Abstract
Elevated expression of the orphan nuclear receptor estrogen-related receptor alpha (ERRα) has been associated with a negative outcome in several cancers, although the mechanism(s) by which this receptor influences the pathophysiology of this disease and how its activity is regulated remains unknown. Using a chemical biology approach it was determined that compounds, previously shown to inhibit canonical Wnt signaling, also inhibited the transcriptional activity of ERRα. The significance of this association was revealed in a series of biochemical and genetic experiments that demonstrate that (a) ERRα, β-catenin (β-cat) and Lymphoid enhancer-binding factor-1 (LEF-1) form macromolecular complexes in cells, (b) ERRα transcriptional activity is enhanced by βcat expression and vice versa, and (c) there is a high level of overlap among genes previously shown to be regulated by ERRα or β-cat. Furthermore, silencing of ERRα and β-cat expression individually or together dramatically reduced the migratory capacity of both breast and prostate cancer cells in vitro. This increased migration could be attributed to the ERRα/β-cat dependent induction of WNT11. Specifically, using (a) conditioned media from cells overexpressing recombinant WNT11 or (b) WNT11 neutralizing antibodies, we were able to demonstrate that this protein was the key mediator of the promigratory activities of ERRα/β-cat. Together, these data provide evidence for an autocrine regulatory loop involving transcriptional upregulation of WNT11 by ERRα and β-cat that influences the migratory capacity of cancer cells.
Keywords: estrogen-related receptor, beta-catenin, Wnt signaling, migration
Introduction
The Estrogen Related Receptor alpha (ERRα, NR3B1) is an orphan member of the nuclear receptor (NR) superfamily of transcription factors whose expression tracks with a negative outcome in breast and ovarian cancers [1–3]. It is unclear whether this ubiquitously expressed receptor requires a canonical small molecule ligand. However, under conditions of metabolic stress, such as fasting, exercise or cold, its expression is rapidly induced in tissues such as the heart, muscle and liver [4–5]. In these tissues, it directs a gene expression program that results in increased mitochondrial number and increased expression of key enzymes required for the tricarboxylic acid cycle (TCA) cycle and β-oxidation of fatty acids [6–8]. Thus, within the confines of normal physiology, ERRα appears to function primarily as a regulator of metabolic function under conditions of high energy demand [9–10]. The extent to which its role as a metabolic regulator is involved in the pathophysiology of cancer, however, remains to be determined [1–3].
In addition to its important role in oxidative metabolism, ERRα can modulate the activity of the estrogen receptor (ER) at some target genes [11]. Indeed, the amino acid homology between ERRα and the classical ERs (ERα and ERβ), particularly in their respective DNA-binding domains, initially suggested that the primary impact of ERRα in hormone-dependent cancers would be to modulate or interface with estrogen signaling pathways in cells. However, we and others have recently determined that only a relatively small percentage of ER target genes are coregulated by ERRα [12–13]. Although the significance of this crosstalk remains to be established, the observation that ERRα knockdown dramatically impacted the in vivo growth and in vitro migration of ER-negative MDA-MB 231 cells highlighted an independent role for this receptor in breast cancer [13].
Crystallographic analysis of the structure of several members of the NR superfamily has indicated that the hormone-binding domain of these proteins is configured in such a way as to create a cavity of between 360–1400A3 that serves to dock small molecule agonists or antagonists [14]. In the case of ERRα however, it has been demonstrated that its potential ligand binding cleft is occupied by the bulky side chains of four phenylalanines and that the remaining space (100A3) available in the pocket is likely to be too small to accommodate a regulatory ligand [15]. Furthermore, by comparison with the structures of other agonist-activated NRs, the apoERRα protein appears to be in the “active” conformation [15–16]. This finding has raised the question as to how the transcriptional activity of this receptor is regulated and whether the processes and pathways that impinge on and activate ERRα can be manipulated for therapeutic advantage. Cofactor availability and activity are likely to be the primary mechanisms by which ERRα activity is regulated [5, 17–18]. It has been shown, for instance, that the generally low basal activity of ERRα in cells can be dramatically upregulated by increasing the expression of either peroxisome proliferator-activated receptor-γ coactivator (PGC)-1 isoform PGC-1α or PGC-1β. Indeed, the expression level and activity of these coactivators are regulated by the physiological stresses that have been shown to activate ERRα transcriptional activity [19–20]. However, the extent to which these proteins regulate ERRα activity in breast cancer remains to be determined. Furthermore, it is unclear what pathological signals regulate the activity/expression of these cofactors, and whether or not there are cofactor-independent pathways that modulate ERRα activity. This goal of this study, therefore, was to identify pathways and processes upstream and downstream of ERRα that impact tumor pathophysiology and may be amenable to therapeutic manipulation.
Materials and Methods
Plasmids
The 3X-ERE-tata-luciferase reporter and pcDNA3-PGC-1α 2X9 were previously described [21]. pCMV-β-Gal (Clontech), pcDNA (Invitrogen) and pBlueScriptII (Statagene) were purchased. pCMX-ΔN89 and TOP-Flash were gifts (Dr. B. Hogan, Duke University). The pMSCV-GFP-hWNT11 plasmid was generated by subcloning WNT11 cDNA (MGC:141946) into pENTR3c (Invitrogen) and recombining into pMSCV-IRES-GFP.
Cell Culture
Cell lines were obtained from the ATCC (2007 to 2009), expanded for two passages, and cryopreserved. All experiments were performed with cells of passage less than 25. These cell lines were authenticated by morphological inspection, short tandem repeat profiling, and mycoplasma testing by the ATCC and cultured in RPMI (Invitrogen) (MDA-MB 436 (HTB-130), SKBR3 (HTB-30), PC-3 (CRL-1435), and HCT-116 (CCL-247)) or DMEM (Invitrogen) (MDA-MB 231 (HTB-26)) supplemented with 8.5% FBS (Sigma), 0.1mM nonessential amino acids, and 1mM sodium pyruvate (Invitrogen). Transient transfections were performed as described previously [22]. Luciferase and β-galactosidase activites were measured using a PerkinElmer Fusion Instrument [22].
Coimmunoprecipitation
Whole cell extracts were prepared using nondenaturing lysis buffer (20mM Tris-HCl, pH 8, 137mM NaCl, 10% glycerol, 1% Nonidet P-40, 2mM EDTA, protease inhibitors (Sigma)). Proteins were immunoprecipitated using antibodies to ERRα [21], β-cat (BD Biosciences), LEF-1 (Santa Cruz Biotechnology), and mouse IgG (Santa Cruz Biotechnology) (5μg antibody/500μg WCE, 16h, 4°C) and Protein-A/G PLUS-Agarose beads (Santa Cruz Biotechnology) (4h, 4°C), washed using lysis buffer 3 times and heat eluted in 2X-sample buffer. Proteins were separated by 10% SDS-PAGE, transferred to nitrocellulose, and detected by Western blotting (ERRα [21]; β-cat (EMD Biosciences); LEF-1 (Santa Cruz Biotechnology); and a light-chain specific secondary antibody (Jackson Immunoresearch)).
Immunoblotting
Whole cell extracts prepared using RIPA buffer (50mM Tris-HCl pH 7.5, 150mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS, 1mM EDTA, protease inhibitors (Sigma)) were separated by 10% SDS-PAGE, transferred onto nitrocellulose membranes and detected using the following antibodies: ERRα [21]; β-cat, (BD Biosciences); WNT11 (AbCam); ATG5 (Cell Signaling); Lamin A (Santa Cruz Biotechnology); or GAPDH, (Santa Cruz Biotechnology).
Adenoviral Transduction
Adenoviruses expressing β-Gal, PGC-1α, PGC-1α 2×9, or PGC-1α L2L3M were generated as described previously [21]. MDA-MB 231 cells were infected at multiplicity of infection of 30 for 48h.
Gene Silencing
Chemical siRNAs (Invitrogen, Qiagen) were used to silence ERRα (siERRα A: HSS103381, siERRα B: HSS103382), β-cat (siβ-cat A: HSS102460, siβ-cat B: HSS102461), WNT11 (siWNT11 A: SI00763378, siWNT11 B: SI03148719), Serum glucocorticoid kinase 1 (SGK1) (siSGK1 A: HSS109684, siSGK1 B: HSS109685) or autophagy protein 5 (ATG5) (siATG5 A: HSS114103, siATG5 B: HSS114104). Control siRNA sequences are listed in Table S2. MDA-MB 231 cells were seeded (250,000 cell per 6-well plate) and siRNAs were transfected using Dharmafect1 (Dharmacon) (100nM, 48h).
RNA preparation and analysis
Total RNA was isolated using the Bio-Rad Aurum RNA purification kit. cDNA was synthesized from 1μg total RNA using iScript (BioRad). qPCR was performed (0.25μl cDNA, 0.3μM primers (Table S1) with iQ SYBRGreen supermix (BioRad)), and results were calculated using the 2−ΔΔCt method.
ChIP Assays
ChIP assays were performed as described previously (additional details in Supplementary Methods) using primer sequences listed in Table S3.
WNT11 Retroviral Overexpression
pMSCV-IRES-GFP-hWNT11 and pMSCV-IRES-GFP-Gal4-DBD (control) were cotransfected (FuGene, Roche Applied Science) with the pCL10A1 packaging vector (Imigenex) into 293TS cells. These viral supernatants were clarified, polybrene supplemented (8μg/ml) and used to infect MDA-MB 231 cells for two serial 24h infections. Positive cells were selected by three rounds of cell sorting for GFP expression, yielding the MDA-MB 231/Control (C) and MDA-MB 231/WNT11 cell lines.
Migration & Viability Assays
For migration assays, cells were serum starved (18h, DMEM (MDA-MB 231) or RPMI (MDA-MB 436, PC-3, and HCT-116) with 0.1% BSA and 10mM HEPES), 2×104 cells (MDA-MB 231, MDA-MB 436, PC-3) or 7.5 ×104 cells (HCT-116) in 100μl were plated (BD Biocoat Control Inserts 8.0 micron, (BD Biosciences)), collagen coated (HCT-116) and migrated towards 8% FBS for 4h (MDA-MB 231, MDA-MB 436, PC-3) or 16h (HCT-116). The membrane was stained (5% crystal violet in 20% methanol) and cells that migrated were counted. Duplicate transwells were used and three high-powered fields (200X) were counted per membrane. Cell viability assays were performed in parallel with the migration studies using the Cell Titer Blue Assay (data not shown, Promega). Cells (2×104 cells in 100μl) were seeded in triplicate on a 96-well plate for 4h followed by addition of Cell Titer Blue dye (20μl) for 12h. Resultant fluorescence at (535nmEx, 620nmEm) was measured using a Perkin-Elmer Fusion Instrument. The fluorescence was calculated as the triplicate average (± SEM) followed by subtraction of background fluorescence from the media. For ERRα, β-cat, and WNT11 silencing, cells were transfected with siRNAs as described (Gene Silencing) 48h prior to the migration assay. For WNT11 overexpression, migration assays were performed using the stable MDA-MB 231 cell lines, Control and WNT11. For conditioned media studies, MDA-MB 231 cells migrated towards conditioned media from the MDA-MB 231/C or MDA-MB 231/WNT11 cells.
Immunodepletion of WNT11
Conditioned media from MDA-MB 231/C and MDA-MB 231/WNT11 was harvested at 80% confluence and was depleted of WNT11 protein by incubation with WNT11 antibody (AbCam) or rabbit IgG (Santa Cruz Biotechnology) (10μg/ml, 4°C for 16h).
Statistical Analyses
Transfection, qPCR, and migration data are represented as mean ± SEM for three biological replicates. Significance was evaluated by ANOVA and the Neumann-Keul’s post-hoc test for migration studies (GraphPad).
Results
Crosstalk between ERRα and Wnt Signaling Pathways
A chemical biology approach was used to define how ERRα is engaged in the regulation of pathways and processes of pathological importance in cancer. Specifically, a high-throughput cell-based assay was used to identify compounds that modulate the transcriptional activity of the PGC-1α/ERRα complex in MDA-MB 436 cells (ERα negative). Among the most interesting compounds identified in this manner were the carbolines (harmol, harmine, harmane and 6-methoxyharmalan), all of which inhibited ERRα transcriptional activity when assayed on a simple reporter (Figure S1). Previously, it has been shown that these carbolines could inhibit the canonical Wnt signaling pathway and, in doing so, enhance PPARγ activity in adipogenesis assays in vitro [23]. Given these data and the established role(s) of Wnt-signaling in cell migration and invasion [24–25], together with our previously published data which implicates ERRα in this biological process [13], it appeared likely that both effectors were components of the same pathway.
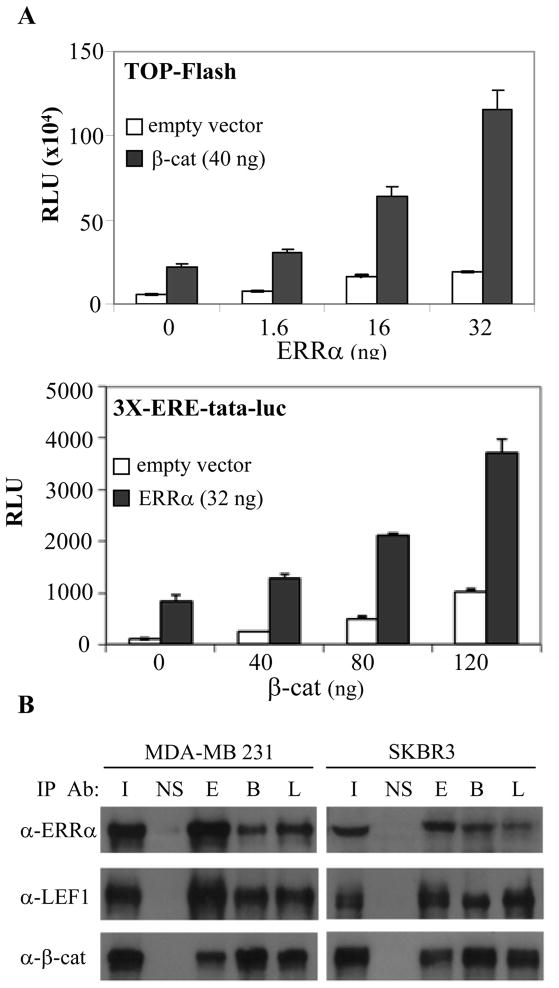
As an initial step in our analysis, we evaluated the impact of ERRα expression on the transcriptional activity of a constitutively active β-cat mutant when assayed on the TOP-Flash (T-cell factor/lymphoid enhancer-binding factor (TCF-LEF)) luc-reporter. As shown in Figure 1A, ERRα expression had a minimal effect on the basal expression of the reporter. However, a robust enhancement of β-cat dependent transcriptional activity was observed upon ERRα expression. Similarly, expression of β-cat significantly enhanced the activity of ERRα when assayed on the 3X-ERE-tata-luc reporter (Figure 1A). As expected, β-cat increased the basal activity of this reporter by enhancing the transcriptional activity of the endogeneous ERRα levels expressed in SKBR3 cells. The pathway crosstalk revealed in these transcriptional assays was further reinforced in a series of biochemical studies which indicated that ERRα, β-cat and LEF-1 physically interact (Figure 1C). Specifically, we were able to demonstrate by co-immunoprecipitation studies performed with endogenously expressed proteins from SKBR3, MDA-MB 231, or MDA-MB 436 (data not shown) breast cancer cells that ERRα interacted with both β-cat and LEF-1. We confirmed the direct nature of these interactions using GST-pull down assays (Figure S2). Thus, using both chemical and biochemical approaches we determined that the Wnt/β-cat and ERRα signaling pathways converge, a finding of likely importance in cancer pathogenesis.
Figure 1.
Crosstalk of ERRα and Wnt signaling pathways. A, ERRα and β-cat potentiate each other’s transcriptional activity when assessed in SKBR3 cells using the TOP-FLASH and 3X-ERE-tata-luc reporters to measure β-cat and ERRα activity, respectively. Similar transcriptional activation was observed in MDA-MB 436 cells. B, Coimmunoprecipitation of endogenous ERRα, β-cat, and LEF-1 from SKBR3 and MDA-MB 231 whole cell extracts was followed by Western blot analysis for the indicated proteins. IP Ab: I, Input; NS, IgG; E, ERRα; B, β-cat; L, Lef-1.
Both ERRα and β-cat regulated signaling pathways influence the migratory capacity of cancer cells
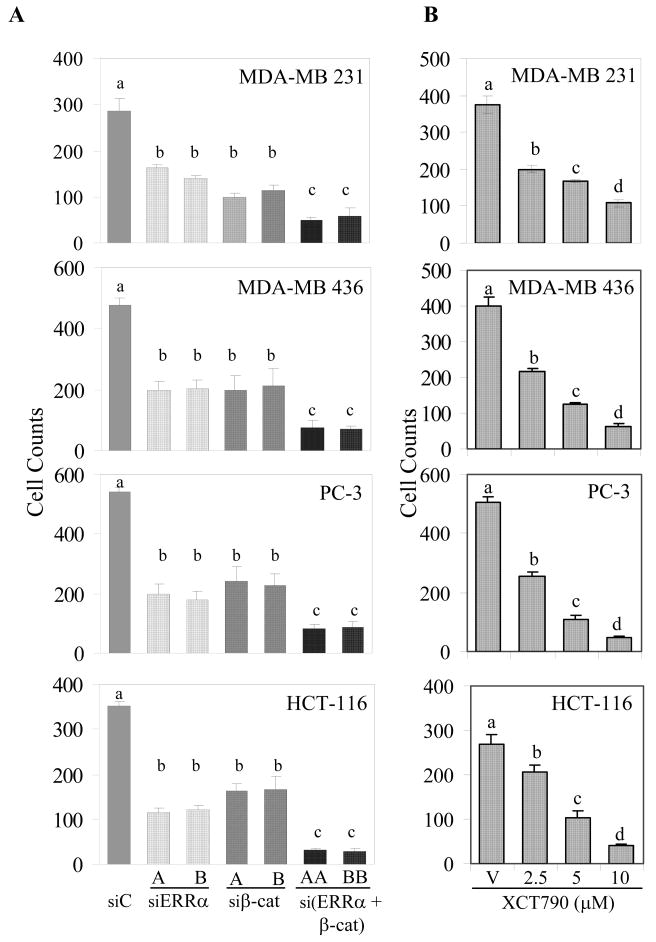
ERRα and β-cat have both been shown in independent studies to function as regulators of cancer cell migration [13, 25]. Thus, given that these proteins physically interact and exhibit functional crosstalk at the level of transcription, it seemed likely that they may also cooperate in the regulation of pathways that regulate cell migration. Thus, we evaluated the impact of siRNA-mediated knockdown of ERRα and β-cat, individually or combined, on the migratory capacity of multiple cancer cell lines including breast (MDA-MB 231 and MDA-MB 436), prostate (PC-3), and colon (HCT-116) cells using a Boyden chamber migration assay. Decreased cell migration was observed after silencing of ERRα or β-cat expression by either of two distinct siRNAs directed against each target, and this activity was further reduced when the expression of both proteins was knocked down simultaneously (Figure 2A). Additionally, migration of MDA-MB 231 and PC-3 cells was not significantly affected by silencing of SGK1 or ATG5, a key component of autophagy with no known function in migration (Figure S3). No significant differences in cell viability were observed in cells treated with the selected siRNAs (data not shown) under the conditions used for the migration assays. The efficacy of each of the siRNAs in reducing the expression of their respective targets was confirmed by western immunoblot analysis (Figure S4A). Furthermore, treatment with the inverse agonist XCT790 resulted in a dose dependent degradation of ERRα and an inhibition of MDA-MB 231, MDA-MB 436, PC-3, and HCT-116 cell migration (Figure 2B, Figure S4B). with no significant changes in cell viability (data not shown). Taken together, these data indicate that both ERRα and β-cat can promote the migratory capacity of several types of cancer cells.
Figure 2.
ERRα and β-cat promote cancer cell migration. A, Silencing of ERRα and/orβ-cat reduced MDA-MB 231, MDA-MB 436, PC-3 & HCT-116 migration. Cells were transfected with two different sequences of siRNAs (A & B) for ERRα, β-cat, scramble (siC) and serum starved for 18h followed by assessment of migratory capacity and viability (data not shown). Mock (transfection reagent), siβ-lactamase, and siATG5 treated cells exhibited similar migration to siC treated cells (Figure S7). B, ERRα degradation by XCT790 impedes MDA-MB 231, MDA-MB 436, PC-3, and HCT-116 migration. Cells were treated with XCT790 (0, 2.5, 5, 10 μM) for 30h and then serum starved with continued drug treatment for another 18h. Migration and cell viability (data not shown) were performed as in panel A. Different letters denote significance (P<0.05).
ERRα regulates the expression of target genes encoding proteins with pro-migratory activities
In light of the crosstalk between the ERRα and β-cat signaling pathways observed, we next asked whether these proteins cooperated in the regulation of genes involved in the migratory response. In previously published work, we utilized classical microarray analysis to identify ERRα target genes that were expressed in HepG2 and MCF-7 cells [6, 13, 26]. Likewise, there are published studies that describe the β-catenin transcriptome in 293T cells [26]. With these datasets in hand, we were able to perform a comparative analysis and identify genes that (a) were regulated in both datasets and (b) were important for cell migration as demonstrated in previous studies [26–31]. In this manner, WNT11, MSX1, and N-cadherin were identified as genes that are likely to be coregulated by ERRα and β-cat.
The expression of WNT11, MSX1, and N-cadherin was next evaluated in MDA-MB 231 cells following the activation of ERRα. While the transcriptional activity of ERRα is regulated by the relative expression and/or activity of cofactors such as PGC-1α and PGC-1β [19–20], other NRs, including PPARγ and HNF-4, can also be coactivated by PGC-1α, which makes it difficult to study the ERRα signaling axis in isolation using this cofactor as an activator [19, 32]. To circumvent this problem, we developed a variant of PGC-1α (PGC-1α 2×9) that interacts in a highly selective manner with ERRα and thus can be used to specifically regulate the activity of this receptor [6]. In addition, when analyzing ERRα target gene expression using PGC-1α as an activator, we also evaluated the impact of an inactive variant of PGC-1α (PGC-1α L2L3M) in parallel.
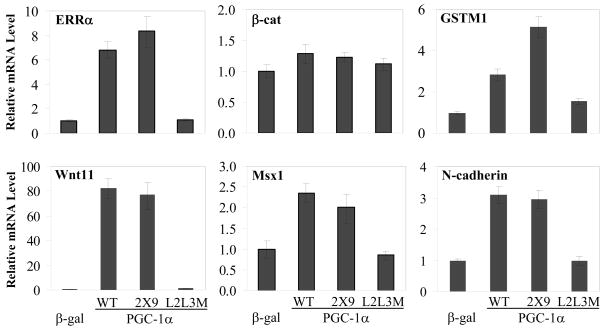
MDA-MB 231 cells were transduced with adenoviruses expressing βgal, PGC-1α, PGC-1α 2×9, or PGC-1α L2L3M, and quantitative PCR was used to assess the resulting changes in mRNA expression of target genes (Figure 3). It was determined that ERRα mRNA levels are increased upon overexpression of PGC-1α (WT or 2×9), consistent with it being an auto-regulated gene [33]. Similarly, GSTM1, an ERRα target gene [12], was also regulated by PGC-1α, whereas β-cat mRNA levels were not significantly affected (Figure 3). Interestingly, WNT11, MSX1, and N-cadherin were all significantly induced by the expression of PGC-1α in MDA-MB 231 cells (Figure 3). Similar results were observed in MDA-MB 436, PC-3, and HCT-116 cells (Figure S5).
Figure 3.
Novel ERRα target genes involved in migration. ERRα activation by PGC-1α and PGC-1α 2X9 induces the expression of WNT11, MSX1, and N-cadherin mRNAs in MDA-MB 231 cells. Cells were infected with adenoviruses expressing β-gal, PGC-1α, PGC-1α 2X9, or PGC-1α L2L3M, followed by qPCR analysis of mRNA levels normalized to 36B4 expression and relative to β-gal.
Coregulation of WNT11, MSX1, and N-cadherin by ERRα and β-cat
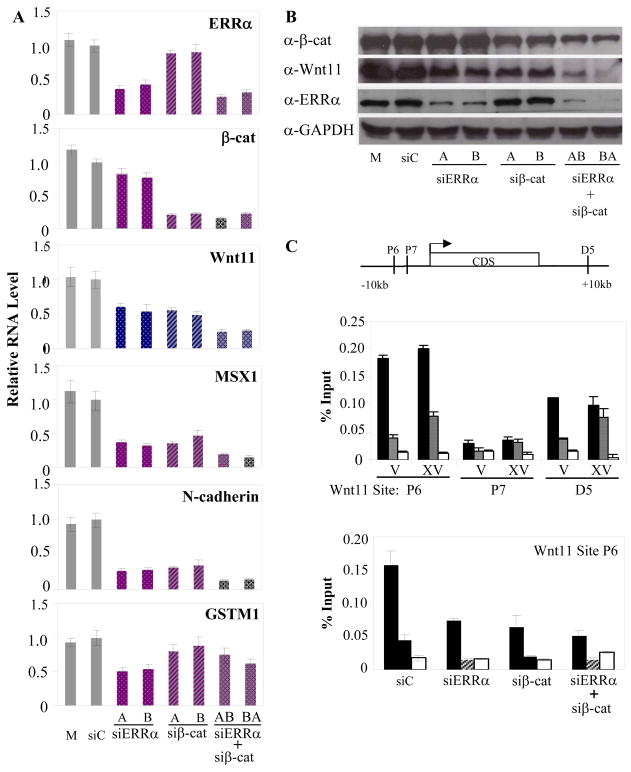
The relative importance of ERRα and β-cat in the regulation of WNT11, MSX1, and N-cadherin was next evaluated. Specifically, the expression of each mRNA and appropriate controls were measured in cells following the introduction of siRNAs directed against ERRα or β-cat. Using two different siRNAs against each target, we observed that silencing of either ERRα or β-cat led to a significant diminution of WNT11, MSX1, and N-cadherin mRNA expression. Simultaneous knockdown of both mRNAs in MDA-MB 231 cells resulted in a further decrease in the expression level of these mRNAs (Figure 4A). Importantly, silencing of β-cat expression did not affect the mRNA level of GSTM1 (Figure 4A), suggesting that not all ERRα target genes are regulated by β-cat (see discussion). Notably, ERRα and β-cat downregulation, either singly or in combination, resulted in a reduction of WNT11 protein levels (Figure 4B). Given the extremely robust regulation of WNT11 mRNA expression by ERRα and β-cat, and its previously described activity as a pro-migratory factor [28–30], we focused the remainder of the studies on defining the mechanism by which this gene is regulated and how it impacts cancer cell biology.
Figure 4.
ERRα and β-cat regulate genes involved in cell migration. A, ERRα and β-cat were silenced individually or in combination using siRNA in MDA-MB 231 cells. Expression of the indicated genes was measured by qPCR and normalized to 36B4 expression and relative to siC. Mock (transfection reagent alone) and two siRNA sequences for ERRα and β-cat were used. B, Western blot analysis of siRNA treated MDA-MB 231 WCE for ERRα, β-cat, and WNT11 to confirm knockdown with GAPDH as loading control. C, ChIP of ERRα and β-cat to WNT11 genomic sequence in MDA-MB 231 cells. Diagram of putative TCF-4 REs and ERREs in WNT11 genomic sequence. ERRα and β-cat recruitment was tested by qPCR. Inhibition of GSK-3β kinase by XV increased β-cat levels and enhanced recruitment of β-cat to both the WNT11 P6 and D5. Downregulation of ERRα and/or β-cat reduces recruitment to the putative ERRα and TCF-4 P6 site. Black bars, ERRα antibody; Striped bars, β-cat antibody; White bars, IgG.
The next step in these studies was to define the mechanism(s) by which ERRα and β-cat regulate WNT11 expression. To this end, we scanned the genomic sequence surrounding the WNT11 gene using Consite for putative TCF and ERRα response elements (Figure S6A). Chromatin immunoprecipitation (ChIP) assays in MDA-MB 231 cells were then used to confirm the functionality of the putative ERRα and/or TCF binding sites. In this manner, significant binding of both ERRα and β-cat was detected at the following two sites: P6 (−3042) and D5 (+27794) (Figure 4C, S6B). Importantly, siRNA-mediated knockdown of ERRα expression significantly reduced β-cat binding at the P6 site and knockdown of β-cat reduced ERRα binding (Figure 4C). These ChIP data provide a molecular explanation for the observed crosstalk that occurs between ERRα and β-cat on the WNT11 gene.
WNT11 acts in an autocrine manner to promote cancer cell migration
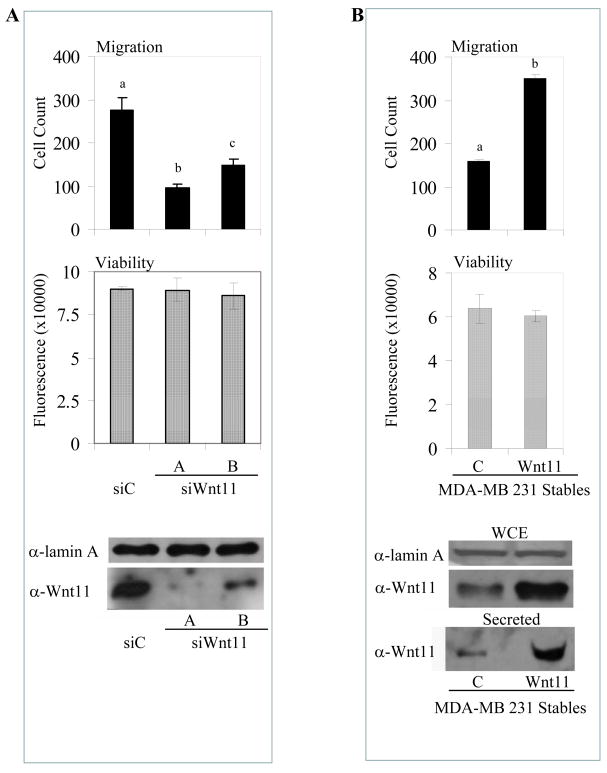
Previous studies have highlighted the pro-migratory activity of WNT11 in biological contexts, including migration of intestinal epithelial cells, neural crest cells, and gastrulation [28–30]; however, little has been done to define the role of WNT11 in cellular models of cancer. To address this issue, we used siRNAs to knock down WNT11 expression in MDA-MB 231 cells and used transwell migration assays to define the impact of this manipulation on cell migration. As shown in Figure 5A, using either of two different siRNAs, we were able to achieve a quantitative knockdown of WNT11 protein expression, a manipulation that resulted in an ~60% reduction in cell migration. We did not observe any effect of WNT11 knockdown on total cell number and/or cell viability under the same experimental conditions (Figure 5A). This effect was not restricted to MDA-231 cells, as we observed that silencing of WNT11 expression also decreased migration of MDA-MB 436, PC-3, and HCT-116 cells when assayed in the same manner (Figure S7). This overexpression data provides strong support that WNT11 is promoting migration, as this could not be a survival effect. To complement these experiments, we created WNT11 overexpressing derivatives of MDA-MB 231 cells and determined that the increased production in both total and secreted WNT11 protein correlated with a significant increase in cell migration (Figure 5B). Together, these experiments suggest that the ERRα/β-cat-dependent target WNT11 acts in an autocrine manner to increase the migration of cancer cells.
Figure 5.
WNT11 promotes MDA-MB 231 migration. A, WNT11 downregulation reduces MDA-MB 231 migration. WNT11 or control (siC) siRNA transfections and migration assays were performed as in Figure 2. B, WNT11 overexpression enhances MDA-MB 231 migration. Stable populations of MDA-MB 231 cells overexpressing WNT11 or empty vector (C) were generated and characterized for migration, viability, and protein expression as previously described. Enhanced secretion of WNT11 was observed by Western blot analysis from MDA-MB 231 cells overexpressing WNT11. Different letters denote significance (P<0.05).
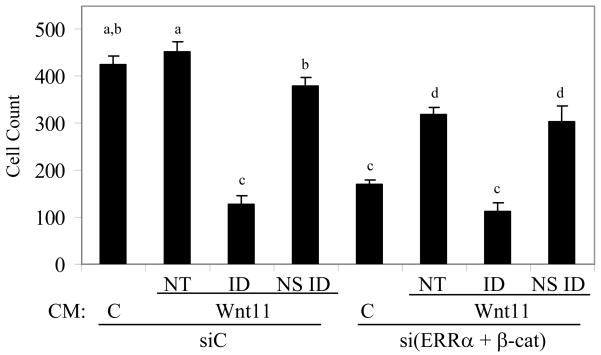
Given that WNT11 is a secreted protein that functions by binding to the extracellular Frizzled Receptor 7 [30], we also tested whether exogenously added WNT11 could restore the reduced migratory phenotype resulting from the silencing of ERRα and β-cat in the MDA-MB 231 parent cell line. For these experiments, we isolated conditioned media (CM) from MDA-MB 231 cells (containing vector alone) or from cells that were engineered to overexpress WNT11. Subsequently, CM was added to the lower chamber of a transwell plate and MDA-MB 231 cells treated with control siRNA (siC) or siERRα and siβ-cat were added to the upper chamber. In cells treated with siC, addition of conditioned media from WNT11 overexpressing cells did not significantly increase cell migration, probably due to the sufficiently high concentration of WNT11 already in the media in control cells. However, immunodepletion of WNT11 from WNT11 overexpressing CM significantly reduced the migration of these cells, indicating that WNT11 is a pro-migratory factor (Figure 6). Importantly, conditioned media from WNT11 overexpressing cells partially reversed the decreased migration resulting from knockdown of ERRα and β-cat. Furthermore, immunodepletion of WNT11 from this conditioned media using a specific antibody, but not with an irrelevant antibody, blocked this restorative capacity. Similar results were obtained using WNT11 and empty vector CM from mouse L cell fibroblasts (Figure S8). Together, these data provide compelling evidence for a regulatory loop involving ERRα,β-cat and WNT11 that influences the migratory capacity in this model of breast cancer.
Figure 6.
WNT11 partially restores reduced MDA-MB 231 migration by silencing of ERRα and β-cat. MDA-MB 231 cells were transfected with siC or siERRα and β-cat and serum starved as previously described. Migration assays were then performed with conditioned media from the MDA-MB 231 derivative cell lines overexpressing WNT11 or empty vector. Immunodepletion of WNT11 from conditioned media reduced migration as compared to mock immunodepletion with rabbit IgG. Different letters denote significance (P<0.05). NT, no treatment; ID, WNT11 immunodepleted; NSID, non-specific immunodepleted
Discussion
Crosstalk between nuclear receptors and the Wnt/β-cat signaling pathway has been demonstrated previously [34]. In the realm of metabolism, it has been shown that crosstalk between PPARγ and Wnt/β-cat signaling enables precise control of adipogenesis resulting from a reciprocal inhibition of each other’s transcriptional activities [35]. It has been shown that β-cat serves as an androgen receptor (AR) coactivator when assessed on endogenous AR target genes such as PSA [36]. Crosstalk between the Wnt pathway and ERα, LRH-1, RXR, and several other NRs has also been shown to occur in a variety of cell-based assays [34, 37]. In this study, we provide compelling evidence that ERRα and β-cat are involved in the regulation of WNT11, MSX1, and N-cadherin expression, genes implicated previously in processes that regulate cell migration. Given the wealth of literature linking WNT11 to cell migration, we focused on evaluating the functional consequences of ERRα/β-cat-mediated regulation of WNT11 [28–30]. In this manner, we were able to determine that WNT11 is important for migration in breast cancer cells and its expression is influenced by both ERRα and β-cat.
Wnt signaling occurs via a canonical or a non-canonical pathway, both of which may interface with ERRα/β-cat [38–39]. In the canonical pathway, Wnt activation of Fzd results in disheveled (dsh)-mediated degradation of axin and GSK-3β inhibition. Inhibition of this destruction complex leads to an increase in the intracellular pool of βcat. The stabilized β-cat has two non-exclusive fates within the cell. Firstly, the protein can translocate to the nucleus, where it regulates target gene expression through its interaction with the T-cell factor/lymphoid enhancer-binding factor family of transcription factors. Alternatively, β-cat can interact with E-cadherin in adherens junctions and stabilize cell-cell interactions. Furthermore, β-cat expression promotes epithelial to mesenchymal transition (EMT), a process that is frequently associated with a loss of E-cadherin. Importantly, it has been shown that inhibition of E-cadherin expression increases nuclear β-cat, resulting in the increased expression of pro-migratory genes [40]. Thus, depending on the relative partitioning of β-cat, it can have either a positive or a negative impact on cell migration. We have established that the ERRα, a karyophilic protein, associates directly with β-cat; thus, it is possible that in addition to cooperating with β-cat in the transcriptional regulation of pro-migratory genes, ERRα may impact migration by regulating the cellular partitioning of β-cat. In many breast cancer cells, including the MDA-MB 231 cells studied herein, E-cadherin expression is extremely low, and thus it is likely that the effects we have observed on migration are a direct consequence of the nuclear action of the ERRα/β-cat complex [41]. Defining the processes that impinge upon and regulate the nuclear versus cytoplasmic actions of the ERRα/β-cat is a focus of our continued efforts in this area.
In this study, we have performed a comprehensive analysis of the mechanisms underlying the ERRα/β-cat dependent regulation of WNT11. Whereas we can show that both of these proteins interact in cells and can bind to the same region in the WNT11 gene, we do not know if they bind to adjacent sites or if a tethering mechanism is involved. Given that other nuclear receptors use both mechanisms to engage target genes, it is likely that both types of interaction, direct and tethering, are utilized. To address this issue, we have performed a preliminary examination of available ChIP-ChIP and ChIP-seq datasets to evaluate the extent to which the binding sites described for ERRα and β-cat converge. In this manner, we found a small but significant overlap in target genes that were found to be enriched for ERRα binding in MCF-7 or SKBR3 breast cancer cells by ChIP-ChIP analysis and those that were found to be enriched for β-catenin binding in HCT116 colon cancer cells by ChIP-seq analysis (Figure S9A) [12, 42]. Furthermore, out of the 547 unique ERRα-regulated genes detected in our previously published MCF-7 microarray study, 39 of these genes were found to contain at least one β-catenin enriched region in the HCT116 ChIP-seq experiment (Figure S9B) [13, 42]. Finally, bioinformatic analyses using Patser revealed that out of the 988 β-catenin target genes identified in the HCT116 ChIP-seq experiment, approximately 17% of the genes also contain at least one putative ERR binding site within the same 600-bp region of DNA that is enriched for β-catenin binding (Figure S9C) [42–44]. The significance and functionality of the convergent sites identified in this manner are currently under investigation.
The identification of WNT11 as a direct transcriptional target of the ERRα/β-cat complex was of particular interest to us as (1) the expression of this protein has previously been associated with increased cell migration [28–30, 45], (2) WNT11 is upregulated in several cancers, including colorectal, prostate, and breast cancer [46–47], and (3) WNT11 induces transformation of mammary epithelial cells [48]. WNT11, initially identified as a non-canonical Wnt, can activate the non-canonical Wnt/Ca2+ pathway, resulting in G-protein-dependent increases in intracellular calcium and subsequent activation of CAMKII (calcium/calmodulin-dependent protein kinase II) and PKC (protein kinase C) [28]. This activity has been shown to increase intestinal epithelial cellular migration [28–30, 46, 48]. Interestingly, both CAMKII and PKC facilitate actin cytoskeleton rearrangements that are critical for cellular migration [49]. Furthermore, WNT11 has been shown to also activate the canonical Wnt pathway [50]. The fact that non-canonical pathway activation of PKC can result in decreased E-cadherin function further underscores the significance of crosstalk between canonical and non-canonical Wnt signaling and the likely importance of the ERRα/β-cat complex in this process.
Taken together, our findings provide evidence for an autocrine regulatory loop involving transcriptional upregulation of WNT11 by ERRα and β-cat, an activity that influences the migratory capacity of cancer cells. Furthermore, these data provide a strong rationale for the development of compounds that inhibit ERRα or the activity of the ERRα/β-cat complex as cancer therapeutics.
Supplementary Material
Acknowledgments
This research was supported by NIH grant DK074652.
We thank Dr. V. Giguere (McGill University) for the ERRα antibody, Dr. B. Hogan (Duke University) for plasmids, and the members of the McDonnell laboratory for insightful discussions.
Footnotes
The authors have nothing to disclose.
References
- 1.Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62(22):6510–8. [PubMed] [Google Scholar]
- 2.Fujimoto J, et al. Clinical implication of estrogen-related receptor (ERR) expression in ovarian cancers. J Steroid Biochem Mol Biol. 2007;104(3–5):301–4. doi: 10.1016/j.jsbmb.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki T, et al. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004;64(13):4670–6. doi: 10.1158/0008-5472.CAN-04-0250. [DOI] [PubMed] [Google Scholar]
- 4.Cartoni R, et al. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. J Physiol. 2005;567(Pt 1):349–58. doi: 10.1113/jphysiol.2005.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreiber SN, et al. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J Biol Chem. 2003;278(11):9013–8. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 6.Gaillard S, et al. Receptor-selective coactivators as tools to define the biology of specific receptor-coactivator pairs. Mol Cell. 2006;24(5):797–803. doi: 10.1016/j.molcel.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 7.Mootha VK, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci U S A. 2004;101(17):6570–5. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreiber SN, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101(17):6472–7. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villena JA, Kralli A. ERRalpha: a metabolic function for the oldest orphan. Trends Endocrinol Metab. 2008;19(8):269–76. doi: 10.1016/j.tem.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29(6):677–96. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 11.Bonnelye E, Aubin JE. Estrogen receptor-related receptor alpha: a mediator of estrogen response in bone. J Clin Endocrinol Metab. 2005;90(5):3115–21. doi: 10.1210/jc.2004-2168. [DOI] [PubMed] [Google Scholar]
- 12.Deblois G, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor alpha as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69(15):6149–57. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- 13.Stein RA, et al. Estrogen-related receptor alpha is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res. 2008;68(21):8805–12. doi: 10.1158/0008-5472.CAN-08-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McEwan IJ. Nuclear receptors: one big family. Methods Mol Biol. 2009;505:3–18. doi: 10.1007/978-1-60327-575-0_1. [DOI] [PubMed] [Google Scholar]
- 15.Kallen J, et al. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (ERRalpha): crystal structure of ERRalpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. J Biol Chem. 2004;279(47):49330–7. doi: 10.1074/jbc.M407999200. [DOI] [PubMed] [Google Scholar]
- 16.Greschik H, et al. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol Cell. 2002;9(2):303–13. doi: 10.1016/s1097-2765(02)00444-6. [DOI] [PubMed] [Google Scholar]
- 17.Huss JM, et al. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol Cell Biol. 2004;24(20):9079–91. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamei Y, et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci U S A. 2003;100(21):12378–83. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon JC, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413(6852):131–8. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1beta coactivation of SREBP. Cell. 2005;120(2):261–73. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 21.Gaillard S, Dwyer MA, McDonnell DP. Definition of the molecular basis for estrogen receptor-related receptor-alpha-cofactor interactions. Mol Endocrinol. 2007;21(1):62–76. doi: 10.1210/me.2006-0179. [DOI] [PubMed] [Google Scholar]
- 22.Norris J, et al. Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J Biol Chem. 1995;270(39):22777–82. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- 23.Waki H, et al. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARgamma expression. Cell Metab. 2007;5(5):357–70. doi: 10.1016/j.cmet.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Demir R, et al. Malignant progression of invasive tumour cells seen in hypoxia present an accumulation of beta-catenin in the nucleus at the tumour front. Exp Mol Pathol. 2009;87(2):109–16. doi: 10.1016/j.yexmp.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Bitler BG, et al. Intracellular MUC1 peptides inhibit cancer progression. Clin Cancer Res. 2009;15(1):100–9. doi: 10.1158/1078-0432.CCR-08-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chamorro MN, et al. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J. 2005;24(1):73–84. doi: 10.1038/sj.emboj.7600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishii M, et al. Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development. 2005;132(22):4937–50. doi: 10.1242/dev.02072. [DOI] [PubMed] [Google Scholar]
- 28.Ouko L, et al. WNT11 signaling promotes proliferation, transformation, and migration of IEC6 intestinal epithelial cells. J Biol Chem. 2004;279(25):26707–15. doi: 10.1074/jbc.M402877200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulrich F, et al. Slb/WNT11 controls hypoblast cell migration and morphogenesis at the onset of zebrafish gastrulation. Development. 2003;130(22):5375–84. doi: 10.1242/dev.00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witzel S, et al. WNT11 controls cell contact persistence by local accumulation of Frizzled 7 at the plasma membrane. J Cell Biol. 2006;175(5):791–802. doi: 10.1083/jcb.200606017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suyama K, et al. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell. 2002;2(4):301–14. doi: 10.1016/s1535-6108(02)00150-2. [DOI] [PubMed] [Google Scholar]
- 32.Rhee J, et al. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A. 2003;100(7):4012–7. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laganiere J, et al. A polymorphic autoregulatory hormone response element in the human estrogen-related receptor alpha (ERRalpha) promoter dictates peroxisome proliferator-activated receptor gamma coactivator-1alpha control of ERRalpha expression. J Biol Chem. 2004;279(18):18504–10. doi: 10.1074/jbc.M313543200. [DOI] [PubMed] [Google Scholar]
- 34.Mulholland DJ, et al. Interaction of nuclear receptors with the Wnt/beta-catenin/Tcf signaling axis: Wnt you like to know? Endocr Rev. 2005;26(7):898–915. doi: 10.1210/er.2003-0034. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Farmer SR. Regulating the balance between peroxisome proliferator-activated receptor gamma and beta-catenin signaling during adipogenesis. A glycogen synthase kinase 3beta phosphorylation-defective mutant of beta-catenin inhibits expression of a subset of adipogenic genes. J Biol Chem. 2004;279(43):45020–7. doi: 10.1074/jbc.M407050200. [DOI] [PubMed] [Google Scholar]
- 36.Taplin ME, et al. Androgen receptor mutations in androgen-independent prostate cancer: Cancer and Leukemia Group B Study 9663. J Clin Oncol. 2003;21(14):2673–8. doi: 10.1200/JCO.2003.11.102. [DOI] [PubMed] [Google Scholar]
- 37.Botrugno OA, et al. Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol Cell. 2004;15(4):499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 39.Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5(3):367–77. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 40.Conacci-Sorrell M, et al. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163(4):847–57. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenny PA, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1(1):84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bottomly D, et al. Identification of {beta}-catenin binding regions in colon cancer cells using ChIP-Seq. Nucleic Acids Res. doi: 10.1093/nar/gkq363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15(7–8):563–77. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 44.Sladek R, Bader JA, Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol. 1997;17(9):5400–9. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uysal-Onganer P, et al. Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells. Mol Cancer. 2010;9:55. doi: 10.1186/1476-4598-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirikoshi H, Sekihara H, Katoh M. Molecular cloning and characterization of human WNT11. Int J Mol Med. 2001;8(6):651–6. doi: 10.3892/ijmm.8.6.651. [DOI] [PubMed] [Google Scholar]
- 47.Zhu H, et al. Analysis of Wnt gene expression in prostate cancer: mutual inhibition by WNT11 and the androgen receptor. Cancer Res. 2004;64(21):7918–26. doi: 10.1158/0008-5472.CAN-04-2704. [DOI] [PubMed] [Google Scholar]
- 48.Christiansen JH, Monkley SJ, Wainwright BJ. Murine WNT11 is a secreted glycoprotein that morphologically transforms mammary epithelial cells. Oncogene. 1996;12(12):2705–11. [PubMed] [Google Scholar]
- 49.Szalay J, et al. Associations of PKC isoforms with the cytoskeleton of B16F10 melanoma cells. J Histochem Cytochem. 2001;49(1):49–66. doi: 10.1177/002215540104900106. [DOI] [PubMed] [Google Scholar]
- 50.Tao Q, et al. Maternal WNT11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120(6):857–71. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.