Abstract
Chromatin coregulators are important players in tumorigenesis and cancer progression. ANCCA is an AAA+ ATPase-containing nuclear coactivator for the estrogen and androgen receptors that is crucial for assembly of chromatin modifying complexes and proliferation of hormone-responsive cancer cells. In this study, we show that ANCCA is overexpressed in > 70% of breast tumors and that its high protein level correlates well with tumor histologic grades (P < 0.0001), highlighting ANCCA as a prognostic factor for poor overall survival and disease recurrence. Strikingly, high level ANCCA correlated with triple-negative tumors that represent highly aggressive disease. Analysis of ANCCA transcript levels in multiple expression profiles of breast cancer identified ANCCA as a common signature gene, indicating that elevated transcripts also strongly correlate with tumor metastasis and poor survival. Biological and mechanistic investigations revealed that ANCCA is crucial for proliferation and survival of triple negative/basal-like cancer cells and that it controls expression of B-Myb, histone methyltransferase EZH2 and an Rb-E2F core program for proliferation, along with a subset of key mitotic kinesins and cell survival genes (IRS2, VEGF and Akt1). In particular, ANCCA overexpression correlated strongly with EZH2 in tumors. Our results suggest that ANCCA may integrate multiple oncogenic programs in breast cancer, serving in particular as a prognostic marker and therapeutic target for triple negative cancers.
Introduction
Functional genomics and gene profiling studies of breast cancer continue to improve our prediction of clinical outcome and selection of therapeutics as well as our understanding of tumor biology, by sub-typing the tumors on their molecular profiles. Among the subtypes, the luminal/estrogen receptorα positive (ER+) tumors present themselves as more differentiated, less aggressive, and highly responsive to endocrine therapy. Tumors overexpressing human epidermal growth factor receptor 2 (HER2), although often more aggressive, are responsive to therapeutics targeting the growth factor receptors. The triple-negative (ER, progesterone receptor, and HER2-negative) breast cancers or TNBCs, however, remain a major challenge for the development of effective therapeutics and identification of risk factors (1). Recent expression array studies (2, 3) characterize most TNBCs as being basal-like due to an expression profile containing high levels of basal cell cytokeratins, similar to those of myoepithelial cells. Although poorly understood, one striking feature of TNBCs is the high expression of proliferation signature genes. Unlike ER-positive or HER2-high subtypes, no molecular markers have been defined that underpin TNBC development or effectively guide their clinical treatment. As a result, conventional chemotherapy remains the mainstay for them. Therefore, there is an urgent need for the identification of risk factors and new treatment options for triple negative tumors.
Chromatin coregulators, particularly in the forms of histone modifying and chromatin remodeling enzymes, have recently emerged as important players in tumorigenesis (4–6). We recently identified a previously uncharacterized gene product dubbed ANCCA (for AAA+ nuclear coregulator cancer associated) as both a direct target and an activator of the proto-oncogene AIB1 (also known as ACTR and SRC-3) (7, 8). ANCCA, a novel member of the AAA+ ATPase proteins, also possesses a bromodomain. We found that high levels of ANCCA are expressed in breast cancer and prostate cancer cells and that RNAi-mediated knockdown of ANCCA strongly inhibits hormone-dependent cancer cell proliferation (8, 9). We also demonstrated that ANCCA acts as a coactivator of ERα and androgen receptor (AR) to mediate estrogen- or androgen-induced expression of specific subsets of genes involved in proliferation and survival of cancer cells. Both the AAA+ ATPase and bromodomains are required for ANCCA to serve as a transcriptional coregulator of the receptors (8). Our further study indicated that one major mechanism of ANCCA function is to facilitate the assembly of a histone modifying protein complex at the chromatin (8).
In this study, we investigated ANCCA expression at both protein and transcript levels in multiple sets of human breast cancer specimens and found that ANCCA is overexpressed in the majority of the tumors. High levels of ANCCA directly correlate with poor survival and disease recurrence in the patients. We also present results that high ANCCA is strongly associated with triple-negative tumors and that aberrant ANCCA expression in TNBC cells controls multiple oncogenic pathways for cancer cell proliferation and survival.
Materials and Methods
Cell culture, siRNA transfection, ChIP assay, RNA analysis and Western blotting
Human breast cancer cell lines were obtained from ATCC in 2002 and 2008. They were tested recently (in 2009–2010) for authenticity by monitoring the cell morphology using microscope, growth rate using cell counting, response to factors or hormones for growth, expression of genes/proteins including the hormone receptors and members of the HER2/ErbB2 family using Western and RT-PCR, and were confirmed to be in line with the ATCC descriptions and with the literature. HMECs were from Lonza/Clonetics. All the cells were used within 4 months after initial receipt or thawing. RNA analysis and Western blotting were performed as previously described (10). For transfection of siRNAs, cells plated at 2.0 × 105 cells per well in six-well plates were transfected using Dharmafect 1 (Dharmacon) according to the manufacturer’s instructions. siRNA targeting ANCCA or control siRNA were used at 100nM concentration. Details of cell lines and their culture conditions, ChIP, antibodies used for Western blotting, and sequences for PCR primers are described in Supplementary Materials & Methods.
Cell proliferation and soft-agar colony formation assays
HCC1937 cells were seeded at density of 1.5 × 105 cells per well of a six-well plate and maintained in full growth medium for 24 hrs before being infected with equal titers of adeno-ANCCA-HA or adeno-vector adenoviruses (8). MDA-MB 468 cells were plated at a density of 2.0 × 105 cells per well in six well plates, and were transfected with siRNA 24 hours later as described above. Medium was changed every other day after transfection and cell proliferation was measured by cell counting of coded samples in triplicates. For colony formation in soft agar, five thousand cells were seeded in each well of six-well plates in medium containing 0.4% SeaPlaqueR Agarose (Lonza) on top of base medium containing 0.8% agarose. Colonies were stained 4 to 5 weeks later with MTT (Sigma Aldrich) and counted using a light microscope.
TUNEL assay
Cells were plated and transfected with siRNA 24 hours later as described above. Cells were plated 72 hrs later on glass chamber slides (Nunc) and processed for TUNEL assay using the In Situ Cell Death Detection kit (Roche Diagnostics) according to manufacturer’s protocol. Slides were mounted with VECTASHIELD Mounting Media (Vector Laboratories). Images were acquired using Olympus BX6. For quantification of TUNEL-positive cells, 10 random fields of the same condition were counted and averaged.
Tumor specimens and immunohistochemistry
Archival paraffin-embedded primary tumors samples were from 225 women diagnosed during 1998–2004 with breast carcinoma and treated at UC Davis Medical Center. Other information on the patients and tumors are in Supplementary Table 1. Five μm sections of the tumor blocks were first subjected to de-paraffinization and then antigen retrieval in 0.01mM sodium citrate buffer (pH 6.0) in a microwave oven at 1000W for 5 minutes and then at 100w for 30 min. Breast tumor tissue microarray or TMAs (BR1002, BR961, and BR208) were obtained from US BioMax, Inc, baked at 60°C for 2 hrs and processed for immunostaining. TMA for EZH2 and ANCCA correlation study has been described (7). Nonspecific immunoglobulin binding was blocked using 10% fetal bovine serum in PBS for 30 min at room temperature. Slides were then incubated at room temperature for 30 min with anti-ANCCA antibody (at 1:300), anti-Ki-67 (NeoMarker, at 1:300), and anti-EZH2 (AC22, Cell Signaling, at 1:50). Anti-ANCCA antibody was raised in rabbit (Covance) and affinity-purified by using GST-ANCCA N-terminus (aa 2-264) expressed and purified from E. coli. Its specificity for IHC was determined using a panel of cell lines and xenograft tumors (9)and is also shown in Supplementary Fig. 1. After incubation with primary antibody, the sections were washed and incubated with biotin-conjugated secondary antibodies for 30 min followed by incubation with avidin DH-biotinylated horseradish peroxidase and developed using diaminobenzidine (DAB) substrate kit (Vector Laboratories) and counterstained using Gill’s Hematoxylin. Images were acquired using an Olympus microscope with DPController software. The percentage of positively stained nuclei was scored as follows: 0–10%, score 0; 11–25%, score 1; 26–50%, score 2; >50%, score 3. The immunoreactivity was evaluated by at least two different investigators with no prior knowledge of patient data.
Statistical analyses and analysis of microarray gene expression data sets
Values of patient age and tumor size are presented as mean ± SE. Association between ANCCA immunoreactivity and other clinico-pathological parameters was evaluated using Pearson χ2 test. Survival curves were generated using the Kaplan-Meier method taking into account censored data. The curves were compared using the log-rank test (Mantel-Cox). For other assays and analysis of gene transcripts in tumor data sets using Oncomine (www.oncomine.org), Student’s (or paired) t-test was used for comparison of experimental groups. Statistic analysis was performed using the SPSS software (version 18, SPSS Inc.). P values of less then 0.05 were considered significant. The method for analysis of microarray gene expression data sets is provided in the Supplementary Methods.
Results
ANCCA, a “signature” gene, is overexpressed in greater than 70% of human breast carcinomas and its overexpression is associated with triple-negative status
To investigate ANCCA expression in breast cancer, we first performed immunohistochemical (IHC) analysis of ANCCA protein expression in a cohort of 225 primary human breast ductal carcinomas and three independent sets of TMAs containing a total of 131 tumor samples and 24 normal breast tissues. Immuno-reactivity for ANCCA was readily detected in the nuclei of a large subset of tumor tissues while little or no staining was observed in the normal breast tissues or tumor-adjacent stroma (Fig. 1 and Supplementary Fig. 2). When compared to normal breast tissue, over 70 % of all tumor samples examined showed increased levels of ANCCA protein expression (Table 1 and Supplementary Table 2). Interestingly, although a large proportion (63%) of ERα-positive tumors displayed ANCCA overexpression, a much stronger association for its overexpression in ERα- and PR-negative tumors was observed (Table 1). More importantly, high levels of ANCCA associate significantly (P = 0.0071) with ERα-, PR- and HER2-negative status, as over 88% of all triple-negative samples showed high expression of ANCCA protein. Moreover, compared to normal human breast epithelial cells (HMECs), elevated levels of ANCCA proteins (~170 kD) was observed in all of the breast cancer cell lines examined (Fig. 2A and Supplementary Fig. 1).
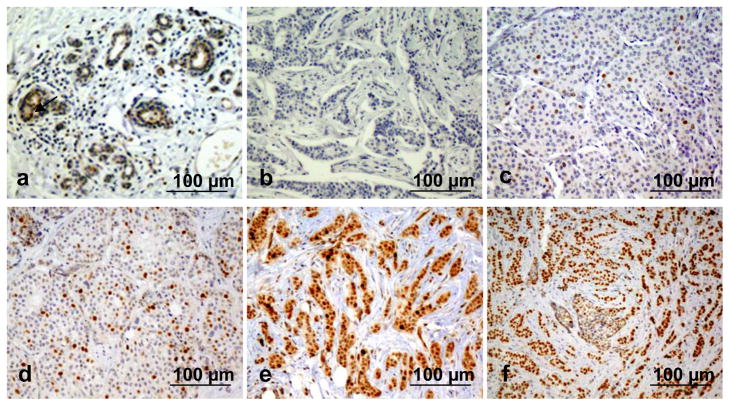
Figure 1.
Immunohistochemistry (IHC) analysis of ANCCA expression in normal or cancerous human breast tissues. Representative images are shown for IHC score 0 with less than 1% of nuclei stained positive, in a histologically normal breast tissue (a) and in a tumor (b); for score 1 with < 25% of positively stained nuclei in a tumor (c); for score 2 with 25–50% of positively stained nuclei a tumor (d); and for score 3 with >50% of positively stained nuclei in an ER-positive tumor (e) and an ER-negative tumor (f).
Table 1.
Tumor characteristics of 200 cases of primary breast carcinoma analyzed by IHC
| ANCCA score | |||
|---|---|---|---|
| 0–1 | 2–3 | P* | |
| N (%) | N (%) | ||
| BC all grade | 49 (22) | 176 (78) | <0.0001 |
| Grade | |||
| I | 13 (45) | 16 (55) | |
| II | 20 (27) | 54 (73) | |
| III | 12 (16) | 63 (84) | <0.0001 |
| Lymph node status | |||
| Positive | 12 (26) | 34 (74) | |
| Negative | 14 (20) | 57 (80) | 0.4 |
| Hormone Receptor status | |||
| ER - positive | 31 (37) | 53 (63) | |
| ER - negative | 16 (14) | 100 (86) | 0.0004 |
| PR - positive | 25 (35) | 46 (65) | |
| PR - negative | 17 (13) | 112 (87) | 0.0005 |
| HER2 status | |||
| Positive | 8 (20) | 31 (80) | |
| Negative | 39 (26) | 112 (74) | 0.63 |
| TN | 8 (11.5) | 62 (88.5) | |
| NTN (all grades) | 41 (31) | 91 (69) | |
| NTN (high grade) | 27 (31.4) | 59 (68.6) | 0.0071 |
| Ki67 (TN) High | 5 (7.5) | 47 (70) | |
| Low | 3 (4.5) | 12 (18) | <0.0001 |
IHC results are presented as frequency with the percentage of cases in parenthesis.
Pearson’s χ2 test, for association with grades and triple negative breast cancer was used.
TN, triple negative tumors; NTN, non-triple negative tumors.
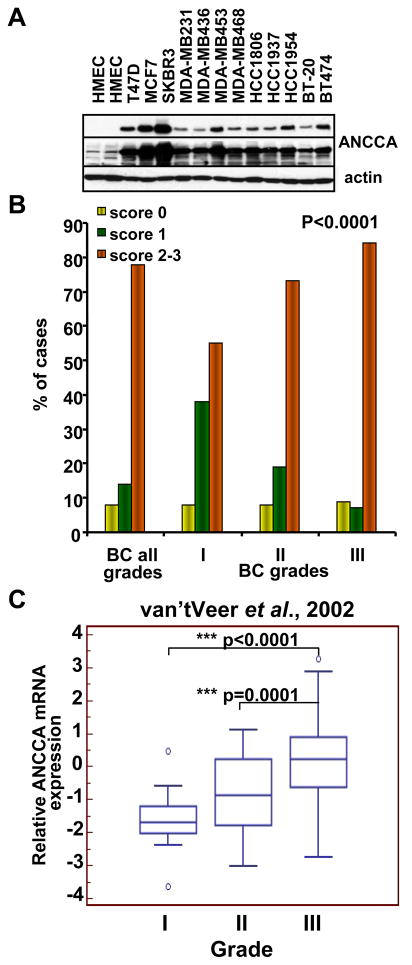
Figure 2.
ANCCA overexpression in breast cancer cell lines and its correlation with tumor grades. A, Western analysis of ANCCA in normal HMEC and different breast cancer cell lines. Top and middle panels, a short and long exposure respectively. B, representative anti-ANCCA IHC images of normal and tumor tissues with different grades. C, distribution of tumors with low (yellow), moderate (green) and high (orange) expression of ANCCA in breast carcinomas of different grades (P < 0.05). D, box-and-whisker plot of ANCCA mRNA expression in tumors of different grades analyzed using www.oncomine.org.
We also interrogated multiple microarray gene expression data sets for ANCCA mRNA change in normal and tumor tissues. Interestingly, ANCCA, listed as pro2000, atad2 or other names, is one of the few genes that overlap between gene signatures identified by several gene profiling studies for tumor classification and prediction of disease outcome (e.g. DCIS to IDC, time to distant metastasis; Supplementary Table 3). Consistent with our IHC analysis, ANCCA transcript examined in multiple studies displayed much higher levels in tumors than normal breast tissues (Supplementary Fig. 3), and its high transcript levels strongly associated with ER-negative or triple negative status (P < 0.009 and P <0.0001 respectively, Supplementary Table 4).
ANCCA overexpression correlates with tumor cell proliferation and disease progression
We next assessed the association of ANCCA expression with other clinical and pathological variables (Table 1). Although no clear association was found between ANCCA level and lymph node status, we observed that high levels of ANCCA expression correlate strongly with higher histologic grades (P < 0.0001). Overall, more differentiated, low-grade tumors showed little to moderate expression of ANCCA, while less differentiated, high-grade tumors had markedly elevated levels of ANCCA protein (Fig. 2B and 2C and Supplementary Fig. 2). Similar results were obtained from analysis of ANCCA transcripts in a cohort of tumors used in a previous study by van’t Veer et al. (11) (Fig. 2C). Consistent with the notion that basal-like/triple-negative tumors tend to display high proliferation index (1), the majority (70%) of ANCCA-overexpressing triple negative tumors also showed high Ki-67 staining (P < 0.0001, Table 1). IHC analysis of ANCCA and Ki-67 expression in adjacent sections of tumors specimens also revealed a high accordance in their expression in individual tumors (Supplementary Fig. 4).
Highly elevated ANCCA predicts poor overall survival and disease recurrence
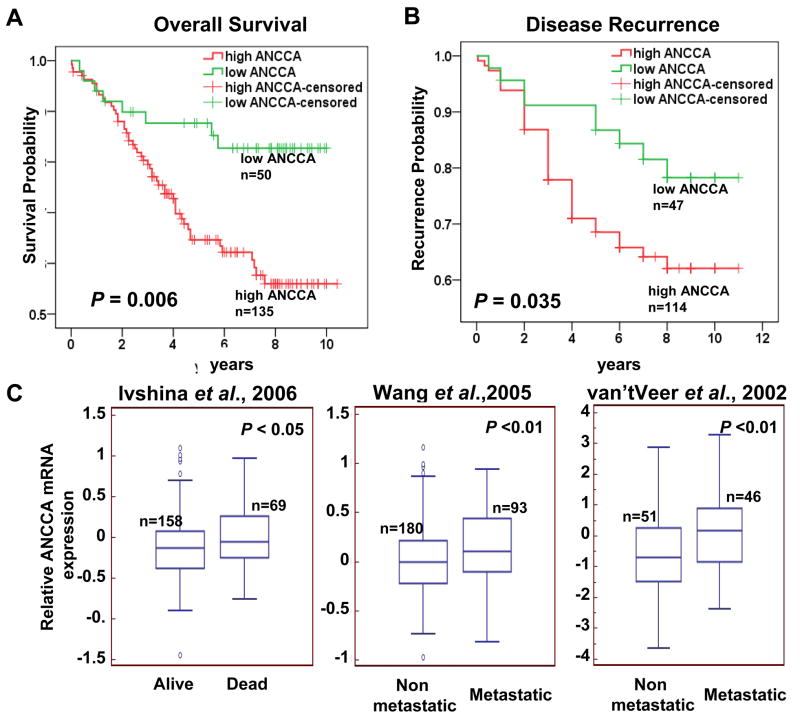
We also examined whether strong ANCCA expression is predictive of disease progression by analysis of a cohort of 185 patients with up to 11 years of follow-up information. Tumors were divided into two groups (low ANCCA and high ANCCA) according to ANCCA IHC scores. As shown in Fig. 3A, patients whose tumors showed high ANCCA expression revealed a significantly shorter overall survival period when compared to those with no or weak ANCCA expression (P = 0.006 by Cox-Mantel log rank test; 80% survival for high ANCCA group, 2.5 years, and for low ANCCA, 5 years). When the relationship between ANCCA levels and survival was assessed with ER status taken into consideration, their association with poor survival can still be observed in patients with ER-negative tumors (P = 0.035, Supplementary Fig. 5). Likewise, patients with high levels of ANCCA protein had an earlier time to disease recurrence compared to patients with low ANCCA expression (Fig. 3B). Moreover, examination of ANCCA transcripts in the datasets of several “gene signature” studies revealed that high ANCCA mRNA levels associate with high probability of death and tumor metastasis (Fig 3C).
Figure 3.
ANCCA overexpression correlates with poor outcomes. A and B, Kaplan-Meier analysis of ANCCA protein expression and disease-free survival (A) and disease recurrence (B). C, high ANCCA transcripts correlates with poor disease-free survival in tumor datasets from Ivshina et al., 2006, and associates with metastasis at five years after diagnosis in tumor datasets from Wang et al., 2005 and van’t Veer et al., 2002. The datasets were analyzed for the association using www.oncomine.org.
ANCCA overexpression promotes anchorage-dependent and-independent proliferation and survival of TNBC cells
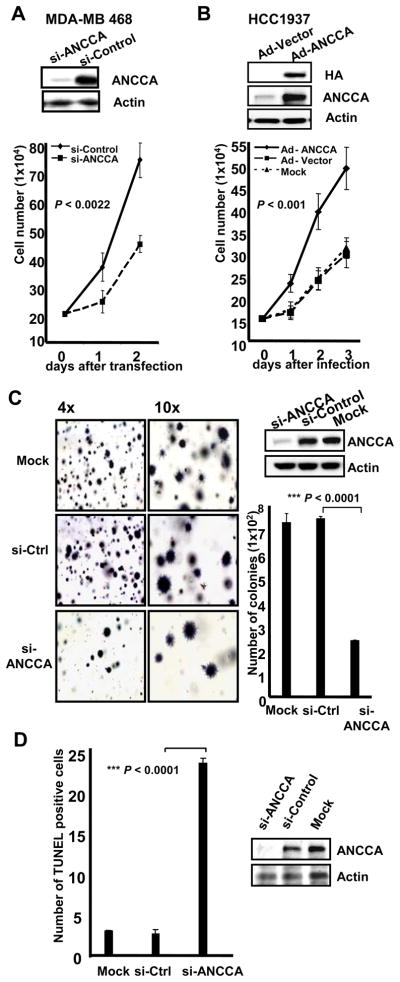
Since expression of ANCCA was particularly high in triple-negative tumors, we examined the role of overexpressed ANCCA in TNBC cells. As shown in Fig. 2A, ANCCA is highly overexpressed in the breast cancer cell lines relative to HMECs. Knockdown of ANCCA via one siRNA in MDA-MB-468 (Fig 4A), -231 (Fig 4C), and -453 (Supplementary Fig 6) cells showed strong inhibitory effects on their proliferation either in regular 2D culture or in a soft-agar, anchorage-independent growth assay. Knockdown of ANCCA using a different siRNA gave a similar effect (Supplementary Fig 7A and 7B). In the BRCA1-defective HCC1937 cells that express a moderate level of ANCCA, adenovirus vector-mediated, ectopic expression of ANCCA led to significantly increased proliferation when compared to cells infected with Adeno-vector control (Fig. 4B).
Figure 4.
Elevated ANCCA is required for proliferation and survival of triple-negative breast cancer (TNBC) cells. A and B, Cells were either infected with equal titers of adeno-ANCCA or adeno-control (HCC1937) or transfected with siRNA targeting ANCCA or control sequence (MDA-MB 468). Western and cell proliferation were performed with cells harvested 48 hrs later (for Western) or at indicated days. C, MDA-MB 231 cells were transfected with siRNA-ANCCA or siRNA-control and plated in soft agar for colony formation or Western. Colonies were stained with MTT four weeks later and counted. Representative images of colonies and their numbers from different treatments are shown. D, MDA-MB 231 cells were processed for TUNEL assay (left panel) or Western, at day 3 after siRNA transfection. Paired samples t-test (for A and B) or independent samples t-test (for C and D) was used.
We next examined whether ANCCA was also important for survival of the triple negative cells. MDA-MB-231 cells were transfected with siRNA and processed for a TUNEL assay. As demonstrated in Fig. 4D and Supplementary Fig. 8, while mock- or control siRNA-transfected cells had very few cells positive for TUNEL staining (<3%), cultures treated with ANCCA siRNA showed a marked increase (approximately 25%) of TUNEL-positive cells. Similar results were obtained with MDA-MB-453 and MDA-MB-468 cells (data not shown). Taken together, these results provide strong evidence that ANCCA overexpression promotes both proliferation and survival of TNBC cells.
ANCCA controls important regulators of cell proliferation and survival pathways including EZH2 and B-Myb
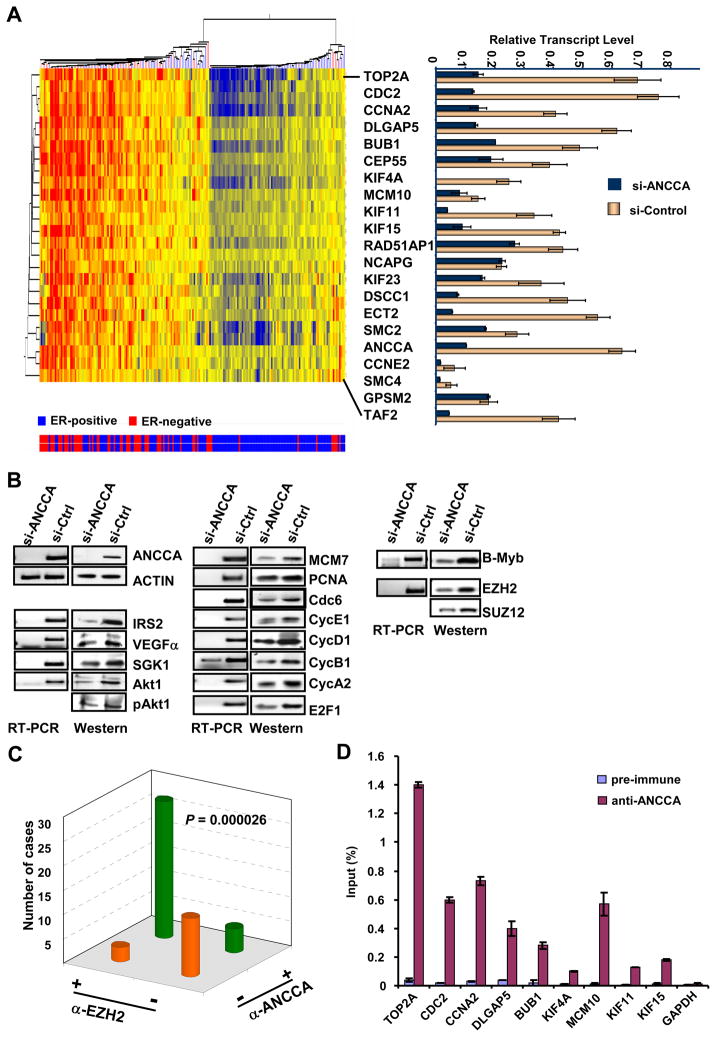
Given the transcriptional coactivator function of ANCCA demonstrated in our previous study (7–9), we analyzed the TRANSBIG Consortium gene expression data set (12), in which ANCCA serves as a prognostic signature gene, for genes having aberrant co-expression associated with overexpressed ANCCA. Strikingly, the analysis identified a group of genes highly enriched in control of cell proliferation and survival with the top 20 genes (25 gene probes) functioning primarily in mitosis (DLGAP5, BUB1, CEP55, KIF4A, KIF11, KIF15, KIF23, DSCC1, ECT2, SMC2, SMC4, GPSM2), DNA replication (MCM10 and Top2A) and cell cycle progression (Fig. 5A, and Supplementary Fig. 9). Intriguingly, consistent with our IHC study, hierarchical clustering demonstrated that high levels of the gene transcripts including ANCCA cluster primarily in ER-negative tumors (Fig. 5A, at the left side of the cluster where tumors are primarily ER-negative indicated by the red bars at the bottom). More importantly, results from ANCCA knockdown in two different TNBC cells showed that most of these genes are indeed controlled by ANCCA because their expression is down regulated upon ANCCA suppression (Fig. 5A, right panel and Supplementary Fig. 10A and 10B). Further analysis showed that additional key cell cycle and DNA replication regulators including cyclin-E1, -B1, -A2, E2F1, Cdc6, MCM7 and PCNA are also regulated by ANCCA (Fig.5B, middle; Supplementary Fig 7C). Moreover, consistent with the critical function of ANCCA in TNBC cell survival, we found that expression of survival pathway proteins such as IRS2, SGK1, VEGFα, Akt and phosphorylated Akt was also significantly affected by ANCCA knockdown (Fig. 5B, left).
Figure 5.
ANCCA controls key regulators of cell proliferation and survival pathways including the oncogene EZH2. A, unsupervised hierarchical clustering of the expression of the top 20 genes (25 probesets, not including ANCCA) found to be co-expressed with ANCCA overexpression in a data set of 198 patients (12). Correlation coefficients (by Spearmann’s test) for the similarity of each gene’s expression to that of ANCCA expression and other information are indicated in Supplementary Fig. 9. Tumor ER status is indicated below by blue bars for ER+ tumors (total of 134) or red bars for ER- tumors (total of 64). Right panel, MDA-MB468 cells were transfected with siRNA targeting ANCCA (dark blue) or control sequence (yellow) and harvested 48 hrs later for real-time RT-PCR analysis. Relative transcript levels were obtained by normalization of expression units for each gene with that of GAPDH. B, MDA-MB 468 cells were treated as above and analyzed by RT-PCR or Western. C. The anti-ANCCA and anti-EZH2 immunoreactivity of a TMA section was scored negative if less than 10% of the epithelial cells displayed any staining, and scored positive if more than 10% of the epithelial cells displayed staining with moderate to high intensity. The association between the different antibody staining was analyzed using Pearson’s χ2 test. D. ChIP with control serum and anti-ANCCA antibody was performed with MDA-MB-468 cells. ChIP DNA was analyzed by real-time PCR. Anti-ANCCA enrichments are presented as percentage of input.
Since overexpression of polycomb group protein (PcG) EZH2 and mitotic regulator B-Myb appear to correlate with high proliferation of basal-like tumors (13, 14), we examined whether ANCCA also controls their expression. Indeed, silencing of ANCCA markedly inhibited EZH2 and B-Myb expression at both the RNA and protein levels in TNBC cells (Fig 5B, right panel), and the inhibition was also observed for Suz12 protein, a critical component of the Polycomb-repressive complex 2 (PRC2). Therefore, these data demonstrate that EZH2 and B-Myb are both downstream targets of ANCCA. To address the disease relevance of ANCCA control of EZH2, we examined by IHC the expression of ANCCA and EZH2 in a cohort of 48 invasive breast carcinoma specimens on a TMA. Consistent with previous findings (15), we found that EZH2 is overexpressed in 62% of the invasive tumors (Fig 5C and Supplementary Fig. 11). Importantly, ANCCA overexpression was significantly correlated to the expression of EZH2 in the primary tumors (P < 0.000026). These results suggest that de-regulated ANCCA may contribute to EZH2 overexpression in the tumors. Finally, to determine whether ANCCA directly controls the expression of any of the 20 genes identified, we performed ChIP assays. With PCR primers that can readily amplify the proximal promoter region of indicated genes, we found that ANCCA occupies the promoter region of Top2A, cdc2, cyclin A2, DLGAP5, Bub1, MCM10, Kif-4A, Kif-11 and Kif-15 in MDA-MB468 cells (Fig 5D). Consistent with our data that ANCCA does not regulate GAPDH, we did not detect any significant anti-ANCCA enrichment at the promoter of GAPDH. Similar ChIP results were obtained from MDA-MB436 cells (Supplementary Fig 10C). Together, our data indicate that ANCCA is directly involved in control of key components of cell proliferation and survival pathways in triple-negative breast cancer cells.
Discussion
TNBCs, particularly those with basal-like gene expression profiles, tend to show highly elevated mitotic and cell proliferation indices and poor prognosis for long-term survival (1). Aberrations of several key transcriptional regulators (e.g., B-Myb, EZH2 or c-Myc overexpression as well as pRb-E2F deficiency) may underlie the elevated expression of proliferation-associated genes (13, 16). Whether their abnormal functions represent independent events in different tumors or are integrated by other mechanisms is unclear. We initially identified ANCCA as a direct target of the oncogene AIB1/ACTR and as a transcriptional coregulator for ERα and AR to promote the expression of genes driving cancer cell proliferation and survival (8, 9). Recent studies from us and others suggest that ANCCA is overexpressed in many human cancers including breast cancer and may act as a coactivator of c-Myc (8, 9, 17). However, its role in breast cancer progression has been poorly understood. In this study, we examined ANCCA expression in several cohorts of human breast cancer specimens and found that its high levels were significantly associated with poor overall survival and disease recurrence. Interestingly, ANCCA overexpression correlated strongly with the triple-negative subtype of breast cancer. The results of in vitro experiments further demonstrated that high levels of ANCCA were required to maintain proliferation and survival of TNBC cells.
Tumor gene expression signatures have been instrumental in advancing our understanding of the molecular mechanisms underlying the diverse biological phenotypes of breast cancers and now have proven utility for prediction of clinical outcomes, such as tumor responsiveness to chemotherapy (2, 11, 18–23). However, only a few genes seem to be shared across all of the different signatures identified by various studies. Interestingly, ANCCA, which was listed as pro2000, ATAD2 or other undefined names, is one of the few genes that overlap frequently between different signatures. For instance, ANCCA is a component of the 231 genes which gave rise to the 70-gene signature that predicts a clinical outcome of short interval to distant metastasis (11) and is one of the 76 genes identified for prediction of distant metastasis of ER-positive but lymph-node-negative, primary breast cancer, which was later validated in a multi-center study (12, 19, 24, 25). Consistent with our IHC results from an independent cohort of tumors which demonstrate that ANCCA protein levels alone predict tumor grades, a high level of ANCCA transcript is a constituent of the genetic grading signature that can reclassify histologic grades of breast cancer (26–28). We also found that ANCCA overexpression strongly correlated with elevated expression of proliferation-associated genes that are often components of the different gene signatures, thereby suggesting that overexpressed ANCCA may either collaborate with them or act upstream to stimulate their expression in the tumors. Intriguingly, ANCCA is not a signature gene for the core serum response signature which largely overlaps with cell proliferation genes (29), but instead is part of the PI3K signature(30), suggesting that, in some cases, elevated ANCCA may function primarily to promote cancer cell survival. Our data from cell culture studies indicate that both pro-proliferation and pro-survival genes are indeed controlled by ANCCA. Thus, ANCCA overexpression may not only constitute part of the gene signature with prognostic value, but also act as a driving force for the altered expression of the signature genes. Additional studies are needed to determine whether ANCCA coordinates the tumor signature gene programs.
It is worth noting that in one of the microarray data sets examined (12), the 20 genes most tightly associated with overexpressed ANCCA (i.e., based upon correlation tests) play important roles in mitosis and/or cell proliferation, the majority of which were found overexpressed in breast cancer or other types of cancers. When de-regulated, Top2A, cdc2, and cyclin E, as well as the other targets of ANCCA identified in this study (e.g., cdc6, B-Myb and EZH2), can display oncogenic activities. Intriguingly, four members of the human kinesin family (total of 45 genes) were co-overexpressed with ANCCA and were validated as ANCCA targets. These kinesins function in different stages of mitosis for spindle assembly and chromosome segregation (31, 32). The aberrant functions of kinesins in cancer render them attractive therapeutic targets (31, 33), or in certain circumstances, may promote cancer cell resistance to taxane-based drugs (34). Together, with the role of ANCCA in mediating expression of pro-survival genes (VEGF, IRS2, SGK and Akt), our data suggest that overexpressed ANCCA may function as a dominant node for integrating and/or eliciting multiple gene expression programs to promote breast cancer progression. De-regulation of the multiple pathways identified here could constitute part of the molecular basis for the observed association between highly-elevated ANCCA protein and poor patient outcome.
Despite our initial identification of ANCCA as a hormone-induced gene (8, 9), we demonstrate here that high levels of ANCCA protein associate most strongly with TNBCs. We also show that high transcript levels of ANCCA tend to associate with ERα-negative or triple-negative status in several data sets from studies of multiple cohorts of tumors (19, 26, 35). Moreover, our unbiased analysis of tumor gene expression datasets points to the association of high levels of ANCCA with the overexpression of other proliferation genes occurring primarily in ERα-negative tumors. How ANCCA is de-regulated in ERα-negative breast cancers is currently unclear. Results obtained by Ciro and colleagues reported that ANCCA is regulated by the pRb-E2F pathway in fibroblasts and osteosarcoma cells (17). Whether this is the major mechanism for ANCCA overexpression in breast cancer awaits further analysis. Given that one salient feature of triple-negative/basal like tumors is their high proliferation index and functional loss of the pRb-E2F pathway (1, 16, 35, 36), de-regulation of ANCCA through loss of pRb-mediated control is an attractive possibility. On the other hand, ANCCA may not be merely a downstream target of pRb-E2F, because ANCCA itself mediates expression of multiple Rb-E2F target genes critical for TNBC cell proliferation.
Several transcriptional coregulators including the ones involved in steroid hormone signaling have been strongly implicated in human malignancies (37–42). AIB1 (also known as ACTR/SRC-3) was initially identified as a gene amplified in breast cancer and a coactivator for ERα (43). Later studies revealed that its overexpression does not correlate with ER and PR status and that its aberrant function may include stimulation of tumor growth of both hormone-dependent and -independent breast cancers via ER-dependent and – independent pathways (44–48). One distinction between ACTR and ANCCA is that while ACTR overexpression correlates strongly with high HER2 expression, high levels of ANCCA do not, but instead associate with triple-negative status. Although such distinction implies a different mechanism for their aberrant expression, our recent studies suggest that ANCCA and ACTR may regulate the expression of each other in certain circumstances (7, 8). EZH2, the histone methyltransferase subunit of PRC2 complex is frequently overexpressed in multiple types of cancer (15, 49). Like ANCCA, EZH2 overexpression in breast cancer correlates with high proliferation and basal-like phenotype and tumor invasiveness (14). ANCCA, however, is unique in that it appears to play a critical role in the control of expression of pro-proliferation genes including EZH2 and B-Myb in triple-negative cancers. Thus, our data suggest that ANCCA may act as an integrator of several oncogenic programs. Given our finding that ANCCA is a transcriptional coregulator, it is reasonable to predict that a major mechanism of overexpressed ANCCA in breast cancer is alteration of multiple gene networks including those of EZH2, pRb-E2F (this study) or c-Myc (17). Our previous study demonstrated that ANCCA is an AAA+ ATPase protein and that its ATPase activity is required for its transcriptional stimulation function (8). ANCCA also possesses a bromodomain that may recognize a distinct histone modification. Given these structure-function features of ANCCA and its aberrant expression in multiple types of human cancers, further studies are warranted to exploit the potential of aberrant ANCCA as a new prognostic marker and a therapeutic target.
Supplementary Material
Acknowledgments
We thank Dr. Laurel Beckett for advice on statistics analysis, and Ping Yang for technical assistance on IHC analysis. This study was supported by NIH grants R01CA113860 and R01DK060019 (HWC). EVK is a recipient of a DoD BCRP Pre-doctoral Fellowship Award (W81XWH-0810689). NPA is a trainee of an NIH T32 training grant.
References
- 1.Schneider BP, Winer EP, Foulkes WD, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14:8010–8. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahusen T, Henke RT, Kagan BL, Wellstein A, Riegel AT. The role and regulation of the nuclear receptor co-activator AIB1 in breast cancer. Breast Cancer Res Treat. 2009;116:225–37. doi: 10.1007/s10549-009-0405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Malley BW, Kumar R. Nuclear receptor coregulators in cancer biology. Cancer Res. 2009;69:8217–22. doi: 10.1158/0008-5472.CAN-09-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–30. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsia EY, Kalashnikova EV, Revenko AS, Zou JX, Borowsky AD, Chen HW. Deregulated E2F and the AAA+ Coregulator ANCCA Drive Proto-Oncogene ACTR/AIB1 Overexpression in Breast Cancer. Mol Cancer Res. 2010;8:183–93. doi: 10.1158/1541-7786.MCR-09-0095. [DOI] [PubMed] [Google Scholar]
- 8.Zou JX, Revenko AS, Li LB, Gemo AT, Chen HW. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc Natl Acad Sci U S A. 2007;104:18067–72. doi: 10.1073/pnas.0705814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou JX, Guo L, Revenko AS, et al. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 2009;69:3339–46. doi: 10.1158/0008-5472.CAN-08-3440. [DOI] [PubMed] [Google Scholar]
- 10.Louie MC, Revenko AS, Zou JX, Yao J, Chen HW. Direct control of cell cycle gene expression by proto-oncogene product ACTR, and its autoregulation underlies its transforming activity. Mol Cell Biol. 2006;26:3810–23. doi: 10.1128/MCB.26.10.3810-3823.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 12.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 13.Thorner AR, Hoadley KA, Parker JS, Winkel S, Millikan RC, Perou CM. In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene. 2009;28:742–51. doi: 10.1038/onc.2008.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez ME, Li X, Toy K, et al. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene. 2009;28:843–53. doi: 10.1038/onc.2008.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10:R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciro M, Prosperini E, Quarto M, et al. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 2009;69:8491–8. doi: 10.1158/0008-5472.CAN-09-2131. [DOI] [PubMed] [Google Scholar]
- 18.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 20.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 21.Deblois G, Hall JA, Perry MC, et al. Genome-wide identification of direct target genes implicates estrogen-related receptor alpha as a determinant of breast cancer heterogeneity. Cancer Res. 2009;69:6149–57. doi: 10.1158/0008-5472.CAN-09-1251. [DOI] [PubMed] [Google Scholar]
- 22.Bonnefoi H, Potti A, Delorenzi M, et al. Validation of gene signatures that predict the response of breast cancer to neoadjuvant chemotherapy: a substudy of the EORTC 10994/BIG 00-01 clinical trial. Lancet Oncol. 2007;8:1071–8. doi: 10.1016/S1470-2045(07)70345-5. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Zou L, Li Q, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. 2010;16:214–8. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foekens JA, Atkins D, Zhang Y, et al. Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J Clin Oncol. 2006;24:1665–71. doi: 10.1200/JCO.2005.03.9115. [DOI] [PubMed] [Google Scholar]
- 25.Vale RD. AAA proteins. Lords of the ring. J Cell Biol. 2000;150:F13–9. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 27.Ma XJ, Salunga R, Tuggle JT, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:5974–9. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teschendorff AE, Naderi A, Barbosa-Morais NL, et al. A consensus prognostic gene expression classifier for ER positive breast cancer. Genome Biol. 2006;7:R101. doi: 10.1186/gb-2006-7-10-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–43. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saal LH, Johansson P, Holm K, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–9. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huszar D, Theoclitou ME, Skolnik J, Herbst R. Kinesin motor proteins as targets for cancer therapy. Cancer Metastasis Rev. 2009;28:197–208. doi: 10.1007/s10555-009-9185-8. [DOI] [PubMed] [Google Scholar]
- 32.Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19:1703–11. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 33.Taniwaki M, Takano A, Ishikawa N, et al. Activation of KIF4A as a prognostic biomarker and therapeutic target for lung cancer. Clin Cancer Res. 2007;13:6624–31. doi: 10.1158/1078-0432.CCR-07-1328. [DOI] [PubMed] [Google Scholar]
- 34.De S, Cipriano R, Jackson MW, Stark GR. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009;69:8035–42. doi: 10.1158/0008-5472.CAN-09-1224. [DOI] [PubMed] [Google Scholar]
- 35.Gauthier ML, Berman HK, Miller C, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–91. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bosco EE, Wang Y, Xu H, et al. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest. 2007;117:218–28. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res. 2009;69:3819–27. doi: 10.1158/0008-5472.CAN-08-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohshiro K, Rayala SK, Kondo S, et al. Identifying the estrogen receptor coactivator PELP1 in autophagosomes. Cancer Res. 2007;67:8164–71. doi: 10.1158/0008-5472.CAN-07-0038. [DOI] [PubMed] [Google Scholar]
- 39.Shen H, Powers N, Saini N, et al. The SWI/SNF ATPase Brm is a gatekeeper of proliferative control in prostate cancer. Cancer Res. 2008;68:10154–62. doi: 10.1158/0008-5472.CAN-08-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Torres-Arzayus MI, de Mora JF, Yuan J, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–74. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 41.Redmond AM, Bane FT, Stafford AT, et al. Coassociation of estrogen receptor and p160 proteins predicts resistance to endocrine treatment; SRC-1 is an independent predictor of breast cancer recurrence. Clin Cancer Res. 2009;15:2098–106. doi: 10.1158/1078-0432.CCR-08-1649. [DOI] [PubMed] [Google Scholar]
- 42.Hsia E, Zou JX, Chen HW. The roles and action mechanisms of p160/SRC coactivators and the ANCCA coregulator in cancer. Prog Mol Biol Transl Sci. 2009;87:261–98. doi: 10.1016/S1877-1173(09)87008-7. [DOI] [PubMed] [Google Scholar]
- 43.Anzick SL, Kononen J, Walker RL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–8. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 44.Yan J, Erdem H, Li R, et al. Steroid receptor coactivator-3/AIB1 promotes cell migration and invasiveness through focal adhesion turnover and matrix metalloproteinase expression. Cancer Res. 2008;68:5460–8. doi: 10.1158/0008-5472.CAN-08-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin L, Liao L, Redmond A, et al. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008;28:5937–50. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–71. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heck S, Rom J, Thewes V, et al. Estrogen-related receptor alpha expression and function is associated with the transcriptional coregulator AIB1 in breast carcinoma. Cancer Res. 2009;69:5186–93. doi: 10.1158/0008-5472.CAN-08-3062. [DOI] [PubMed] [Google Scholar]
- 48.Fereshteh MP, Tilli MT, Kim SE, et al. The nuclear receptor coactivator amplified in breast cancer-1 is required for Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis in mice. Cancer Res. 2008;68:3697–706. doi: 10.1158/0008-5472.CAN-07-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.