a robust transcriptional response that promotes the expression of stress response and virulence genes [6,7]. The ability of CA-MRSA strains to escape immune clearance may in part explain why these strains cause more clinical pathology [7,8].
The agr system of S. aureus is a global regulator of virulence factors in culture [9] and is implicated in promoting greater disease severity in animal models of staphylococcal infection [10]. The agr chromosomal locus has 2 promoters, P2 and P3, that control the expression of the divergent RNAII and RNAIII transcripts, respectively. RNAII encodes the core components of the agr system, including the AgrB, AgrD, AgrC, and AgrA proteins. AgrD is the precursor of the autoinducing peptide (AIP), which is processed by a transmembrane endopeptidase, AgrB, before being secreted from the cell. When the concentration of extracellular AIP exceeds a critical threshold, it binds and activates the AgrC histidine kinase that phosphorylates regulator AgrA, which in turn promotes transcription from the P2 and P3 promoters. The RNAIII transcript is itself the main effector for the agr system and thereby coordinates the upregulation of secreted virulence factors and the downregulation of cell surface proteins [11].
Linking the observed severe clinical phenotype with known features of staphylococcal virulence factors, we hypothesized that CA-MRSA responds to the hostile environment of the PMN phagosome by engaging the agr system in order to maximize the chances of a favorable outcome in the form of bacterial survival or PMN destruction. To test this prediction, we followed the fates of CA-MRSA USA300 (LAC) after ingestion by PMN. We quantitated agr-dependent RNAIII expression in the ingested wild-type USA300 (LAC) and compared the relative abilities of the wild-type and isogenic agr deletion mutant USA300 (LAC agr KO) to lyse PMN. In addition, we evaluated the contribution of the agr-regulated toxin α-hemolysin in mediating the downstream effects of the agr system by studying the effect of hla deletion on USA300 (LAC)-mediated PMN lysis.
Phagocytosis of USA300 (LAC) by PMN rapidly activated the bacterial agr system, and the α-hemolysin generated within the PMN phagosome contributed in part to the destruction of PMN within 2 h after ingestion. Our data suggest that α-hemolysin may be part of a broader agr-mediated defensive response employed by USA300 (LAC) to counter the toxic contents of PMN phagosomes.
Materials and Methods
Ficoll-Paque PLUS was purchased from GE Healthcare (Piscataway, N.J., USA). Clinical-grade Dextran T500 (molecular weight 500,000 Da) was purchased from Pharmacosmos A/S (Holbaek, Denmark); patient-grade endotoxin-free 0.9% sodium chloride and endotoxin-free patient-grade H2O were purchased from Baxter Healthcare (Deerfield, Ill., USA). HEPES, Hank's buffered salt solution (HBSS), and Dulbecco's PBS with and without divalent cations were purchased from Mediatech (Manassas, Va., USA). Heparin sodium (1,000 USP units/ml) was obtained through the University of Iowa pharmacy and purchased from APP Pharmaceuticals (Schaumburg, Ill., USA). The HEMA-3 staining kit was obtained from Fisher Diagnostics (Pittsburgh, Pa., USA). Chloramphenicol, poly-L-lysine-coated Poly-Prep slides, dihydrocytochalasin B (DHCB), and diphenylene iodonium (DPI) were purchased from Sigma-Aldrich (St. Louis, Mo., USA). Human serum albumin (HSA) was obtained from Talecris Biotherapeutics (Research Triangle Park, N.C., USA). Tryptic soy broth (TSB) and tryptic soy agar (TSA) were purchased from BD Biosciences (San Jose, Calif., USA). Ninety-six-well optical plates and SYBR Green PCR Master Mix were purchased from Applied Biosystems (Foster City, Calif., USA), whereas Costar 96-well plates were purchased from Corning Incorporated (Corning, N.Y., USA). An LDH Cytotoxicity Detection Kit (LDH), random hexamers, dNTPs, and AMV reverse transcriptase were all purchased from Roche (Indianapolis, Ind., USA), whereas the CellTiter-Glo assay was purchased from Promega (Madison, Wisc., USA). Primers for real time PCR analysis were purchased from Integrated DNA Technologies (Coralville, Iowa, USA). RNA extraction kits were obtained from Qiagen (Valencia, Calif., USA), DNase was purchased from Ambion (Austin, Tex., USA), and polypropylene tubes were obtained through Becton Dickinson (Franklin Lakes, N.J., USA). X-ray film was purchased from Research Products International Corp. (Mt. Prospect, Ill., USA). Sheep anti-α-hemolysin IgG conjugated to horseradish peroxidase was purchased from Toxin Technology (Sarasota, Fla., USA). SuperSignal West Pico chemiluminescent substrate was purchased from Thermo Scientific (Rockford, Ill., USA). Nitrocellulose membranes were purchased from Whatman (Dassel, Germany). Rabbit erythrocytes were purchased from Hemostat Laboratories (Dixon, Calif., USA).
Bacterial Strains and Growth Conditions
Strains of Escherichia coli were grown in Luria-Bertani broth or on Luria agar plates, and the growth medium was supplemented with ampicillin (100 μg/ml) for the maintenance of plasmids. Strains of S. aureus were grown in TSB or on TSA, and the growth medium was supplemented with chloramphenicol (10 μg/ml) as needed for the maintenance of plasmids. Several different strains of S. aureus, expressing 2 different forms of green fluorescent protein (GFP), were employed in these studies (table 1). ALC1435, a derivative of RN6390, constitutively expressing the GFP-encoding plasmid pALC1420 was provided by Dr. Ambrose Cheung (Department of Microbiology, Dartmouth Medical School, Hanover, N.H., USA). As a representative CA-MRSA isolate, USA300-0114 (LAC), which was first isolated in Los Angeles, Calif., was obtained from Dr. Frank DeLeo (Laboratory of Human Bacterial Pathogenesis, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institute of Health, Hamilton, Mont., USA). To express GFP in USA300 (LAC), plasmid pALC1420 was electroporated into strain USA300 (LAC). For comparison, we developed an improved GFP-expressing plasmid called pCM29 (details below) and transformed this plasmid into USA300 (LAC). To assess the role of the agr system, we utilized 2 different USA300 (LAC) agr knockouts. A complete deletion of the agr system USA300 (LAC agr KO) was constructed previously [12] and plasmids pALC1420 or pCM29 were transformed into this mutant to express GFP (table 1). USA300 (LAC agrA KO) was constructed in order to have an agr mutant that could be complemented (details below). To assess the contribution of α-toxin, USA300 (LAC hla KO) and the complemented strains used in our study were provided by Dr. Olaf Schneewind (Department of Microbiology and Pediatrics, University of Chicago, Chicago, Ill., USA). The MRSA strain USA300 (18811) was obtained from Dr Barry Kreiswirth (Public Health Research Institute and the University of Medicine and Dentistry of New Jersey, Newark, N.J., USA).
Table 1.
S. aureus strains used in this study
| Strain name | Plasmid | Background, relevant features | Origin, recent study | GFP | FACS |
|---|---|---|---|---|---|
| 18811 | _ | USA300-0247 MRSA | [26] | − | − |
| ALC1435 | pALC1420 | RN6390 Wt, Cmr | [43] | − | − |
| LAC | – | USA300-0114MRSA | [7] | − | − |
| LAC | pALC1420 | USA300-0114 MRSA, Cmr | This study | + | − |
| LAC agr KO | pALC1420 | USA300-0114 MRSA, Cmr | This study | + | − |
| LAC | pCM29 | USA300-0114 MRSA, Cmr | This study | + | + |
| LAC agr KO | pCM29 | USA300-0114 MRSA, Cmr | This study | + | + |
| LAC Ma KO | – | USA 300-0114 MRSA | [44] | − | − |
| LAC Ha KO | phla | USA300-0114 MRSA, Cmr | [44] | − | − |
| LAC agr A KO | - | USA300-0114MRSA | This study | − | − |
| LAC agr A KO | pAgrClagrA | USA300-0114 MRSA, Cmr | This study | − | − |
The plasmids that were used to transform some strains are shown. + in the GFP column indicates that a strain expresses GFP. + in the FACS column indicates that the strain was used in flow cytometry experiments to study HOCl-mediated bleaching of GFP.
Cmr = Chloramphenicol resistant.
Bacteria were grown overnight at 37°C with shaking at 200 rpm in TSB and supplemented when necessary with 10 μg/ml chloramphenicol to retain GFP plasmid pALC1420 or superfolded highly expressed GFP (sGFP) plasmid pCM29, respectively. Overnight cultures were diluted to an optical density (OD) of 0.05 at 550 nm in fresh TSB media supplemented with 0.01% HSA and appropriate antibiotic at a final concentration of 10 μg/ml and incubated for 2.5 h to reach midlogarithmic growth (OD550 ∼0.1–0.2). Bacteria were recovered (5,000 g for 5 min in a tabletop microcentrifuge) and resuspended in 20 mM HEPES-buffered HBSS containing divalent cations Ca2+ and Mg2+. Bacteria were used immediately or held on ice until opsonization.
Construction of GFP Plasmid pCM29
Plasmid pDB59 [13] was used as the backbone for the GFP reporter construction. Initially restriction sites were built into the pDB59 plasmid for subsequent cloning. Oligonucleotides CLM433 (5′-GATCC GCTAGCATTCGATCATG-3′) and CLM434 (5′-AATTCATGATCGAATGCTAGCG-3′) were combined to 50 μM each, boiled for 5 min and slowly cooled to room temperature. The agr promoter and YFP gene on pDB59 were removed with BamHI and EcoRI digest. To incorporate the new restriction sites, the oligonucleotide mixture was diluted 100-fold and 0.5 pmol were ligated into the digested pDB59. The resulting plasmid was verified by restriction analysis and designated pCM28. To build the new GFP reporter, a DNA fragment encompassing the sarAP1 promoter driving the sGFP gene was PCR amplified from plasmid pCM11 [14] using oligonucleotides CLM435 (5′-GTTGTTGCTAGCCTGATATTTTTGACTAAACCAAATG-3′) and CLM332 (5′-GTTGTTGAATTCTTAGTGGTGGTGGTG-3′). The PCR product was digested by NheI and EcoRI and ligated into pCM28 digested by the same enzymes. The resulting plasmid was verified by restriction analysis and was designated pCM29.
USA300 (LAC agrA KO) Strain Construction
Strains containing the agrA::tn551(Erm) insertion were constructed by phage transduction with α80 phage as previously reported [15]. The phage lysate was prepared from strain RN6112 [16] containing the agrA::tn551(Erm) insertion and transduced into strain USA300 (LAC). Transductants were selected on TSA containing erythromycin 150 μg/ml (to overcome background resistance). Successful crosses of the agrA::tn551(Erm) insertion were confirmed by PCR on purified genomic DNA using oligonucleotides MJT063 (5′-GGTATACAAAGAGACGTTACAT-3′) and MJT064 (5′-GAAATTGCTGATAATGCAGACAATGT-3′). A complemented version of the USA300 (LAC agrA KO) was constructed by transformation with plasmid pAgrC1agrA [17].
PMN Isolation
PMN were isolated from normal healthy volunteers and purified from peripheral blood as described by Nauseef [18]. Informed consent was obtained from each individual following a protocol approved by the Institutional Review Board for human subjects at the University of Iowa. Isolated PMN were suspended in sterile endotoxin-free HBSS without divalent cations at a final density no greater than 20 × 106/ml. A manual differential was performed after staining (HEMA-3 staining kit); the leukocyte preparation was 95–98% PMN. PMN were held on ice until use.
Intraphagosomal Killing of S. aureus
PMN were challenged with opsonized S. aureus at varied multiplicities of infection (MOI; CFU of S. aureus: PMN). Bacteria were suspended (5 × 106/ml) in 20 mM HEPES buffered HBSS containing divalent cations, 1% HSA, and 10% pooled human serum and opsonized by tumbling for 20 min at 37°C. Opsonized bacteria were pelleted (5,000 g for 5 min), washed, resuspended at 5 × 106/ml in 20 mM HEPES-buffered HBSS containing divalent cations, and immediately used in the phagocytosis assay.
PMN and opsonized S. aureus were mixed at the desired MOI in a 5-ml round-bottom polypropylene tube and tumbled for 10 min at 37°C. As indicated in some experiments, PMN were pretreated with DPI (10 μM) or DHCB (2.5 mg/ml) prior to challenge with S. aureus to inhibit the NADPH oxidase or phagocytosis, respectively. After 10 min the samples were removed and the PMN were pelleted (500 g for 5 min in a Beckman Coulter Allegra 6R centrifuge) and extracellular bacteria were collected by aspiration. PMN containing ingested bacteria were resuspended in the original volume of 20 mM HEPES buffered HBSS with divalent cations and 1% HSA. An aliquot was removed immediately after resuspension to assess the intraphagosomal viability of S. aureus at experimental time zero, and additional aliquots were obtained at subsequent time points to quantitate bacterial killing within PMN over time (up to 120 min). Each sample at each time point was diluted 1/50 into pH 11 H2O, incubated at room temperature for 5 min, vortexed, and serially diluted into a 96-well plate containing 0.9% saline, as previously described [19]. Several dilutions of each sample were plated dropwise on TSA plates and placed overnight at 37°C for enumeration of colonies. The CFU per milliliter from replicate plates were averaged and data presented as the percent of viable organisms relative to the number at experimental time zero.
Flow Cytometry
In parallel with the determination of the bacterial viability of phagocytosed S. aureus, the persistence of GFP fluorescence in internalized bacteria was assessed. We modified our previously reported method for the evaluation of hypochlorous acid (HOCl)-dependent bleaching of GFP that employed fluorescence microscopy [20] to a more rapid throughput assay that utilizes fluorescence-activated cell sorting (FACS) for quantitation. Essential for the success of this modification was the development of the sGFP plasmid pCM29 (see above), which resulted in much brighter signals in individual organisms to allow FACS detection; the previously used construct was not bright enough to be used in FACS. Using organisms expressing the sGFP, we were able to accurately monitor the fate of ingested organisms within PMN phagosomes over an extended period of time after phagocytosis.
At specific intervals after the initiation of phagocytosis (30, 60, and 120 min), the fluorescence of paired samples (normal vs. DPI-treated PMN) was measured immediately on an Accuri C6 flow cytometry system (Accuri Cytometers, Inc., Ann Arbor, Mich., USA). In addition, aliquots of PMN without S. aureus were analyzed and gated as the GFP-negative control. All cells outside of this gate on an FITC histogram plot were considered GFP positive. Samples with DPI-treated PMN were analyzed at each time point in parallel to provide a reference for the total amount of cell-associated viable S. aureus. For each sample, the geometric mean and percent of GFP-positive cells were determined (FCS Express C3 software; Ann Arbor, Mich., USA) and a mean fluorescence intensity was calculated as the product of the geometric mean and the percent of fluorescent cells. The fraction of ingested bacteria that remained fluorescent at any time point is expressed relative to the fluorescence of DPI-treated PMN. Triplicate samples at each time point were assessed in each experiment, and each experiment was repeated on ≥3 separate occasions. For a visual confirmation of phagocytosis and to validate the bleaching quantitated by FACS with the results of a direct visual inspection of ingested organisms, the persistence of phagocytosed GFP S. aureus was also measured using fluorescence microscopy. Slides were prepared at various time points as described by Schwartz et al. [20]. For each sample 100 PMN were counted and intracellular cocci remaining fluorescent were reported as the percent fluorescent at time x. Each experiment was repeated on 3 separate occasions.
Real Time PCR
To examine the transcriptional responses of phagocytosed S. aureus, PMN were pelleted (500 g for 5 min in a Beckman Coulter Allegra 6R centrifuge) at specific time points after being fed S. aureus, resuspended in Qiagen RNeasy mini kit buffer RLT containing β-mercaptoethanol, and added to lysing matrix B silica tubes (MP Biomedicals). Samples were homogenized using a FASTprep 24-bead beater (MP Biomedicals) and mRNA was prepared from the homogenate using Qiagen RNeasy kit protocol. Samples were treated with DNase to remove remaining PMN and bacterial DNA, analyzed for purity using a NanoDrop spectrophotometer, and stored at −80°C. cDNA from both RNA preps was generated, and the resulting samples were used for real time PCR. Oligonucleotide primers were designed using Primer Express software and synthesized by Integrated DNA Technologies. SYBR Green fluorescent probes were used to continuously monitor the formation of PCR products during amplification. Reactions were performed in 96-well optical plates with a 7700 Sequence Detector (Applied Biosystems). The PCR process was performed as follows: initial denaturation at 50°C for 2 min, hot start at 95°C for 10 min, thermal cycling at 95°C for 15 s, and primer anneal and extension at 60°C for 1 min; this was repeated for 40 cycles. Standard curves were determined for each target gene using purified genomic DNA at concentrations of 0.156–2.5 ng/ml. Each experiment was performed in triplicate and the results were normalized to the amount of GAPDH transcripts. The GAPDH primers amplified S.aureus GAPDH but did not amplify human GAPDH (data not shown). The primer sequences were as follows: GAPDH forward primer 5′-TAC ACA AGA CGC ACC TCA CAG A-3′, GAPDH reverse primer 5′-ACC TGT TGA GTT AGG GAT GAT GTT T-3′; RNAIII forward primer 5′-AGT CAC CGA TTG TTG AAA TGA TAT CT-3′, RNAIII reverse primer 5′-AGG AAG GAG TGA TTT CAA TGG C-3′. The wash step after phagocytosis removed extracellular and cell-associated bacteria to an extent similar to that achieved by treatment with lysostaphin prior to PMN recovery for RNA extraction (data not shown).
Assessment of PMN Integrity
PMN integrity was assessed in 2 ways by measuring the release of cytoplasmic lactate dehydrogenase (LDH) or ATP. A Roche Cytotoxicity Detection Kit was used to measure spectrophotometrically the amount of LDH released from PMN according to the manufacturer's protocol. PMN were collected at time zero and at specified time points subsequently. The amount of LDH detected in media containing uninfected PMN served as a measure of background (low control), and the amount of LDH present in lysed PMN served as a measure of the total amount of LDH present in PMN (high control). Measuring the amount of LDH detected at different time points, the percent of PMN lysis was calculated as per the manufacturer's protocol. All assays were done in triplicate, and the relative percent of lysis was calculated for each condition. Results are presented as the percent of lysis relative to PMN spontaneous LDH release.
In addition to measuring the release of LDH from PMN, we assessed PMN integrity by quantitating ATP release using a Promega CellTiter-Glo luminescent assay according to manufacturer's protocol. PMN were collected at time zero and subsequently at intervals, and the loss of PMN ATP was quantitated as the decrease in luminescence over time.
Immunoblots for α-Hemolysin in Culture Supernatants
S. aureus strains were grown overnight in TSB containing appropriate antibiotics. Overnight cultures were diluted 1:200 in 5 ml TSB containing antibiotics and grown with shaking at 37°C for 15 h. Cells were removed by centrifugation, and the supernatant was clarified using a 0.2-μm syringe filter. Filtered supernatants were mixed 1:1 with SDS-PAGE loading buffer, and 5 μl of each sample was electrophoresed on a 15% polyacrylamide gel. The proteins were transferred to nitrocellulose membranes using a PROTEAN II device (Bio-Rad Laboratories, Hercules, Calif., USA). Membranes were blocked overnight at 4°C with 5% milk in Tris-buffered saline (20 mM Tris-HCl, pH 7.0, with 137 mM NaCl) containing 0.1% Tween 20 (TBST). Sheep anti-α-hemolysin IgG conjugated to horseradish peroxidase was diluted 1:1,000 in 5% milk in TBST and incubated with the membranes at room temperature for 2 h. Membranes were washed with agitation for 15 min and then twice for 5 min with TBST. SuperSignal West Pico chemiluminescent substrate was added for 5 min at room temperature followed by exposure to X-ray film.
Red Blood Cell Lysis
Filtered culture supernatants were prepared as described above for α-hemolysin immunoblots and were serially diluted in 2-fold steps in TSB. Rabbit erythrocytes were prepared by washing 3 times with PBS and resuspended at 3% v/v in PBS. Seventy microliters of erythrocytes was placed into each well of a 96-well plate. Thirty microliters of diluted culture supernatant was added to the erythrocytes and mixed thoroughly, and the microtiter plates were incubated statically at 37°C. Hemolysis was detected by the loss of turbidity as measured by OD at 630 nm using a Tecan Infinite M200 plate reader (Tecan, Switzerland). To determine relative hemolytic activity, the concentration of supernatant necessary for 50% cell lysis at 40 min was established by adjusting the data to a 4-parameter logistic fit using KaleidaGraph software (version 3.08d; Synergy Software, Reading, Pa., USA). Hemolytic activity is reported relative to wild-type USA300 strain (LAC).
Statistical Analysis
All results are expressed as means ± standard error of the mean (SEM). Significant differences (p < 0.05) were identified using a paired or unpaired Student's t test as appropriate.
Results
Impact of the agr System on the Phagosomal Fate of S. aureus
CA-MRSA USA300 (LAC) was selected as a model strain for assessing the role of the agr regulatory system in human PMN because it has been the subject of extensive recent genomic, transcriptional, and proteomic studies [7,21]. To assess the role of the agr system and the downstream effector α-hemolysin (hla) in this study, we used strain USA300 (LAC) and isogenic deletion mutants in the agr operon and hla. We will refer to the deletion mutants henceforth as USA300 (LAC agr KO) and USA300 (LAC hla KO). USA300 (18811), a clinically relevant MRSA strain was also used as a comparison. Unlike USA300 (LAC), USA300 (18811) has a missense mutation in agrA. Consequently, USA300 (18811) is reported to be deficient in both α- and δ-hemolysin (see table 1 for reference).
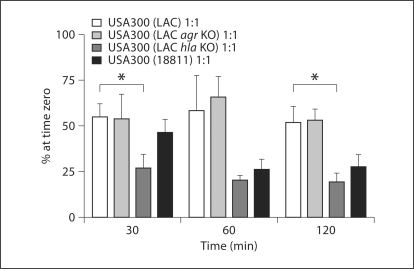
When bacteria were fed to PMN at an MOI of 1:1, killings of USA300 (LAC), USA300 (LAC agr KO), and USA300 (18811) were similar at 30, 60, and 120 min (fig. 1). USA300 (LAC hla KO) suffered a greater loss in viability at 30 and 120 min relative to the USA300 (LAC). As demonstrated previously [20], the PMN-mediated killing of staphylococci requires a functional NADPH oxidase as a source of oxidants; pretreatment of PMN with DPI resulted in the complete survival of both USA300 (LAC) and USA300 (LAC agr KO) over the course of the experiment (data not shown).
Fig. 1.
Viability of S. aureus USA300 (LAC), USA300 (LAC agr KO), USA300 (LAC hla KO), and USA300 (18811) recovered from PMN. PMN were fed opsonized S. aureus strains, either USA300 (LAC), USA300 (LAC agr KO), USA300 (LAC hla KO), or USA300 (18811), at an MOI of 1: 1. The viability of the intracellular bacteria for each strain was determined by colony plating. Samples were analyzed at 30, 60, and 120 min after phagocytosis. The data are presented as means ± SEM (n ≥ 3). The USA300 (LAC) strain was compared against each of the other strains using an unpaired t test. ∗ p < 0.05 between USA300 (LAC) and USA300 (LAC hla KO) at 30 and 120 min.
We recently reported that the intraphagosomal fate of S. aureus can be assessed by monitoring the time-dependent loss of fluorescence of GFP expressed in the bacterial cytoplasm [20]. Bleaching of GFP requires a functional phagocyte NADPH oxidase as a source of H2O2, the azurophil granule myeloperoxidase, and chloride to generate HOCl, which chlorinates tyrosine 66 in GFP and ablates the fluorescence of the chromophore [22]. Overall, the bleaching of cytoplasmic GFP parallels the killing of ingested S. aureus[20]. We compared the kinetics of the bleaching of the GFP in USA300 (LAC) and USA300 (LAC agr KO) within PMN. At an MOI of 1:1, both USA300 (LAC agr KO) and its isogenic control were bleached at an identical rate and extent when fluorescence was quantitated by FACS (see online supplementary figure 1, www.karger.com/doi/10.1159/000319855). In a similar fashion, the rate and extent of the loss of fluorescence were identical for the pair of strains when examined using fluorescence microscopy (data not shown). Taken together, the initial damage to and killing of ingested USA300 (LAC) by PMN were the same, regardless of the presence of a functional agr system, when exposed to the same levels of HOCl in the phagosome. At higher MOIs, there was much less bleaching and greater survival, as previously described [20].
RNAIII Expression by S. aureus in PMN Phagosomes
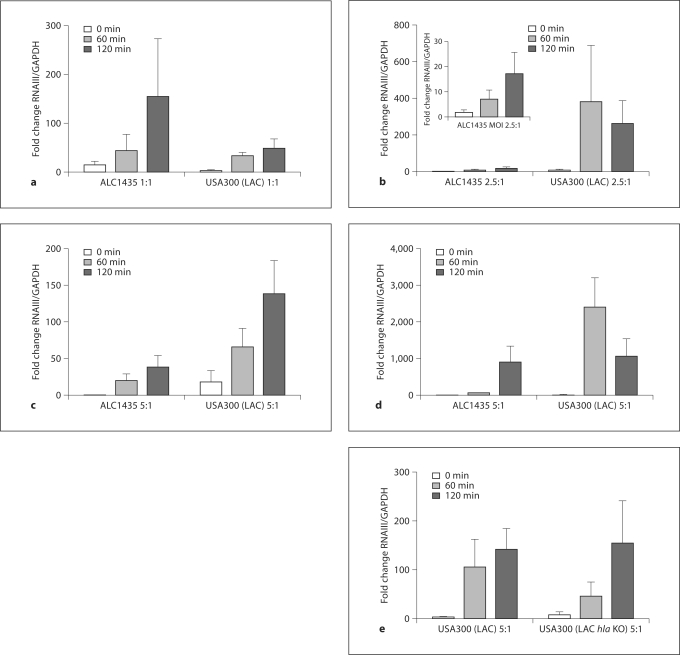
Based on the persistent survival of the USA300 (LAC) strain inside PMN despite exposure to HOCl stress, we reasoned that ingested S. aureus may respond to the phagosomal environment by increasing the expression of factors that promote recovery from sublethal damage and eventual survival. To gain insight into the expression of virulence factors by phagocytosed USA300 (LAC), we used real-time RT-PCR to quantitate transcripts of RNAIII, the major effector of the agr system, at varied MOIs and at different time points after ingestion by PMN. For comparison with wild-type USA300 (LAC), we studied in parallel RNAIII expression by ingested ALC1435 and USA300 (LAC hla KO) (fig. 2). RNAIII expression was rapidly induced in all 3 strains of S. aureus after ingestion by PMN, although the kinetics and the magnitude of RNAIII upregulation were notably different between the USA300 (LAC) and ALC1435 strains over a range of MOIs (1:1, 2.5:1, and 5:1). Whereas RNAIII expression was greater in ALC1435 at an MOI 1:1 (fig. 2a), RNAIII expression by USA300 (LAC) at an MOI of 2.5:1 was >380-fold and >250-fold higher than the postopsonization control at 60 and 120 min, respectively (fig. 2b). At an MOI of 5:1, RNAIII expression was ∼65-fold and ∼140-fold higher in USA300 (LAC) than the postopsonization control at 60 and 120 min, respectively (fig. 2c). Furthermore, USA300 (LAC) upregulated RNAIII expression dramatically (2,408 ± 795-fold and 1,061 ± 476-fold greater than the control at 60 and 120 min, respectively) when fed to PMN that were incapable of generating oxidants due to pretreatment with DPI (fig. 2d). At an MOI of 5:1 there was no significant difference in RNAIII expression between the USA300 (LAC hla KO) and its isogenic control (fig. 2e).
Fig. 2.
Upregulation of RNAIII expression in S. aureus after ingestion by PMN. Normal PMN were fed opsonized S. aureus, either strain ALC1435, USA300 (LAC), or USA300 (LAC hla KO), at MOIs of 1: 1 (a), 2.5: 1 (b), and 5: 1 (c, e). In parallel, PMN pre-treated with DPI prior to microbial challenge were fed S. aureus (d) at an MOI of 5: 1. At 0, 60, and 120 min after phagocytosis, RNAIII expression in recovered S. aureus was quantified by real-time RTPCR using GAPDH as the reference gene. The fold change for each sample relative to the postopsonized S. aureus and prior to exposure to PMN was calculated and is shown as the mean ± SEM (n ≥ 3). The inset in b shows the data for the ALC1435 strain where the y-axis has been expanded between 0 and 30 for clarity.
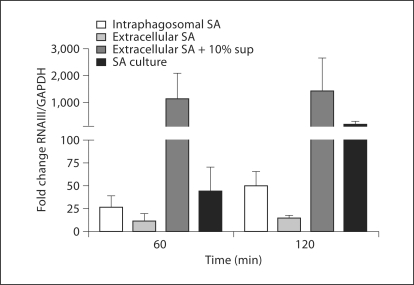
To assess whether internalization of S. aureus by PMN was necessary for the induction of RNAIII that we observed, bacteria opsonized in pooled human serum were presented to PMN that were pretreated with DHCB, an inhibitor of phagocytosis. Opsonized bacteria were PMN-associated but not ingested in the presence of DHCB-treated PMN, as judged by fluorescence microscopy (data not shown). Despite being in the presence of activated PMN, extracellular USA300 (LAC) did not upregulate RNAIII. To determine if the extracellular bacteria were able to upregulate RNAIII in response to AIP, the organisms were exposed to 10% of AIP-rich culture supernatant added exogenously. The AIP-challenged extracellular bacteria upregulated RNAIII >1,400-fold relative to the control at 120 min (fig. 3), thus demonstrating that the failure of extracellular USA300 (LAC) to increase RNAIII transcription was not a consequence of being incapable of responding or of inhibition mediated by apolipoprotein B or other antagonizing serum proteins present during opsonization; rather, the lack of response in extracellular bacteria resulted from the absence of a sufficient stimulus to initiate RNAIII expression. Taken together, these findings suggest that the confining phagosomal environment promoted the rapid activation of the agr system in S. aureus in vitro. The heightened response of USA300 (LAC) ingested by DPI-treated PMN extends the previous observations that NADPH-derived reactive oxygen species oxidize AIP and thereby disrupt AIP signaling in the agr pathway [23]; the absence of an operational oxidase in DPI-treated PMN would leave AIP functional and allow positive feedback through the agr operon.
Fig. 3.
RNAIII expression by intracellular and extracellular USA300 (LAC). Opsonized S. aureus USA300 (LAC) were incubated at an MOI of 2.5: 1 with either PMN, in which phagocytosis was inhibited by pretreatment with DHCB, or normal PMN in the absence or presence of 10% AIP-rich culture supernatant (10% sup). RNAIII expression at 60 and 120 min after phagocytosis was quantified from bacteria recovered from the phagosomes of normal PMN, nonphagocytosed bacteria outside DHCB-treated PMN, and nonphagocytosed bacteria in the presence of AIP. A subculture of USA300 (LAC) grown in TSB was used as a positive control. GAPDH was used as the reference gene and the fold change relative to the postopsonized control was calculated. Data are presented as means ± SEM (n ≥ 3). The mean values for intraphagosomal S. aureus, extracellular S. aureus, extracellular S. aureus + 10% supernatant, and S. aureus in TSB at 60 min were 26.0, 11.4, 1,135.3, and 44.3, respectively; at 120 min the values were 50.0, 14.2, 1,440.1, and 182.9, respectively. sup = Supernatant; SA = S. aureus.
PMN Lysis by S. aureus: Dependence on the agr System
RNAIII controls the expression of several staphylococcal virulence factors, including α-hemolysin, a toxin that forms heptameric pores in the membranes of many host cells [24]. To determine whether the activation of the agr operon in PMN phagosomes and its downstream effectors induces PMN lysis, PMN were fed opsonized S. aureus at 3 MOIs (1:1, 5:1, and 10:1) and PMN lysis, reflected as the extracellular release of cytosolic LDH, was measured.
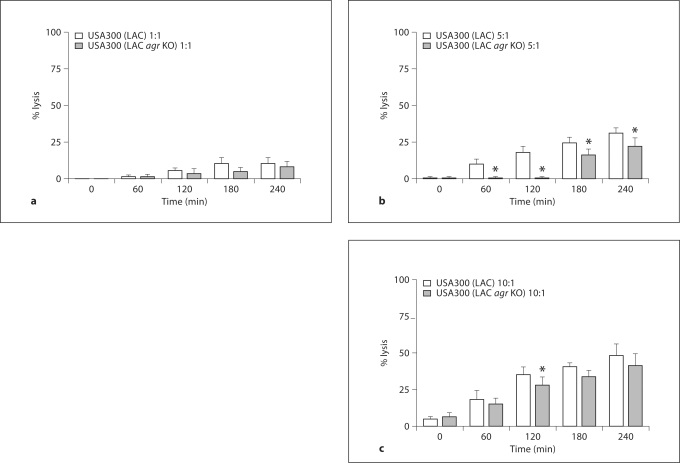
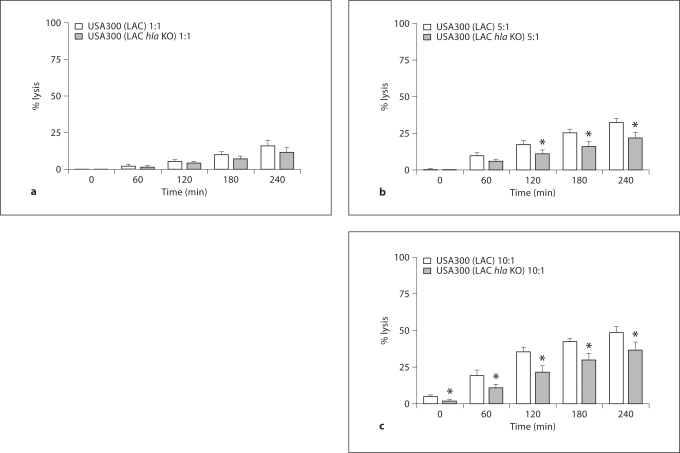
To assess the contribution of the agr system to the lysis of PMN, we compared the capacity of ingested USA300 (LAC) and USA300 (LAC agr KO) to lyse PMN. PMN lysis correlated with bacterial burden; minimal lysis was seen at an MOI of 1:1 and greater lysis occurred at MOIs of 5:1 and 10:1 in both strains (fig. 4a–c). At an MOI of 5:1 the differences between the 2 strains reached statistical significance for all time points between 60 and 240 min. At an MOI of 10:1, extensive PMN lysis masked the significance at all time points except 120 min although the same trend prevailed. The extensive PMN lysis at the highest MOI may reflect the overwhelming bacterial burden per PMN rather than toxin-mediated lysis. Similar correlations between PMN health, bacterial burden, and the impact of the agr were observed in assays of PMN integrity using the Promega CellTiter-Glo reagent (data not shown). Furthermore, USA300 (LAC) lysed DPI-treated PMN in an MOI-dependent manner (data not shown), indicating that PMN oxidant production was not a prerequisite for USA300 (LAC)-induced cytotoxicity.
Fig. 4.
PMN lysis after ingestion of USA300 (LAC) and USA300 (LAC agr KO). PMN were fed opsonized S. aureus, either strain USA300 (LAC) or USA300 (LAC agr KO), at an MOI of 1: 1 (a),5: 1 (b), or 10: 1 (c). The percent of cell lysis was calculated using the LDH release assay at 0, 60, 120, 180, and 240 min after phagocytosis and the results are presented as means ± SEM (n ≥ 4). ∗ p < 0.05, using a paired t test, between USA300 (LAC) and USA300 (LAC agr KO) at the indicated time points and MOIs.
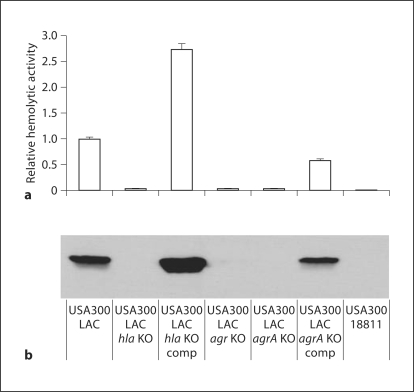
Since the agr system is a global regulator of pathogenesis, we shifted our attention to virulence factors that act downstream of the RNAIII effector. To this end, we assessed the contribution of the pore-forming toxin α-hemolysin to PMN lysis. Considering the genetic variability among S. aureus strains, we first tested whether the expression and activity of hla was indeed controlled by agr in our strains. Since agr regulation of α-hemolysin occurs predominantly at the posttranscriptional level, we performed immunoblots and assessed functional activity with rabbit erythrocyte lysis. As anticipated from reports on MSSA strains [25], α-hemolysin was secreted in USA300 (LAC) but was absent in USA300 (LAC agr KO) (fig. 5). Given the challenge of complementing the complete locus deletion in USA300 (LAC agr KO), we evaluated the expression of α-hemolysin in a strain engineered to be deficient in only agrA, USA300 (LAC agrA KO), encoding the response regulator of the system in parallel with the genetically complemented strain. Similar to the results with USA300 (LAC agr KO), α-hemolysin expression was abolished in USA300 (LAC agrA KO) and complementation of the mutant with the pAgrC1agrA plasmid (table 1) restored α-hemolysin expression and activity. These data demonstrate a direct link between functional Agr signaling and α-hemolysin production and activity in our USA300 (LAC) stains. Additionally, as anticipated based on Kennedy et al. [26], USA300 (18811) did not produce α-hemolysin (fig. 5). With the genetic circuitry established, we examined the fate of PMN after the ingestion of USA300 (LAC hla KO). USA300 (LAC hla KO) elicited less PMN lysis than did USA300 (LAC) both at an MOI of 5:1 between 120 and 240 min and at an MOI of 10:1 at all time points studied (fig. 6b, c), whereas little effect was observed at an MOI of 1:1 (fig. 6a). These results were strikingly similar to those obtained with the USA300 (LAC agr KO) (fig. 4), which did not secrete α-hemolysin (fig. 5). Taken together, these data suggest that the agr-dependent lysis of PMN by USA300 (LAC) at MOIs >1 was in part mediated by α-hemolysin.
Fig. 5.
α – Hemolysin expression and activity in USA300 (LAC), the isogenic KO strains, and USA300 (18811). Clarified overnight culture medium was prepared from the following S. aureus strains: USA300 (LAC), USA300 (LAC hla KO), USA300 (LAC hla KO) complemented mutant, USA300 (LAC agr KO), USA300 (LAC agrA KO), USA300 (LAC agrA KO) complemented mutant, and USA300 (18811). a Hemolytic activity present in the culture medium from each strain was determined by the rabbit RBC lysis assay, and the results were normalized to the USA300 (LAC) strain and reported as mean fold changes ± SEM (n = 3). b Proteins in each sample were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with an anti-α-hemolysin-HRP conjugated antibody. The USA300 (LAC hla KO) and USA300 (LAC hla KO) complemented strains serve as negative and positive reference controls, respectively. comp = Complemented.
Fig. 6.
PMN lysis after ingestion of USA300 (LAC) and USA300 (LAC hla KO). As in figure 4, PMN were fed opsonized S. aureus and USA300 (LAC), and LDH release was monitored over time. Independently, PMN were fed USA300 (LAC hla KO) at an MOI of 1: 1 (a), 5: 1 (b), or 10: 1 (c). The percent of cell lysis was calculated using the LDH release assay at 0, 60, 120, 180, and 240 min after phagocytosis and the results are presented as means ± SEM (n ≥ 7). ∗ p < 0.05, using a paired t test, between strains USA300 (LAC) and USA300 (LAC hla KO) at the indicated time points and MOIs.
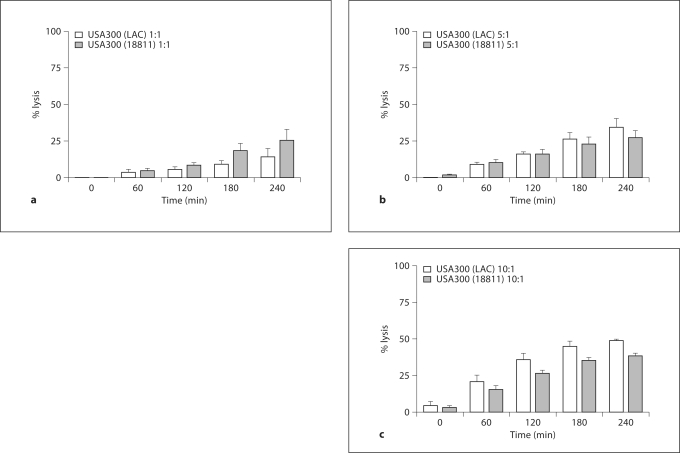
To further determine the importance of the agr system between clinically relevant strains of MRSA, we compared the relative abilities of USA300 (LAC) and USA300 (18811) to lyse PMN. There were no significant differences between the 2 strains in their ability to lyse PMN; however, USA300 (18811) trended to be less lytic at MOIs >1 at the later time points (fig. 7).
Fig. 7.
PMN lysis after ingestion of USA300 (LAC) versus USA300 (18811). PMN were fed opsonized USA300 (LAC) or USA300 (18811) at MOIs of 1: 1 (a), 5: 1 (b), or 10: 1 (c), and LDH release was monitored over time. The percent of cell lysis was calculated using the LDH release assay at 0, 60, 120, 180, and 240 min after phagocytosis and the results are presented as means ± SEM (n = 3). No significant differences were observed between strains at the chosen time points using a paired t test.
Discussion
PMN constitute a critical element of cellular defense against pyogenic infections, including those caused by staphylococci. Within the PMN phagosome, a dynamic interplay between host and microbial elements occurs, generating a broad array of potent antimicrobial and degradative agents while the entrapped organism reacts rapidly in an attempt to survive. Within the context of the optimal PMN attack on ingested staphylococci, the oxidants derived from the phagocyte NADPH oxidase provide H2O2 which in turn serves as substrate for the myeloperoxidase-catalyzed generation of the potently microbicidal HOCl [27]. Under conditions most favorable for the host, HOCl and its derivatives synergize with granule-derived antimicrobial peptides and proteolytic enzymes to kill and degrade the majority of ingested staphylococci [5]. Despite the generation of potent microbicides in phagosomes, significant numbers of ingested staphylococci survive their encounter with human PMN, with 20% or more of ingested organisms still viable 2 h after phagocytosis [7,20]. Although the efficiency of killing that we observed [51.7 ± 8.7% survived at 120 min for USA300 (LAC)] may appear to be less than that reported previously, it is important to note that these data are expressed as a fraction of the number of viable organisms immediately following a wash step, included to eliminate extracellular bacteria from microscopy analysis, 10 min after the initiation of phagocytosis. Given the relatively rapid killing of staphylococci by human PMN, a significant loss of viability occurs in the first 10 min, thus decreasing the apparent rate of killing at subsequent time points when expressed in this fashion. When compared to the inoculum at the onset of phagocytosis, our data are consistent with those previously reported [23].
Given the capacity of staphylococci to survive in PMN both in vitro and in vivo [28], ingested bacteria must respond to the hostile environment of the phagosome to evade, endure, or repair structural damage mediated by the antimicrobial attack. We observed at an MOI >1:1 that phagocytosed S. aureus USA300 (LAC) rapidly upregulated the expression of RNAIII, the major effector that is produced upon agr activation and controls the expression of the transcription and translation of several key virulence factors. Because the expression of virulence genes is energetically costly, the agr operon tightly regulates their expression. At the heart of the agr system is AIP, a small peptide secreted by S. aureus that diffuses into the extracellular space and is thought to mediate chemical communication among bacterial cells in a process known as quorum sensing [29]. At a specific cell density, or quorum, the concentration of AIP reaches a level sufficient to activate the agr system via binding to its receptor, AgrC, on the bacterial surface. As a consequence of activating the agr operon, positive feedback signaling through the 2-component system occurs resulting in an increased production of the RNAIII effector.
We observed an increased expression of RNAIII by staphylococci in the PMN phagosome, a membrane-bound compartment that approximates the size of the ingested organism and has an estimated volume of ∼1.2 fl [30]. Each phagosome contains ≥1 bacteria that have the capacity to secrete AIP. Since bacteria sense the local concentration of AIP, the activation of the agr system and expression of RNAIII in ingested S. aureus likely reflect the increase in accumulated AIP given its restricted diffusion and the spatial constraints of the phagosome. Consistent with this idea, we demonstrate that S. aureus that interact with but are not ingested by DHCB-treated PMN do not upregulate RNAIII. AIP secreted from bacteria that remain outside PMN readily diffuses away, resulting in local concentrations of AIP that remain under the threshold level required for AgrC signaling. Trapping AIP within a host intracellular compartment is not unique to the PMN phagosome; S. aureus internalized within epithelial cell endosomes in vitro activate the agr system and express higher levels of its downstream effectors α- and β-hemolysin [31,32]. These are thought to facilitate the escape of bacteria from the endosomal compartment and allow reproduction in the cytoplasm [33]. Considered together, these data suggest that the intraphagosomal upregulation of RNAIII reflects the activation of agr through diffusion sensing [34] rather than quorum sensing per se. Consistent with our findings, Carnes et al. [35] recently reported that bacteria trapped as individual cells or small groups within a synthetic lipid sphere, conditions that model those in phagosomes or endosomes, are sufficient to activate the agr operon, including RNAIII and hla. S. aureus ingested by PMN in which the NADPH oxidase had been inhibited by DPI exhibited greater agr activation than did staphylococci in fully functional PMN. This effect reflects in part the ability of HOCl to disrupt AIP signaling in normal but not in DPI-treated PMN. The agrD in USA300 (LAC) encodes an agr group I AIP peptide, which is an 8-amino acid peptide with a pentapeptide thiolactone macrocyclic structure. The methionine residue at its C terminus is critical for its function, and its oxidation to a sulfoxide by HOCl abolishes its ability to activate the agr system [23]. In the absence of a functional NADPH oxidase, as is the case in DPI-treated PMN, HOCl is not generated in the phagosome [8] and the vulnerable methionine in AIP remains unmodified and active. As a consequence, the higher concentrations of functional AIP in the phagosomes of DPI-treated PMN fuel the positive feedback signaling of the agr operon and increased RNAIII expression.
Given the rapidity of agr activation after phagocytosis, we reasoned that this response might contribute to the self-defense of ingested bacteria against the PMN-mediated attack. However, activation of the agr response did not confer a survival advantage to ingested S. aureus USA300 (LAC) relative to the isogenic agr mutant. Improved survival of ingested staphylococci, indirectly assessed by the resistance to bleaching of cytoplasmic GFP, was most clearly demonstrated at high MOIs, i.e. conditions that tax the phagosomal antimicrobial activity; killing and HOCl-mediated bleaching of GFP-expressing staphylococci are inversely proportional to the MOI, which is in part a reflection of insufficient oxidant generation at the higher MOI [20]. In addition, ingested staphylococci at an MOI ≥5:1 lysed PMN through a mechanism in part dependent on α-hemolysin. In S. aureus, RNAIII positively regulates both the transcription and translation of α-hemolysin [25], a monomeric toxin that assembles into a heptameric pore on susceptible host cells and causes host cell lysis [24,36]. Consistent with the dependence of the translation of hla on the presence of RNAIII, the USA300 (LAC agr KO) did not express RNAIII and lacked α-hemolysin activity.
Valeva et al. [37] demonstrated that exogenously added α-hemolysin fails to insert its pore-forming domain into the plasma membrane of intact PMN and consequently does not promote PMN lysis. However, the target for α-hemolysin secreted by ingested staphylococci is the phagosomal membrane and not the plasma membrane. The former is a mosaic made up of components of secretory and granule membranes that fuse with the invaginating plasma membrane during phagocytosis [38,39]. The distinct phospholipid compositions of plasma and phagosomal membranes contribute to the selective recruitment of p40phox, an element of the phagocyte NADPH oxidase, to the phagosome during phagocytosis [40]. S. aureus invading epithelial cells escape the endosome by disrupting the membranes of the vesicular compartment through an α-hemolysin-dependent mechanism [32]. We speculate that the accumulated intraphagosomal α-hemolysin facilitated the breakdown of the phagosome compartment in PMN in a fashion similar to that seen in epithelial endosomes. In addition, the potent toxic agents that reside in the phagosomal compartment, including reactive oxygen species as well as proteases such as elastase and cathepsins, would be present in the phagosome lumen and thereby contribute to toxicity to and lysis of PMN.
Although they have been the subject of extensive study, the determinants underlying the lysis of PMN by ingested staphylococci are multifactorial, reflecting in part the redundancy in the repertoire of virulence factors that S.aureus expresses. Our studies of USA300 (LAC) and isogenic mutants implicate the agr system and α-hemolysin in contributing to the lysis of PMN, but the relationship is complex and does not necessarily translate to all strains of S.aureus. USA300 (18811) maintained the ability to lyse PMN despite the lack of a functional agr system and the absence of α-hemolysin; these findings are in agreement with a recent study by Kobayashi et al. [41]. In addition to α-hemolysin, S. aureus expresses many other toxins that have been implicated in contributing to PMN lysis during infection. Those that have received recent attention in the context of CA-MRSA infections include the Panton-Valentine leukocidin (PVL) toxin and phenol soluble modulins (PSMs). Although the contribution of PVL to the pathology associated with S. aureus infections remains a contentious issue [42], PSMs have been found more recently to have potent effects on PMN both in vitro and in murine models of bacteremia, peritonitis, and soft tissue infections [8]. Because the contributions of PVL and PSMs were not assessed in our studies, we cannot exclude the possibility that these toxins may also contribute to findings in our model system. In fact, given the extensive number of virulence factors that agr regulates, it is likely that S. aureus responds to ingestion by PMN with the expression of multiple factors in addition to α-hemolysin in an effort to survive in the phagosome [8]. However, the complex biology of PMN lysis extends beyond the direct consequences of staphylococcal toxins and includes host-driven responses to ingested S. aureus. Kobayashi et al. [41] have recently demonstrated that ingested USA300 initiate a form of programmed cellular necrosis in PMN that culminates in lysis, is strain-dependent, and is influenced by serum factors. Taken collectively, these data demonstrate that PMN lysis may be driven by the pathogen as well as the host cell and that the mode of PMN lysis may not be the same in all experimental settings or S. aureus infections.
Experimental data strongly suggest that the cooperative effects of a variety of virulence factors expressed by S. aureus soon after ingestion by PMN coordinate both direct and indirect actions that culminate in the destruction of PMN. Together these events contribute to the successful evasion of PMN killing by some staphylococci. Evidence of such a dynamic interaction between the host cell and the bacteria emphasizes the need for and importance of studying host-microbe interactions in concert. Further dissection of downstream effectors of the agr system in response to challenges presented in the PMN phagosome will provide important insight into some of the mechanisms that S. aureus employs to survive elimination by the host innate immune system and cause clinical disease.
Supplementary Material
PMN-mediated bleaching of S. aureus USA300 (LAC) and USA300 (LAC agr KO) PMN were fed opsonized S. aureus strains, either USA300 (LAC) (black) or USA300 (LAC agr KO) (grey) at an MOI of 1:1. The percent of fluorescent intracellular bacteria for each strain was determined by FACS. Samples were analyzed at 30 minute time intervals up to 120 minutes after phagocytosis. The data are represented as the mean standard error of the mean (SEM) (n = 3).
Acknowledgements
We acknowledge C. Malone for the plasmid constructions, B. Kreiswirth for strain USA300 (18811), F. DeLeo for USA300 (LAC), O. Schneewind for USA300 (LAC hla KO) and the phla-complemented strain, and A. Cheung for strain ALC1435. This work was supported by National Institutes of Health grants T32 AI 7511 (M.T.), RO1 AI078921 (A.R.H.), and RO1 AI07958 (W.M.N.), and by a Merit Review Grant (W.M.N.) from the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, with facilities and resources of the Veterans Administration in Iowa City, Iowa, USA.
Footnotes
Yun Yun Pang and Jamie Schwartz contributed equally to this work.
References
- 1.Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, Fosheim G, McDougal LK, Chaitram J, Jensen B, Fridkin SK, Killgore G, Tenover FC. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J Infect Dis. 2006;193:172–179. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 2.Klevens RM, Edwards JR, Tenover FC, McDonald LC, Horan T, Gaynes R. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992–2003. Clin Infect Dis. 2006;42:389–391. doi: 10.1086/499367. [DOI] [PubMed] [Google Scholar]
- 3.Fridkin SK, Hageman JC, Morrison M, Sanza LT, Como-Sabetti K, Jernigan JA, Harriman K, Harrison LH, Lynfield R, Farley MM. Methicillin-resistant Staphylococcus aureus disease in three communities. N Engl J Med. 2005;352:1436–1444. doi: 10.1056/NEJMoa043252. [DOI] [PubMed] [Google Scholar]
- 4.Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA. 2009;106:5883–5888. doi: 10.1073/pnas.0900743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nauseef WM. How human neutrophils kill and degrade microbes: an integrated view. Immunol Rev. 2007;219:88–102. doi: 10.1111/j.1600-065X.2007.00550.x. [DOI] [PubMed] [Google Scholar]
- 6.Palazzolo-Ballance AM, Reniere ML, Braughton KR, Sturdevant DE, Otto M, Kreiswirth BN, Skaar EP, DeLeo FR. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J Immunol. 2008;180:500–509. doi: 10.4049/jimmunol.180.1.500. [DOI] [PubMed] [Google Scholar]
- 7.Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 8.Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A, DeLeo FR, Otto M. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 9.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 10.Bubeck WJ, Patel RJ, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun. 2007;75:1040–1044. doi: 10.1128/IAI.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 12.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malone CL, Boles BR, Lauderdale KJ, Thoendel M, Kavanaugh JS, Horswill AR. Fluorescent reporters for Staphylococcus aureus. J Microbiol Methods. 2009;77:251–260. doi: 10.1016/j.mimet.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res. 2010;28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- 15.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 16.Recsei P, Kreiswirth B, O'Reilly M, Schlievert P, Gruss A, Novick RP. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 17.Jensen RO, Winzer K, Clarke SR, Chan WC, Williams P. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J Mol Biol. 2008;381:300–309. doi: 10.1016/j.jmb.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 18.Nauseef WM. Isolation of human neutrophils from venous blood. Methods Mol Biol. 2007;412:15–20. doi: 10.1007/978-1-59745-467-4_2. [DOI] [PubMed] [Google Scholar]
- 19.Decleva E, Menegazzi R, Busetto S, Patriarca P, Dri P. Common methodology is inadequate for studies on the microbicidal activity of neutrophils. J Leukoc Biol. 2006;79:87–94. doi: 10.1189/jlb.0605338. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz J, Leidal KG, Femling JK, Weiss JP, Nauseef WM. Neutrophil bleaching of GFP-expressing staphylococci: probing the intraphagosomal fate of individual bacteria. J Immunol. 2009;183:2632–2641. doi: 10.4049/jimmunol.0804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burlak C, Hammer CH, Robinson MA, Whitney AR, McGavin MJ, Kreiswirth BN, DeLeo FR. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol. 2007;9:1172–1190. doi: 10.1111/j.1462-5822.2006.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espey MG, Xavier S, Thomas DD, Miranda KM, Wink DA. Direct real-time evaluation of nitration with green fluorescent protein in solution and within human cells reveals the impact of nitrogen dioxide versus peroxynitrite mechanisms. Proc Natl Acad Sci USA. 2002;99:3481–3486. doi: 10.1073/pnas.062604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothfork JM, Timmins GS, Harris MN, Chen X, Lusis AJ, Otto M, Cheung AL, Gresham HD. Inactivation of a bacterial virulence pheromone by phagocyte-derived oxidants: new role for the NADPH oxidase in host defense. Proc Natl Acad Sci USA. 2004;101:13867–13872. doi: 10.1073/pnas.0402996101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 25.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci USA. 2008;105:1327–1332. doi: 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klebanoff SJ. Myeloperoxidase: friend and foe. J Leukoc Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 28.Rogers DE, Tompsett R. The survival of staphylococci within human leukocytes. J Exp Med. 1952;95:209–230. doi: 10.1084/jem.95.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji G, Beavis RC, Novick RP. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: implications for microbial killing. J Biol Chem. 2006;281:39860–39869. doi: 10.1074/jbc.M605898200. [DOI] [PubMed] [Google Scholar]
- 31.Qazi SN, Counil E, Morrissey J, Rees CE, Cockayne A, Winzer K, Chan WC, Williams P, Hill PJ. Agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect Immun. 2001;69:7074–7082. doi: 10.1128/IAI.69.11.7074-7082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shompole S, Henon KT, Liou LE, Dziewanowska K, Bohach GA, Bayles KW. Biphasic intracellular expression of Staphylococcus aureus virulence factors and evidence for Agr-mediated diffusion sensing. Mol Microbiol. 2003;49:919–927. doi: 10.1046/j.1365-2958.2003.03618.x. [DOI] [PubMed] [Google Scholar]
- 33.Kahl BC, Goulian M, van WW, Herrmann M, Simon SM, Kaplan G, Peters G, Cheung AL. Staphylococcus aureus RN6390 replicates and induces apoptosis in a pulmonary epithelial cell line. Infect Immun. 2000;68:5385–5392. doi: 10.1128/iai.68.9.5385-5392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- 35.Carnes EC, Lopez DM, Donegan NP, Cheung A, Gresham H, Timmins GS, Brinker CJ. Confinement-induced quorum sensing of individual Staphylococcus aureus bacteria. Nat Chem Biol. 2010;6:41–45. doi: 10.1038/nchembio.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGee MP, Kreger A, Leake ES, Harshman S. Toxicity of staphylococcal alpha toxin for rabbit alveolar macrophages. Infect Immun. 1983;39:439–444. doi: 10.1128/iai.39.1.439-444.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valeva A, Walev I, Pinkernell M, Walker B, Bayley H, Palmer M, Bhakdi S. Transmembrane beta-barrel of staphylococcal alpha-toxin forms in sensitive but not in resistant cells. Proc Natl Acad Sci USA. 1997;94:11607–11611. doi: 10.1073/pnas.94.21.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirsch JG. Cinemicrophotographic observations on granule lysis in polymorphonuclear leucocytes during phagocytosis. J Exp Med. 1962;116:827–834. doi: 10.1084/jem.116.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segal AW, Dorling J, Coade S. Kinetics of fusion of the cytoplasmic granules with phagocytic vacuoles in human polymorphonuclear leukocytes: biochemical and morphological studies. J Cell Biol. 1980;85:42–59. doi: 10.1083/jcb.85.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Gaffney PR, Coadwell J, Chilvers ER, Hawkins PT, Stephens LR. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox) Nat Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi SD, Braughton KR, Palazzolo-Ballance AM, Kennedy AD, Sampaio E, Kristosturyan E, Whitney AR, Sturdevant DE, Dorward DW, Holland SM, Kreiswirth BN, Musser JM, DeLeo FR. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. J Innate Immun. 2010 doi: 10.1159/000317134. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009;119:2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheung AL, Nast CC, Bayer AS. Selective activation of sar promoters with the use of green fluorescent protein transcriptional fusions as the detection system in the rabbit endocarditis model. Infect Immun. 1998;66:5988–5993. doi: 10.1128/iai.66.12.5988-5993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O. Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat Med. 2007;13:1405–1406. doi: 10.1038/nm1207-1405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PMN-mediated bleaching of S. aureus USA300 (LAC) and USA300 (LAC agr KO) PMN were fed opsonized S. aureus strains, either USA300 (LAC) (black) or USA300 (LAC agr KO) (grey) at an MOI of 1:1. The percent of fluorescent intracellular bacteria for each strain was determined by FACS. Samples were analyzed at 30 minute time intervals up to 120 minutes after phagocytosis. The data are represented as the mean standard error of the mean (SEM) (n = 3).