Abstract
Neutrophils are key effectors of the host innate immune response against bacterial infection. Staphylococcus aureus is a preeminent human pathogen, with an ability to produce systemic infections even in previously healthy individuals, thereby reflecting a resistance to effective neutrophil clearance. The recent discovery of neutrophil extracellular traps (NETs) has opened a novel dimension in our understanding of how these specialized leukocytes kill pathogens. NETs consist of a nuclear DNA backbone associated with antimicrobial peptides, histones and proteases that provide a matrix to entrap and kill various microbes. Here, we used targeted mutagenesis to examine a potential role of S. aureus nuclease in NET degradation and virulence in a murine respiratory tract infection model. In vitro assays using fluorescence microscopy showed the isogenic nuclease-deficient (nuc-deficient) mutant to be significantly impaired in its ability to degrade NETs compared with the wild-type parent strain USA 300 LAC. Consequently, the nuc-deficient mutant strain was significantly more susceptible to extracellular killing by activated neutrophils. Moreover, S. aureus nuclease production was associated with delayed bacterial clearance in the lung and increased mortality after intranasal infection. In conclusion, this study shows that S. aureus nuclease promotes resistance against NET-mediated antimicrobial activity of neutrophils and contributes to disease pathogenesis in vivo.
Key Words: Neutrophil extracellular traps, Staphylococcus aureus, Nuclease, Innate immunity, Virulence factor
Introduction
Since 1882, numerous clinical and laboratory studies have defined the importance of Staphylococcus aureus as the causative agent for a wide spectrum of human and veterinary infections [1,2], including its role in sepsis and abscess formation. Nowadays, infections caused by antibiotic-resistant strains of S. aureus, such as community-acquired methicillin-resistant S. aureus (CA-MRSA), have reached epidemic proportions globally. In addition to their increasing prevalence and incidence, CA-MRSA strains appear to be especially virulent, leading to overwhelming and tissue-destructive infections, such as necrotizing fasciitis and fulminant, necrotizing pneumonia [3,4].
S. aureus pathogenesis is complex and multifactorial as the organism expresses numerous virulence factors which can act either alone or in concert to induce various pathogenic conditions [2]. Among its virulence factors, S. aureus produces a wide variety of exoenzymes, including nucleases, proteases, lipases, hyaluronidase and collagenase [5]. These enzymes have the ability to generate bacterial nutrients by host tissue breakdown and thereby promote bacterial growth and increase invasive disease potential [6,7].
Although expression and secretion of an extracellular nuclease by S. aureus have been documented for a long time [8,9,10], the specific role of S. aureus nuclease in pathogenesis is poorly understood. Recently, its contribution in biofilm formation was investigated [11]. The authors showed that a nuclease-deficient (nuc-deficient) mutant formed a thicker biofilm containing increased levels of matrix-associated released DNA. Therefore, one potential contribution of nuclease expression by S. aureus is involvement in biofilm dispersal and subsequent promotion of bacterial spreading [11].
A major new paradigm regarding host innate immune defense against bacterial pathogens is the function of neutrophil extracellular traps (NETs). Released at sites of infection by activated neutrophils, NETs consist of nuclear or mitochondrial DNA as a backbone with embedded antimicrobial peptides, histones and cell-specific proteases and thereby provide an extracellular matrix to entrap and kill various microbes [12]. Recently, the ability of the important Gram-positive bacterial pathogens group A Streptococcus and Streptococcus pneumoniae to resist NET-dependent killing has been linked to their ability to secrete nucleases, a phenotype that contributes to the pathogenesis of necrotizing soft tissue infection (group A Streptococcus) or pneumonia (S. pneumoniae) in corresponding animal models of infection [13,14,15].
Here we investigated whether S. aureus nuclease expression could promote NET evasion and thereby contribute to disease pathogenesis, using isogenic bacterial mutant strains, fluorescence microscopy-based in vitro assays and a murine model of S. aureus respiratory tract infection.
Materials and Methods
Bacterial Strains and Mutants
S. aureus strain LAC (pulsed-field type USA300), a community-acquired CA-MRSA strain, was used in this study [16]. The USA300 clone of MRSA is epidemic in the United States and is associated with skin infection [17,18], necrotizing fasciitis [19] and severe pneumonia [20], often in previously healthy individuals. We used a panel of nuc-deficient mutants and control strains (table 1) [21] to study the effect of nuclease expression on virulence. Bacteria were grown in Brain-Heart Infusion (BHI) medium at 37°C with shaking. Fresh overnight cultures were diluted 1:100 in BHI and then grown to mid-logarithmic growth phase (OD600 = 0.7) for use in in vitro and in vivo experiments. Bacterial suspensions were used directly for in vitro experiments by diluting the bacteria in respective cell culture media to the desired concentration. For in vivo experiments, the bacteria were centrifuged at 4,000 rpm for 10 min and the pellet was resuspended in sterile phosphate-buffered saline (PBS) to reach the desired bacterial concentration.
Table 1.
Bacterial strains
| Strain | Name | Genotype |
|---|---|---|
| AH 1785 | wt + pCM28 | Wild type + pCM28 (empty vector control) |
| AH 1787 | nuc + pCM28 | nuc:ltrB + pCM28 (empty vector control) |
| AH 1773 | nuc + pCM28nuc | nuc:ltrB + pCM28nuc (complementing vector) |
| AH 1263 | wt | CA-MRSA strain LAC, USA300-0114 PFGE type, Erm sensitive |
| AH 1680 | nuc | S. aureus LAC nuc:LtrB |
Nuclease Assays
To measure nuclease activity, supernatants from the panel of S. aureus strains (table 1) were harvested from mid-logarithmic growth cultures (OD600 = 0.7) after centrifugation for 10 min at 4,000 rpm. A volume of 2.5 μl of the supernatant or sterile BHI (negative control) was incubated with 7.5 μl calf thymus DNA (1 mg/ml, Sigma) and 40 μl DNase buffer (3 mM MgCl2, 3 mM CaCl2, 300 mM Tris; pH 7.4) for 60 min at 37°C. The nuclease reaction was stopped with 12.5 μl 0.33 M EDTA (pH 8.0), then 12.5 μl 6× loading dye was added and 20 μl of each sample was run on a 1% agarose gel for visual examination of DNA degradation.
Neutrophil Killing Assays
Human neutrophils were isolated from healthy donors by using the PolymorphPrep system (Axis-Shield). Neutrophils were resuspended in RPMI containing 2% nuclease-free (heat-inactivated at 70°C [22]) fetal calf serum (FCS) and plated in nontreated tissue culture plates (Greiner Bio-One, Cellstar®) at a concentration of 2 × 106 cells/ml. The cells were treated with 10 μg/ml cytochalasin D (Sigma-Aldrich) to block phagocytosis and with 25 nM phorbol 12-myristate 13-acetate (PMA) to stimulate NET formation. After incubation for 20 min at 37°C in 5% CO2, the neutrophils were infected with bacteria at a multiplicity of infection (MOI) of 2. The plates were centrifuged at 1,600 rpm for 5 min and incubated for 30 and 90 min at 37°C in 5% CO2. Serial dilutions in sterile PBS were plated on Todd-Hewitt agar plates for enumeration of surviving bacteria. The percentage of surviving bacteria was calculated in comparison to bacterial growth control grown under the same conditions in the absence of cells.
NET Entrapment Assays
A volume of 10 ml bacteria at OD600 = 0.7 was incubated in the presence of 0.33 mg/ml fluorescein-5-isothiocyanate (FITC isomer I, Invitrogen) for 30 min on ice. Human neutrophils were resuspended in RPMI without phenol red containing 2% nuclease-free FCS and plated in nontreated tissue culture plates at a concentration of 2 × 106 cells/ml. Cells were stimulated with 25 nM PMA for 20 min at 37°C in 5% CO2, then infected with bacteria at a MOI of 200, and incubated for 90 min at 37°C in 5% CO2. After incubation, the plate was centrifuged at 800 rpm for 5 min. From the wells containing neutrophils, the supernatant was carefully aspirated and fresh, prewarmed medium was added. This washing procedure was done twice to remove bacteria that were not entrapped within the NETs. Fluorescence was measured with a fluorescence reader at 485/538 nm. The percentage of entrapped bacteria was calculated as (A458/538 of wells containing neutrophils)/(A458/538 of wells without neutrophils) × 100.
NET Visualization and Quantification
Neutrophils were seeded on poly-L-lysine-coated cover slides and stimulated with 25 nM PMA for 20 min at 37°C in 5% CO2. The cells were infected with S. aureus at a MOI of 2, centrifuged at 1,600 rpm for 5 min and incubated for 90 min at 37°C in 5% CO2. After incubation, cells were fixed with 4% paraformaldehyde, washed with PBS and blocked with 2% whole goat serum (MP Biomedicals) in PBS + 2% BSA for 45 min at room temperature. To visualize NETs, the slides were incubated for 1 h at room temperature with antibodies against myeloperoxidase (MPO, rabbit anti-MPO, 1:300 diluted, Dako) and against histone H2A-H2B-DNA complex (mouse monoclonal anti-H2A-H2B-DNA, PL2–6, stock 2.65 mg/ml, 1:3,000 diluted) [23]. After incubation, the slides were washed 3 times with PBS and incubated for 45 min at room temperature with secondary antibodies, Alexa fluor 488 goat anti-rabbit IgG (1:500, Invitrogen) or Alexa fluor 488 rabbit anti-mouse IgG (1:500, Invitrogen). After washing, the slides were mounted on glass slides using Prolong Gold with 4’,6’-diamidino-2-phenylindole (DAPI) (Invitrogen). Washing steps were conducted with PBS and the antibodies were diluted in 2% BSA-PBS (for MPO staining) and 2% BSA-PBS + 0.2% Triton X-100 (for H2A-H2B-DNA complex staining). Mouse IgG2b (Thermo Scientific) and rabbit IgG (Jackson) were used as respective isotype control antibodies. Images were recorded using a Zeiss Axiolab microscope (Zeiss 20×/0.5 Plan-Neofluar objective) with an attached Sony Digital Photo Camera DKC-5000. The total amount of neutrophils and the amount of neutrophils releasing NETs per field of view were counted in 6 individual images per sample. The proportion of NETs per total amount of neutrophils was calculated for comparison of NET degradation by different bacterial strains.
Microscopy to Determine Killing of Bacteria in NETs
To determine the viability of S. aureus entrapped within NETs, neutrophils were seeded, stimulated and infected as described above. After 30 min of incubation, the cells were washed and the Live/Dead BacLight™ Bacterial Viability Kit (Invitrogen) used according to the manufacturer's recommendations. After staining for 15 min at room temperature, the slides were washed with PBS and fixed with 1% paraformaldehyde for 5 min at room temperature. Then the slides were washed again 3 times with PBS and mounted in Prolong Gold with DAPI (Invitrogen). The samples were analyzed using a confocal laser-scanning 2-photon microscope: Fluoview FV1000 with FluoView™ Spectral Scanning technology (Olympus). Images were recorded using 20×/0.7 UPlanSAp, 40×/1.30 oil UPlanFLN or 60×/1.42 oil PlanApoN objectives at calibrated magnifications.
In vivo Mouse Model
An established mouse model of S. aureus respiratory tract infection [24,25] was used to analyze the in vivo impact of S. aureus nuclease. Outbred, 8-week old-female CD1 mice were obtained from Charles River Laboratories and housed for a minimum of 3 days after shipping prior to the start of each experiment. Mice were anesthetized with an intraperitoneal injection of 50 mg/kg ketamine and 5 mg/kg rompun and then infected intranasally with wild-type or nuc-mutant bacteria. For histological studies and to determine bacterial load in lung tissue, the animals were infected with a sublethal dose of 2 × 108 colony-forming units (CFU) in 30 μl PBS. Twenty-four hours after infection, the lungs were inflated via the trachea with 10% paraformaldehyde and fixed overnight for histological examination. For determination of bacterial load, the lungs were removed 6 and 24 h after infection and homogenized in PBS for 2 × 1 min using zirconia beads (1 mm, Biospec) in a Mini-Beadbeater (BioSpec Products). Serial dilutions in sterile PBS were plated on Todd-Hewitt Agar plates for enumeration of CFU. For determinations of survival curves, the animals were infected with 3 or 4 × 108 CFU in 30 μl PBS and mortality was recorded.
Immunohistochemistry
Lung samples were kept in 70% ethanol prior to embedding in paraffin. Sections (7 μm thick) were deparaffinized by immersing successively in 3 changes of xylene for 10 min each and rehydrated by immersing in decreasing concentrations of ethanol (100, 95, 70%, each twice for 5 min). After washing 3 times with PBS, antigen retrieval was performed by microwave-heating the slides for 2 × 5 min in citrate buffer (10× Antigen retrieval solution, Dako). After cooling down for 20 min at room temperature, the slides were washed with PBS, and immunostained overnight at 4°C with primary antibodies against rabbit anti-murine cathelicidin (CRAMP, stock 1.55 mg/ml, 1:300 diluted, provided by Richard Gallo, UCSD, Calif., USA [26]) and mouse anti-H2A-H2B-DNA complex antibody (stock 2.65 mg/ml, 1:3,000 diluted [23]) or respective isotype controls (rabbit IgG whole molecule, Jackson ImmunoResearch, and mouse IgG2b, Thermo Scientific). After washing 3 times with PBS, slides were incubated with secondary antibodies Alexa 568 goat anti-rabbit (Invitrogen, 1:500) and Alexa 488 goat anti-mouse (Invitrogen, 1:500) for 1 h at room temperature. After washing 3 times, the sections were mounted on glass slides using Prolong Gold with DAPI (Invitrogen).
Statistical Analysis
Data were analyzed by using Excel 2003 (Microsoft) and GraphPad Prism 5.0 (GraphPad Software). Experiments were performed in triplicate or quadruplicate in at least 3 independent experiments. Differences between 2 groups in vitro were analyzed by using a paired, one-tailed Student's t test. For in vivo data, significant outliers were excluded using the Grubbs’ test (GraphPad software) and normal distribution of data was verified by D'Agostino and Pearson omnibus normality test (GraphPad software) prior to statistical analysis. Differences between 2 groups were then analyzed using an unpaired, one-tailed Student's t test. Analysis of survival curves was performed by the use of the Gehan-Breslow-Wilcoxon test (GraphPad software). p < 0.05 was considered to be significant.
Results
S. aureus Nuclease Promotes Degradation of Calf Thymus DNA
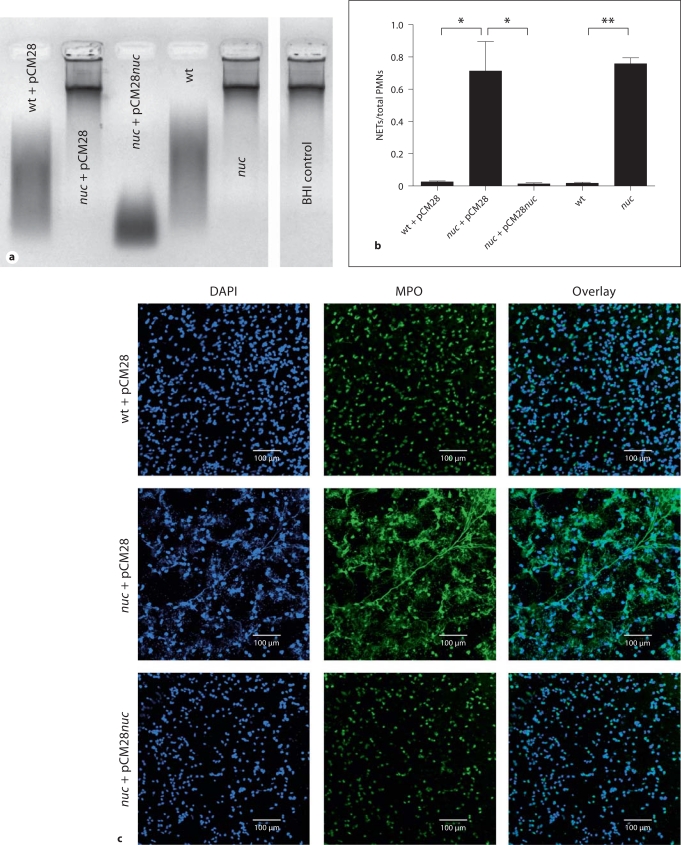
Using targeted mutagenesis, nuclease gene mutants of the CA-MRSA strain LAC were generated as described elsewhere [21]. To assess interaction with host DNA, bacterial supernatant was collected and the nuclease activity was tested in an agarose-gel-based DNA degradation assay. As shown in figure 1a, the nuc-mutant strains (nuc and nuc + pCM28 empty vector control) exhibited no nuclease activity. Sterile BHI medium alone was used as negative control. In contrast, supernatants from the wild-type strain (wt) and its respective vector control (wt + pCM28) or complemented strain (nuc + pCM28nuc) efficiently degraded calf thymus DNA (fig. 1a).
Fig. 1.
S. aureus nuclease degrades calf thymus DNA and NETs. a Representative agarose gel of bacterial culture supernatants after incubation with calf thymus DNA to detect nuclease activity. Samples with nuclease activity show a smear of degraded DNA on the gel as is seen in the lanes representing the S. aureus LAC wild-type empty vector control (wt + pCM28), complemented nuc-mutant strain (nuc + pCM28nuc) and wild-type (no vector, wt). In the lanes with samples from nuc-mutant empty vector control (nuc + pCM28), nuc-mutant (no vector, nuc) and sterile BHI medium (negative control), the DNA is not degraded, indicating that no nuclease activity is present. b Quantification of NETs after coincubation of PMA-stimulated neutrophils with S. aureus LAC wild-type empty vector control (wt + pCM28), nuc-mutant empty vector control (nuc + pCM28) or complemented mutant strain (nuc + pCM28nuc), wild-type (no vector, wt) or nuc-mutant (no vector, nuc) at a MOI of 2 for 90 min. After coincubation, the slides were fixed and stained for MPO to visualize the NETs and mounted in DAPI to stain DNA. The amount of neutrophils that release NETs was counted per field of view and compared with the total amount of neutrophils. The results of 4 (wt + pCM28, nuc + pCM28, nuc + pCM28nuc) or 3 (wt, nuc) independent experiments were analyzed using a paired, one-tailed Student's t test. ∗ p < 0.05; ∗∗ p < 0.01. c Representative immunofluorescent micrograph of PMA-stimulated neutrophils coincubated with S. aureus LAC wild-type empty vector control (wt + pCM28), nuc-mutant empty vector control (nuc + pCM28) or complemented mutant strain (nuc + pCM28nuc). NETs were visualized with a primary rabbit-anti-MPO antibody and a secondary Alexa 488-labeled goat-anti-rabbit antibody (green). DNA is stained with DAPI (blue).
S. aureus Nuclease Promotes Degradation of NETs
To test whether S. aureus is able to degrade NETs via its nuclease activity, neutrophils were stimulated with PMA for NET induction and then incubated in the presence of wild-type or nuc-mutant bacteria (table 1). After 90 min of coincubation, neutrophils infected with wild-type bacteria showed no intact NETs whereas approximately 70% of the neutrophils treated with nuc mutant were associated with intact NETs (fig. 1b, c). Control experiments analyzing the release of lactate dehydrogenase as marker for cytotoxicity and elastase as marker for neutrophil activation did not reveal significant differences between the strains (online supplementary figure 1, www.karger.com/doi/10.1159/000319909), indicating that S. aureus nuclease has no effect on cytotoxicity or activation of neutrophils. We conclude that the nuclease is efficiently degrading NETs.
S. aureus Nuclease Mediates Resistance against Entrapment and Killing within NETs
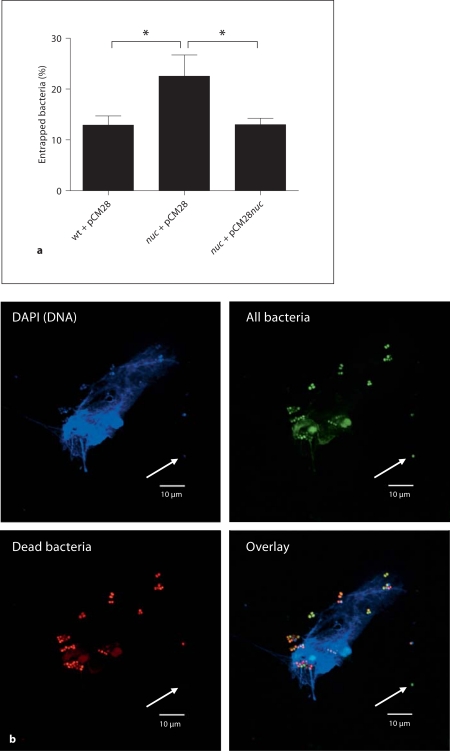
NETs have been shown to mediate their antimicrobial activity through (1) bacterial entrapment and (2) subsequent direct bacterial killing by antimicrobial peptides and histones which are embedded in the NETs or accumulated in close proximity to the cell [27]. As shown in figure 2a, significantly more nuc-mutant bacteria are found to be entrapped by NETs compared with wild-type bacteria, which have the ability to degrade the NETs (fig. 1b, c). The few remaining NETs that are not eliminated by the wild-type strain have the same capability to capture bacteria as NETs coincubated with the nuc-mutant strain (online suppl. fig. 2). However, a nuclease-dependent reduction in total NET surface area, as shown in figure 1b, c, allows the wild-type bacteria to largely avoid entrapment (fig. 2a) and subsequent killing within NETs. As confirmed in figure 2b, bacteria that are entrapped by NETs can be efficiently killed through accumulating concentrations of antimicrobial peptides and histones within NETs or surrounding the cell.
Fig. 2.
S. aureus nuclease facilitates evasion from NET entrapment. a Quantitative analysis of bacterial entrapment by activated neutrophils. FITC-labeled bacteria [S. aureus LAC wild-type empty vector control (wt + pCM28), nuc-mutant empty vector control (nuc + pCM28) or complemented mutant strain (nuc + pCM28nuc)] were coincubated with PMA-stimulated neutrophils at a MOI of 200 for 90 min at 37°C in 5% CO2. After incubation, the plates were centrifuged and the wells were carefully washed twice with fresh medium to remove bacteria that were not entrapped within the NETs. The percentage of entrapped bacteria was calculated as (A458/538 nm of wells containing neutrophils)/(A458/538 of wells without neutrophils) × 100. The results of 5 independent experiments were analyzed using a paired, one-tailed Student's t test. ∗ p < 0.05. b Representative fluorescent micrograph showing viability of S. aureus LAC nuc-mutant (nuc + pCM28) entrapped by or in close proximity to NETs. Live/Dead Bac-Light™ Bacterial Viability Kit (Invitrogen) was used to determine the viability of bacteria after coincubation with stimulated neutrophils. Similar bacterial killing within remaining NETs has been detected in case of the wild-type strain (data not shown). The green dye (SYTO 9) generally labels all bacteria – bacteria with intact as well as damaged membranes. In contrast, the red dye (propidium iodide) penetrates only bacteria with damaged membranes, causing a reduction in the green (SYTO 9) fluorescence stain. Note that bacteria entrapped by or in close proximity to the NETs are dead (red + green) whereas bacteria that are further away from the NETs remain alive (green, white arrow).
Finally, the overall bacterial viability of wild-type and nuc-mutant bacteria was quantified by enumerating the surviving bacteria after coincubation with activated neutrophils for 30 or 90 min. Using a total neutrophil-killing assay, the nuc-mutant exhibited a slight but significant increased susceptibility to neutrophil killing compared with the parental strain. The difference between wild-type and complemented strain was not significant (online suppl. fig. 3). However, when neutrophils were treated with cytochalasin D to block phagocytosis, but not the formation of NETs (fig. 3a), the differences between wild-type, nuc-mutant and complemented strains became more distinct. As shown in figure 3b, the nuc-mutant strain is significantly more susceptible to the extracellular antimicrobial activity of neutrophils compared with the wild-type and complemented strains (fig. 3b), demonstrating that nuclease efficiently mediates resistance against entrapment and subsequent killing within NETs.
Fig. 3.
S. aureus nuclease mediates resistance against extracellular killing by neutrophils. a Representative immunofluorescent micrograph of neutrophils coincubated with S. aureus LAC nuc + pCM28 in the presence or absence of 10 μg/ml cytochalasin D (to block phagocytosis) showing that NETs are produced in the presence of cytochalasin D. NETs were visualized with a primary rabbit-anti-MPO antibody and a secondary Alexa 488-labeled goatanti-rabbit antibody (green). DNA is stained with DAPI (blue). b Bacterial growth inhibition after coincubation of S. aureus LAC wild-type empty vector control (wt + pCM28), nuc-mutant empty vector control (nuc + pCM28) or complemented mutant strain (nuc + pCM28nuc) with PMA-stimulated neutrophils. Phagocytosis was blocked by treatment of the cells with 10 μg/ml cytochalasin D, 20 min prior to infection. Data are presented as percentage of surviving bacteria compared with the respective bacterial growth control (100%). The results of 4 independent experiments were analyzed using a paired, one-tailed Student's t test. ∗∗ p < 0.01.
Role of S. aureus Nuclease in Respiratory Tract Infection
Nearly 3 decades ago, Nugent and Pesanti [28] reported that S. aureus is cleared from the lungs of infected mice by an unidentified extracellular killing mechanism. Based on this discovery, we decided to investigate the presence and role of NETs in an established murine model for respiratory tract infection described by Bubeck Wardenburg et al. [24,25]. Using immunofluorescence microscopy, we identified NET formation in the alveolar space of mouse lungs 24 h after intranasal infection with 2 × 108 CFU of S. aureus strain LAC (fig. 4). PBS-treated mice were used as negative control and exhibited no NET formation in the lung tissue (fig. 4).
Fig. 4.
Formation of NETs in S. aureus-infected lungs in vivo. Representative immunofluorescent micrograph showing the presence of NETs in the alveolar space of murine lung sections 24 h after intranasal infection with 2 × 108 CFU of S. aureus LAC wild-type. In the right panel of S. aureus-infected lung tissue, a cell in the alveolar space is visible (white arrow), which releases a mixture of CRAMP and DNA-histone complexes (NETs) into the surrounding (alveolar space at the top of the DAPI-stained nucleus). Those NETs are absent in lungs from the PBS control mice. NETs were visualized using a triple-staining of DAPI to stain DNA (blue), monoclonal mouse anti-H2A-H2B-DNA complex antibody followed by an Alexa 488-goatanti-mouse antibody (green) and rabbitanti-CRAMP antibody followed by Alexa 568-goat-anti-rabbit antibody (red).
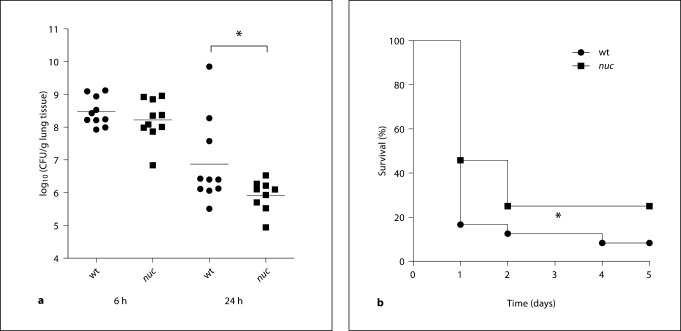
Having confirmed the presence of NETs in vivo, we analyzed the effect of nuclease-mediated NET degradation on S. aureus pathogenesis using the above-mentioned animal model. Mice were intranasally infected with a sublethal dose of 2 × 108 CFU wild-type or nuc-mutant bacteria and bacterial load in the lung tissue was quantified at 6 and 24 h after infection. Control experiments confirmed that the nuc mutation is stable in vivo (online suppl. fig. 4), as shown by a nuclease degradation assay with bacteria recovered at 24 h after infection. At 6 h after infection we found similar amounts of wild-type and nuc bacteria in the lungs of infected mice (fig. 5a). However, after 24 h, nearly 1 log-fold more bacteria were recovered from the lungs of wild-type infected mice compared with mice infected with the nuc-mutant strain (fig. 5a). These results suggest nuclease production impairs S. aureus clearance from lung tissue.
Fig. 5.
S. aureus nuclease expression mediates pathogenesis in vivo. a Bacterial load (CFU) in lung tissue of 10 mice (pooled from 2 individual experiments with 5 mice each) at 6 and 24 h after intranasal infection with 2 × 108 CFU S. aureus LAC wild-type (wt) or nuc-mutant (nuc) strain. Differences between the 2 groups were analyzed by using a unpaired, one-tailed Student's t test (∗ p < 0.05). b Survival of female CD-1 mice (n = 24, pooled from 4 individual experiments with 6 mice each) after intranasal infection with 3 or 4 × 108 CFU of S. aureus LAC wild-type (wt) or nuc-mutant (nuc) strain. Comparison of survival curves was performed with the Gehan-Breslow-Wilcoxon test (∗ p < 0.05).
Finally, to examine the overall virulence contribution of nuclease production, mice were infected with a lethal dose of 4 × 108 CFU wild-type or nuc-mutant bacteria to score mortality within 5 days after intranasal infection. As shown in figure 5b, wild-type mice exhibited a slightly, but significantly (p = 0.037) faster mortality rate compared with mice infected with nuc-mutant bacteria.
In conclusion our data revealed that S. aureus nuclease facilitates escape from NET-mediated killing by neutrophils and thereby impairs an important host immune defense mechanism required for efficient clearance of the pathogen.
Discussion
Neutrophils are the principal phagocytic cells of humans and other mammals and have been shown to be essential in host immune defense against staphylococcal infections [29]. Classically, two mechanisms were considered to mediate the direct antimicrobial activity of neutrophils. Neutrophils can recognize, bind, engulf and subsequently inactivate the invading microbes within phagolysosomes. Alternatively, neutrophil degranulation can release antimicrobial factors of the cell into the surroundings. Only recently has a third and novel mechanism of antimicrobial activity of neutrophils been recognized, originally described in the landmark study of Brinkmann et al. [27], namely the formation of NETs. Evidence is now accumulating indicating that extracellular trap formation is a feature of other immune cells including mast cells [30] and eosinophils [31]. Extracellular traps develop after stimulation with mitogens, cytokines, or pathogens themselves, in a process dependent upon induction of a reactive-oxygen-species-mediated signaling cascade. Extracellular traps can consist of nuclear or mitochondrial DNA as a backbone with associated antimicrobial peptides, histones and cell-specific proteases, which provide a matrix to entrap and kill microbes [12].
Recently, in vitro analyses have shown that NETs are effective in entrapment and killing of S. aureus [27,32] (fig. 3b). Using immunofluorescence microscopy, we have confirmed the presence of NETs in the lung tissue of S. aureus-infected mice, indicating that NETs are produced in vivo in response to S. aureus infection and may therefore contribute to host immune defense [27,32] (fig. 3b). Indeed, these may now be considered to represent an important component of the nonphagocytic, extracellular clearance of S. aureus from lungs observed by Nugent and Pasani [28] in an earlier study.
Certain leading bacterial pathogens have evolved mechanisms to avoid NET-based immune clearance, either through NET degradation [13,14,15], resistance to the intrinsic antimicrobial effectors within NETs [33,34], or the suppression of NET production [35]. In fact, experiments based on the manipulation of the microbial side of the host-pathogen interaction have provided one of the best demonstrations of a critical role of NETs in innate host defense [12].
In the present study, we have used an isogenic nuclease mutant of S. aureus to show that the pathogen uses nuclease expression to reduce its entrapment by NETs and build up relative resistance against killing by NETs. Whole-blood killing experiments using opsonized bacteria did not reveal any significant differences between the wild-type and mutant strains (data not shown), suggesting that nuclease is only mediating resistance to extracellular NET-mediated killing but not to intracellular killing within phagocytes.
S. aureus nuclease production was found to be associated with delayed bacterial clearance in the lung and significantly increased mortality using a well-established model for S. aureus respiratory tract infections [24,25]. Accordingly, S. aureus joins other leading Gram-positive pathogens, such as group A Streptococcus and S. pneumoniae, which facilitate their own NET evasion through nuclease production. Drugs that inhibit nuclease activity, induce or stabilize NET formation may support host immune defense and help to improve the outcome of bacterial infection with S. aureus and other common pathogens.
Supplementary Material
Nuclease-independent activation and cytotoxicity of neutrophils after co-incubation with S. aureus. Percentage of LDH release (as marker for cytotoxicity) and elastase release (as marker for activation) by PMA-stimulated neutrophils after co-incubation with S. aureus LAC wild type empty vector control (wt + pCM28), nuc-mutant empty vector control (nuc + pCM28) or complemented mutant strain (nuc + pCM28nuc). The results of n = 3 independent experiments were analyzed using a paired, one-tailed Student's t-test. P values of < 0.05 were considered to be significant. No significant differences were detected between the three different bacterial strains.
Entrapment of bacteria by NETs. Representative immuno-fluorescent micrograph showing entrapment of FITC-labelled bacteria within NETs. S. aureus LAC wild type empty vector control (wt + pCM28) or nuc-mutant empty vector control (nuc + pCM28) were co-incubated with PMA-stimulated neutrophils at a MOI of 2 for 90 min at 37°C in 5% CO2. After washing and fixation, NETs were visualized with DNA-intercalating dye Dapi (blue). Bacteria are shown in green. Note that the remaining NETs that are not eliminated by the wild type strain have the same capability to capture bacteria compared to those NETs co-incubated with the nuc-mutant strain.
Total (extra- and intracellular) antimicrobial activity of neutrophils against S. aureus strains. Bacterial growth inhibition after co-incubation of S. aureus LAC wild type empty vector control (wt + pCM28), nuc-mutant empty vector control (nuc + pCM28) or complemented mutant strain (nuc + pCM28nuc) with PMA-stimulated neutrophils. Data are presented as percentage surviving bacteria compared to the respective bacterial growth control (100%). The results of n = 4 independent experiments were analyzed using a paired, one-tailed Student's t-test. P values of < 0.05 were considered to be significant (*** p<0.005).
Nuc-mutation remains stable in vivo. To confirm stability of the nuc-mutant (nuc) in vivo, bacteria were recovered from murine lungs at 24 h after infection. Harvested bacteria were grown overnight in BHI and culture supernatants were tested for nuclease activity in an agarose-gel-based nuclease assay. A representative agarose gel is shown, which demonstrates that bacteria recovered from wild type (wt)-infected mice show nuclease activity (DNA degradation), whereas the bacteria recovered from lungs after infection with the nuc-mutant (nuc) do not exhibit nuclease activity.
Elastase and LDH assays According to the literature, elastase-release was used as marker for neutrophil activity and degranulation [i] and LDH-release was used as marker for cytotoxicity [ii]. Human neutrophils were resuspended in RPMI without phenol red containing 2% nuclease-free FCS (70°C heat-inactivated) and plated in non-treated tissue culture plates (Greiner Bio-One, CELLSTAR®) at a concentration of 2 × 106 cells/ml. Neutrophils were stimulated with 25nM PMA for 20 min at 37°C in 5% CO2. Then, neutrophils were infected with bacteria at MOI of 2, the plates were centrifuged at 1600 rpm for 5 min and incubated for 90 min at 37°C in 5% CO2. After incubation, micrococcal nuclease (Worthington) was added at a concentration of 500 mU/ml to degrade NETs and to release elastase from NETs. The reaction was stopped with 5 mM EDTA and the plate was centrifuged at 1000 rpm for 8 min. For elastase measurement, 50 μl of the supernatant was incubated with 50 μl of 200 μM elastase substrate (N-(Methoxysuccinyl)-Ala-Ala-Pro-Val 4-nitroanilide, Sigma) for 30 min at room temperature. Optical density was measured at 405nm (VersaMax Tunable Microplate Reader, Molecular Devices). For LDH measurement, the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega) was used according to the manufacturer's recommendations. The percentage of LDH or elastase release was calculated compared to 100% cell lysis control (cells lysed with 0.25 % Triton X-100 for 10 min).
Neutrophil killing assays Human neutrophils were resuspended in RPMI containing 2% nuclease-free FCS and plated in non-treated tissue culture plates at a concentration of 2 × 106 cells/ml. Neutrophils were stimulated with 25 nM PMA for 20 min at 37°C in 5% CO2. Then, neutrophils were infected with bacteria at MOI of 2, the plates were centrifuged at 1600 rpm for 5 min and incubated for 30 and 90 min at 37°C in 5% CO2. After incubation, cells were lysed with 0.25 % Triton X-100 by pipetting up and down. Serial dilutions in sterile PBS were plated on THA plates for enumeration of surviving cfu. The percentage of surviving bacteria was calculated in comparison to bacterial growth control grown under the same conditions in the absence of cells.
Acknowledgements
The authors wish to thank Suzan H.M. Rooijakkers and Daphne A.C. Stapels for helpful scientific discussions, and the UCSD Microscopy Facility (supported by UCSD Neuroscience Microscopy Shared Facility Grant P30 NS047101) for assistance. This work was supported by NIH Grants AI077780 to V.N. and AI083211 to A.R.H. M.v.K.-B. was supported through a fellowship from the Deutsche Akademie der Naturforscher Leopoldina (BMBF-LPD 9901/8-187), N.M.H. through the Ruth L. Kirschstein National Research Service Award, NIH 1 F31 GM90658-01 and E.T.M.B. through the K.F. Hein Fonds, Studie en Indivuele Noden and Dr. Hendrik Muller's Vaderlandsch Fonds.
References
- 1.Ogston A. Micrococcus poisoning. J Anat Physiol. 1882;16:526–567. [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Chambers HF, DeLeo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi SD, Braughton KR, Palazzolo-Ballance AM, Kennedy AD, Sampaio E, Kristosturyan E, Whitney AR, Sturdevant DE, Dorward DW, Holland SM, Kreiswirth BN, Musser JM, DeLeo FR. Rapid neutrophil destruction following phagocytosis of Staphylococcus aureus. J Innate Immun. 2010 doi: 10.1159/000317134. E-pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronner S, Monteil H, Prévots G. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev. 2004;28:183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Dinges MM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung AL. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40:1–9. doi: 10.1016/S0928-8244(03)00309-2. [DOI] [PubMed] [Google Scholar]
- 8.Cuatrecasas P, Fuchs S, Anfinsen CB. Catalytic properties and specificity of the extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1967;242:1542–1547. [PubMed] [Google Scholar]
- 9.Okabayashi K, Mizuno D. Surface-bound nuclease of Staphylococcus aureus: purification and properties of the enzyme. J Bacteriol. 1974;117:222–226. doi: 10.1128/jb.117.1.222-226.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis A, Moore IB, Parker DS, Taniuchi H. Nuclease B, a possible precursor of nuclease A, an extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1977;252:6544–6553. [PubMed] [Google Scholar]
- 11.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Köckritz-Blickwede M, Nizet V. Innate immunity turned inside-out: antimicrobial defense by phagocyte extracellular traps. J Mol Med. 2009;87:775–783. doi: 10.1007/s00109-009-0481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, DeLeo FR, Musser JM. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci USA. 2005;102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 15.Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol. 2006;16:401–407. doi: 10.1016/j.cub.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 16.Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Saïd-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol. 2005;175:3907–3919. doi: 10.4049/jimmunol.175.6.3907. [DOI] [PubMed] [Google Scholar]
- 17.Begier EM, Frenette K, Barrett NL, Mshar P, Petit S, Boxrud DJ, Watkins-Colwell K, Wheeler S, Cebelinski EA, Glennen A, Nguyen D, Hadler JL. A high-morbidity outbreak of methicillin-resistant Staphylococcus aureus among players on a college football team, facilitated by cosmetic body shaving and turf burns. Clin Infect Dis. 2004;39:1446–1453. doi: 10.1086/425313. [DOI] [PubMed] [Google Scholar]
- 18.Kazakova SV, Hageman JC, Matava M, Srinivasan A, Phelan L, Garfinkel B, Boo T, McAllister S, Anderson J, Jensen B, Dodson D, Lonsway D, McDougal LK, Arduino M, Fraser VJ, Killgore G, Tenover FC, Cody S, Jernigan DB. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N Engl J Med. 2005;352:468–475. doi: 10.1056/NEJMoa042859. [DOI] [PubMed] [Google Scholar]
- 19.Miller LG, Perdreau-Remington F, Rieg G, Mehdi S, Perlroth J, Bayer AS, Tang AW, Phung TO, Spellberg B. Necrotizing fasciitis caused by community-associated methicillin-resistant Staphylococcus aureus in Los Angeles. N Engl J Med. 2005;352:1445–1453. doi: 10.1056/NEJMoa042683. [DOI] [PubMed] [Google Scholar]
- 20.Francis JS, Doherty MC, Lopatin U, Johnston CP, Sinha G, Ross T, Cai M, Hansel NN, Perl T, Ticehurst JR, Carroll K, Thomas DL, Nuermberger E, Bartlett JG. Severe community-onset pneumonia in healthy adults caused by methicillin-resistant Staphylococcus aureus carrying the Panton-Valentine leukocidin genes. Clin Infect Dis. 2005;40:100–107. doi: 10.1086/427148. [DOI] [PubMed] [Google Scholar]
- 21.&Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR: Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus (submitted for publication). [DOI] [PMC free article] [PubMed]
- 22.von Köckritz-Blickwede M, Chow OA, Nizet V. Fetal calf serum contains heat-nucleases that degrade neutrophil extracellular traps. Blood. 2009;114:5245–5246. doi: 10.1182/blood-2009-08-240713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losman MJ, Fasy TM, Novick KE, Monestier M. Monoclonal autoantibodies to subnucleosomes from a MRL/Mp(–)+/+ mouse. Oligoclonality of the antibody response and recognition of a determinant composed of histones H2A, H2B, and DNA. J Immunol. 1992;148:1561–1569. [PubMed] [Google Scholar]
- 24.Bubeck Wardenburg J, Scheeweind O. Vaccine protection against Staphylococcus aureus pneumonia. J Exp Med. 2008;205:287–294. doi: 10.1084/jem.20072208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bubeck Wardenburg J, Patel RJ, Schneewind O. Surface proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus pneumonia. Infect Immun. 2007;75:1040–1044. doi: 10.1128/IAI.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, Agerberth B, Gudmundsson GH, Gallo RL. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J Invest Dermatol. 2001;117:91–97. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- 27.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 28.Nugent KM, Pesanti EL. Nonphagocytic clearance of Staphylococcus aureus from murine lungs. Infect Immun. 1982;36:1185–1191. doi: 10.1128/iai.36.3.1185-1191.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Köckritz-Blickwede M, Rohde M, Oehmcke S, Miller LS, Cheung AL, Herwald H, Foster S, Medina E. Immunological mechanisms underlying the genetic predisposition to severe Staphylococcus aureus infection in the mouse model. Am J Pathol. 2008;173:1657–1668. doi: 10.2353/ajpath.2008.080337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Köckritz-Blickwede M, Goldmann O, Thulin P, Heinemann K, Norrby-Teglund A, Rohde M, Medina E. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood. 2008;111:3070–3080. doi: 10.1182/blood-2007-07-104018. [DOI] [PubMed] [Google Scholar]
- 31.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, Schmid I, Straumann A, Reichenbach J, Gleich GJ, Simon HU. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 32.Jann NJ, Schmaler M, Kristian SA, Radek KA, Gallo RL, Nizet V, Peschel A, Landmann R. Neutrophil antimicrobial defense against Staphylococcus aureus is mediated by phagolysosomal but not extracellular trap-associated cathelicidin. J Leukoc Biol. 2009;86:1159–1169. doi: 10.1189/jlb.0209053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong W, Juneau RA, Pang B, Swords WE. Survival of bacterial biofilms within neutrophil extracellular traps promotes nontypeable Haemophilus influenzae persistence in the chinchilla model for otitis media. J Innate Immun. 2009;1:215–224. doi: 10.1159/000205937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauth X, von Köckritz-Blickwede M, McNamara CW, Myskowski S, Zinkernagel AS, Beall B, Ghosh P, Gallo RL, Nizet V. M1 protein allows group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 2009;1:202–214. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zinkernagel AS, Timmer AM, Pence MA, Locke JB, Buchanan JT, Turner CE, Mishalian I, Sriskandan S, Hanski E, Nizet V. The IL-8 protease SpyCEP/ScpC of group A Streptococcus promotes resistance to neutrophil killing. Cell Host Microbe. 2008;4:170–178. doi: 10.1016/j.chom.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nuclease-independent activation and cytotoxicity of neutrophils after co-incubation with S. aureus. Percentage of LDH release (as marker for cytotoxicity) and elastase release (as marker for activation) by PMA-stimulated neutrophils after co-incubation with S. aureus LAC wild type empty vector control (wt + pCM28), nuc-mutant empty vector control (nuc + pCM28) or complemented mutant strain (nuc + pCM28nuc). The results of n = 3 independent experiments were analyzed using a paired, one-tailed Student's t-test. P values of < 0.05 were considered to be significant. No significant differences were detected between the three different bacterial strains.
Entrapment of bacteria by NETs. Representative immuno-fluorescent micrograph showing entrapment of FITC-labelled bacteria within NETs. S. aureus LAC wild type empty vector control (wt + pCM28) or nuc-mutant empty vector control (nuc + pCM28) were co-incubated with PMA-stimulated neutrophils at a MOI of 2 for 90 min at 37°C in 5% CO2. After washing and fixation, NETs were visualized with DNA-intercalating dye Dapi (blue). Bacteria are shown in green. Note that the remaining NETs that are not eliminated by the wild type strain have the same capability to capture bacteria compared to those NETs co-incubated with the nuc-mutant strain.
Total (extra- and intracellular) antimicrobial activity of neutrophils against S. aureus strains. Bacterial growth inhibition after co-incubation of S. aureus LAC wild type empty vector control (wt + pCM28), nuc-mutant empty vector control (nuc + pCM28) or complemented mutant strain (nuc + pCM28nuc) with PMA-stimulated neutrophils. Data are presented as percentage surviving bacteria compared to the respective bacterial growth control (100%). The results of n = 4 independent experiments were analyzed using a paired, one-tailed Student's t-test. P values of < 0.05 were considered to be significant (*** p<0.005).
Nuc-mutation remains stable in vivo. To confirm stability of the nuc-mutant (nuc) in vivo, bacteria were recovered from murine lungs at 24 h after infection. Harvested bacteria were grown overnight in BHI and culture supernatants were tested for nuclease activity in an agarose-gel-based nuclease assay. A representative agarose gel is shown, which demonstrates that bacteria recovered from wild type (wt)-infected mice show nuclease activity (DNA degradation), whereas the bacteria recovered from lungs after infection with the nuc-mutant (nuc) do not exhibit nuclease activity.
Elastase and LDH assays According to the literature, elastase-release was used as marker for neutrophil activity and degranulation [i] and LDH-release was used as marker for cytotoxicity [ii]. Human neutrophils were resuspended in RPMI without phenol red containing 2% nuclease-free FCS (70°C heat-inactivated) and plated in non-treated tissue culture plates (Greiner Bio-One, CELLSTAR®) at a concentration of 2 × 106 cells/ml. Neutrophils were stimulated with 25nM PMA for 20 min at 37°C in 5% CO2. Then, neutrophils were infected with bacteria at MOI of 2, the plates were centrifuged at 1600 rpm for 5 min and incubated for 90 min at 37°C in 5% CO2. After incubation, micrococcal nuclease (Worthington) was added at a concentration of 500 mU/ml to degrade NETs and to release elastase from NETs. The reaction was stopped with 5 mM EDTA and the plate was centrifuged at 1000 rpm for 8 min. For elastase measurement, 50 μl of the supernatant was incubated with 50 μl of 200 μM elastase substrate (N-(Methoxysuccinyl)-Ala-Ala-Pro-Val 4-nitroanilide, Sigma) for 30 min at room temperature. Optical density was measured at 405nm (VersaMax Tunable Microplate Reader, Molecular Devices). For LDH measurement, the CytoTox 96® Non-Radioactive Cytotoxicity Assay (Promega) was used according to the manufacturer's recommendations. The percentage of LDH or elastase release was calculated compared to 100% cell lysis control (cells lysed with 0.25 % Triton X-100 for 10 min).
Neutrophil killing assays Human neutrophils were resuspended in RPMI containing 2% nuclease-free FCS and plated in non-treated tissue culture plates at a concentration of 2 × 106 cells/ml. Neutrophils were stimulated with 25 nM PMA for 20 min at 37°C in 5% CO2. Then, neutrophils were infected with bacteria at MOI of 2, the plates were centrifuged at 1600 rpm for 5 min and incubated for 30 and 90 min at 37°C in 5% CO2. After incubation, cells were lysed with 0.25 % Triton X-100 by pipetting up and down. Serial dilutions in sterile PBS were plated on THA plates for enumeration of surviving cfu. The percentage of surviving bacteria was calculated in comparison to bacterial growth control grown under the same conditions in the absence of cells.