Summary
Host cell invasion by the Apicomplexa critically relies on regulated secretion of transmembrane micronemal proteins (TM-MICs). Toxoplasma gondii possesses functionally non-redundant MICs complexes that participate in gliding motility, host cell attachment, moving junction formation, rhoptry secretion and invasion. The TM-MICs are released onto the parasite’s surface as complexes capable of interacting with host cell receptors. Additionally, TgMIC2 simultaneously connects to the actomyosin system via binding to aldolase. During invasion these adhesive complexes are shed from the surface notably via intramembrane cleavage of the TM-MICs by a rhomboid protease. Some TM-MICs act as escorters and assure trafficking of the complexes to the micronemes. We have investigated the properties of TgMIC6, TgMIC8, TgMIC8.2, TgAMA1 and the new micronemal protein TgMIC16 with respect to interaction with aldolase, susceptibility to rhomboid cleavage and presence of trafficking signals. We conclude that several TM-MICs lack targeting information within their C-terminal domains, indicating that trafficking depends on yet unidentified proteins interacting with their ectodomains. Most TM-MICs serve as substrates for a rhomboid protease and some of them are able to bind to aldolase. We also show that the residues responsible for binding to aldolase are essential for TgAMA1 but dispensable forTgMIC6 function during invasion.
Keywords: Toxoplasma gondii, trafficking, polytopic, rhomboid, protease, Golgi, microneme
Introduction
Toxoplasma gondii is an obligate intracellular parasite of the phylum Apicomplexa, which also includes the deadly agent of malaria, Plasmodium falciparum. Host cell invasion by these parasites is a multi-step process (Carruthers & Boothroyd, 2007) propelled by the gliding motility machinery. It is initiated by apical attachment of the parasite to the host cell, followed by reorientation, formation of a junction between the parasite and host cell membranes, penetration and, finally, sealing of the parasitophorous vacuolar membrane.
Some of the proteins implicated in invasion are sequentially released from two types of secretory organelles, named micronemes and rhoptries (Carruthers & Sibley, 1997). In T. gondii, four complexes composed of soluble and transmembrane microneme proteins or including rhoptry neck proteins (RONs) have been investigated and shown to perform non-overlapping functions during invasion (Figure 1A). More complexes are, however, likely to contribute to invasion since additional uncharacterized transmembrane microneme proteins (TM-MICs) are encoded in the genome.
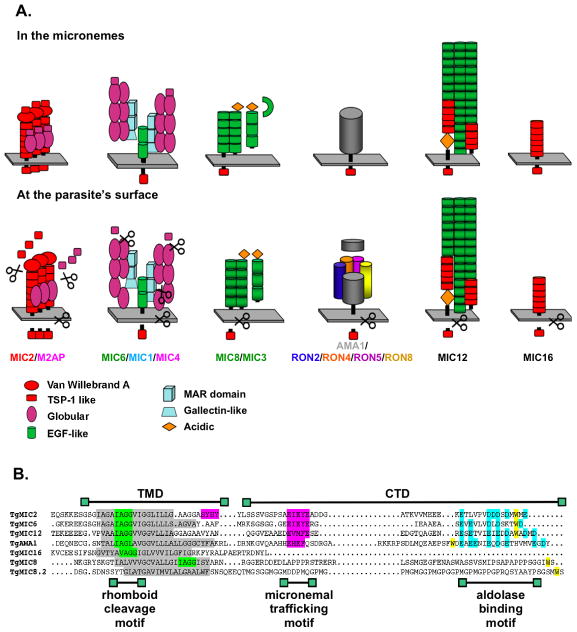
Figure 1.
A. Schematic representation of the four major microneme complexes in T. gondii, as well as of TgMIC12 and TgMIC16, when in the micronemes (top) and on the parasite’s surface (bottom). Represented is the currently known composition of the complexes, including the various proteolytic cleavage events as demonstrated in (Carruthers et al., 2000, Harper et al., 2006) for TgMIC2/M2AP, in (Opitz et al., 2002, Meissner et al., 2002, Sawmynaden et al., 2008) for TgMIC6/MIC1/MIC4, in (Meissner et al., 2002, Cerede et al., 2005) for TgMIC8/MIC3, in (Hehl et al., 2000, Alexander et al., 2005, Straub et al., 2008, Besteiro et al., 2009) for TgAMA1/RON2/RON4/RON5/RON8, in (Opitz et al., 2002) for TgMIC12 and in this study for TgMIC16.
B. Amino acid sequence alignment of the TMDs and CTDs of TgMIC2, TgMIC6, TgMIC12, TgAMA1, TgMIC16, TgMIC8 and TgMIC8.2. Boxed in grey are the TMDs, in green the putative rhomboid cleavage sites and in pink the motifs for traffic to the micronemes. The tryptophan signature residue boxed in yellow and the acidic residues boxed in blue are both involved in binding to aldolase.
The selective participation of each of the four complexes in the invasion process has been uncovered by generating conventional or conditional knockouts of the genes encoding components of the complexes. The TM-MIC TgMIC2 forms a multimeric complex with the soluble partner TgM2AP (Rabenau et al., 2001, Jewett & Sibley, 2004). Parasites depleted in TgMIC2 are markedly deficient in host-cell attachment, motility and hence unable to invade host cells (Huynh & Carruthers, 2006). Another complex, composed of the transmembrane protein TgMIC6, is interacting with two soluble molecules TgMIC1 and TgMIC4. Genetic disruption of any of the three encoding genes is still compatible with parasite survival (Reiss et al., 2001) even if the complex has been demonstrated to play an important role in invasion in vitro and to contribute to virulence in vivo (Blumenschein et al., 2007, Cerede et al., 2005, Sawmynaden et al., 2008). A third complex is composed of the TM-MIC TgMIC8 and the soluble protein TgMIC3. Genetic disruption of TgMIC8 interferes with rhoptries secretion and, consequently, prevents formation of the moving junction (MJ) and completion of invasion (Kessler, 2008). A fourth complex, which uniquely localizes to the MJ (Alexander et al., 2005), contains the rhoptry proteins TgRON2, TgRON4, TgRON5 and TgRON8 (Alexander et al., 2005, Besteiro et al., 2009, Straub et al., 2008) and the TM-MIC TgAMA1, which anchors the complex to the parasite plasma membrane. Parasites lacking TgAMA1 efficiently attach to host cells but are defective in rhoptry secretion, fail to create a MJ and are consequently unable to invade host cells (Mital et al., 2005). Gliding motility is not significantly altered in the absence of TgAMA1 or TgMIC8 (Mital et al., 2005, Kessler, 2008).
So far, TgMIC2 and TgMIC6 are the only TM-MIC shown to play a crucial role as force-transducer during motility and invasion. TgMIC2 binds to receptor(s) on the host cell surface and establishes simultaneously a connection, via its C-terminal cytoplasmic domain (CTD), with the parasite’s actomyosin system, hence powering parasite motility. The CTD of TgMIC2, TgMIC6 and other members of the TRAP family in Plasmodium (TRAP, CTRP and TLP) interact with aldolase, a glycolytic enzyme also capable of binding to filamentous-actin (F-actin) (Buscaglia et al., 2003, Jewett & Sibley, 2003, Heiss et al., 2008, Zheng et al., 2009). It is unknown whether other T. gondii TM-MICs, that are part of adhesive complexes and exhibiting crucial functions in invasion, can as well interact with aldolase and thus act as bridge molecules.
At the end of the penetration process, the tight interactions formed between the different MIC complexes and the host cell receptors have to be disengaged to let the parasite freely replicate. This has been proposed to occur by proteolytic shedding of the MIC complexes from the parasite’s surface. Cell-based cleavage assays and studies on parasites have demonstrated that one of these critical cleavage events takes place at a conserved motif within the luminal part of the transmembrane domains of TgMIC2, TgMIC6, TgMIC12 and TgAMA1. The protease responsible for this intramembrane cleavage was named microneme protein protease 1 (MPP1) and likely corresponds to a plasma membrane rhomboid-like protease (Opitz et al., 2002, Brossier et al., 2003, Urban & Freeman, 2003, Zhou et al., 2004, Howell et al., 2005). Prime candidates for this shedding activity are TgROM4 and TgROM5, which are found at the plasma membrane of the parasite (Brossier et al., 2005, Dowse et al., 2005). More recently, parasites depleted in TgROM4 indicate that this protease acts as sheddase for TgMIC2 and TgAMA1 and hence critically contributes to the creation of an apical-posterior gradient of adhesins necessary for an apical orientation of the parasite during invasion (Buguliskis et al.). At the end of the penetration process, the tight interactions formed between the different MIC complexes and the host cell receptors have to be disengaged to let the parasite freely replicate. This has been proposed to occur by proteolytic shedding of the MIC complexes from the parasite’s surface by the TgROM5 activity.
A prerequisite for successful invasion is the correct trafficking of the MIC complexes, from the endoplasmic reticulum (ER), where they are pre-assembled, to the micronemes, where they are stored prior to invasion. Similar to other eukaryotic sorting mechanisms, some TM-MICs are accurately targeted to the micronemes via recognition of a tyrosine-based motif in the cytoplasmic CTDs (Sheiner & Soldati-Favre, 2008). TgMIC2 and TgMIC6 CTDs contain such a microneme targeting motif (EIEYE) and have been shown to serve as escorters for the soluble MICs that are part of the respective complexes (Di Cristina et al., 2000, Opitz et al., 2002, Reiss et al., 2001). Recent studies have also revealed an important contribution of some of the soluble MICs to trafficking. TgMIC1 was shown to promote folding of TgMIC6 by serving as a quality control mechanism (Saouros et al., 2005) and other soluble MICs contain pro-peptides that act as luminal forward targeting elements and are indispensable for correct trafficking of the entire complex (Brydges et al., 2008, El Hajj et al., 2008, Harper et al., 2006).
To gain insight into the mechanistic contribution of each of the MIC complexes to invasion, we have undertaken a detailed analysis of the TM-MICs currently identified in T. gondii and included a new member, TgMIC16. We have searched for the presence of trafficking determinants, assessed their susceptibility to intramembrane cleavage and their ability to interact with aldolase. The results indicate that in contrast to TgMIC2, TgMIC6 and TgMIC12, the CTDs of TgMIC8, TgMIC8.2, TgAMA1 and TgMIC16 do not carry the information for proper trafficking to the micronemes and cannot therefore be considered as escorters. All these TM-MICs, apart from TgMIC8.2, appear to be susceptible to intramembrane cleavage and the CTDs of TgMIC6, TgMIC12 and TgAMA1 can bind to aldolase in pull down assays. Additionally, we have identified specific residues within the CTD of TgAMA1 that are required for both association with aldolase and host cell invasion. Collectively these data support a model describing the involvement of TM-MICs, as part of complexes with distinct and non-overlapping functions during invasion.
Results
TgMIC16 is a conserved Coccidia TM-MIC containing 6 TSR domains
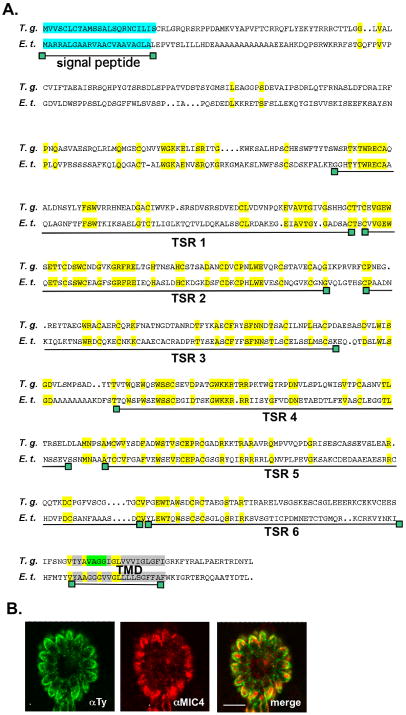
A search in the T. gondii genome database for putative new microneme proteins containing TRAP-family-like transmembrane sequences led to the identification of a gene encoding a hypothetical protein (TGME49_089630) of 669 amino acids. This gene model (80.m00085) has also been identified by a recent in silico screen for secretory proteins and was proposed to reside in an apical compartment (Chen et al., 2008). The amino acid sequence of the protein includes an N-terminal predicted signal peptide, six putative TSR type 1 domains (Figure S1) and one TMD (transmembrane domain). This TMD is located close to the C-terminal end, contains a motif reminiscent of a rhomboid cleavage site and delimits a very short C-terminal tail (Figures 2A and 2B). Another TMD is also predicted at the N-terminal end of the protein (TMHMM prediction program), but with a low probability and therefore it is not depicted as such in the schemes. A search of the available apicomplexan genomes revealed that homologues of TGME49_089630 are present in the genomes of N. caninum and E. tenella but are absent in Hemosporidia, suggesting that this gene is restricted to the Coccidia. Alignment of the amino acid sequences of these genes (Figure 2A) uncovered a very similar domain structure.
Figure 2.
A. Amino acid sequences alignment of the T. gondii TgMIC16 (EU791458) and the homologous gene in E. tenella (SNAP00000003913). The predicted annotations of N. caninum and E. tenella genes await experimental confirmation. Boxed in light blue is the putative signal peptide, in grey the putative TMD and in light green the rhomboid cleavage motif. The strictly conserved residues are boxed in yellow. The six putative TSR domains are indicated. The TMD prediction was performed with the program TMHMM.
B. Double immunofluorescence analysis by confocal microscopy of intracellular parasites transiently expressing MIC16-Ty carried out with anti-Ty-1 (green) and anti-MIC4 (red), a micronemal marker. Scale bars indicates 10μm.
Transient expression of this new protein carrying a Ty epitope at the C-terminus revealed a micronemal localization in T. gondii tachyzoites (Figure 2B). Reflective of this localization and the nomenclature status, this protein has been named TgMIC16 (accession number EU791458). Given its predicted domain structure and the presence of a putative rhomboid cleavage site in the TMD, this protein was included in this study along with the TM-MICs that are part of the four major T. gondii MIC complexes (TgAMA1, TgMIC2, TgMIC6 and TgMIC8). We also included one of the homologues of TgMIC8, TgMIC8.2, previously known as MIC8-like 1 (Kessler, 2008). A chimera of the TgMIC8 ectodomain fused to the TM-CTD portion of TgMIC8.2 was able to functionally complement mic8ko parasites, indicating that the TM-CTDs of these proteins are functionally equivalent (Kessler, 2008). Finally, TgMIC12, the homologue of the Eimeria TM-MICs EtMIC4 and EmTFP250 (Witcombe et al., 2004, Periz et al., 2009), shown before to be susceptible to rhomboid cleavage (Opitz et al., 2002), was also included in the comparative analysis.
Multiple motifs and functions are conserved in the TM-CTDs of the TM-MICs
Several lines of evidence indicate that the TM-CTDs of the TM-MICs play an essential role in supporting the functionality of their respective complexes (information regarding the composition and susceptibility to proteolytic cleavage of these complexes is recapitulated in Figure 1A), and therefore an alignment of the amino acid sequences of these domains was performed and carefully examined (Figure 1B).
Previous studies on the aldolase binding capacity of the CTDs of TgMIC2 and other TRAP-related TM-MICs have demonstrated the importance of both a stretch of acidic residues and a penultimate tryptophan residue in the extreme C-terminal sequence (Buscaglia et al., 2003, Starnes et al., 2006). TgMIC6 and TgMIC12 possess both the acidic stretch and a tryptophan residue near the C-terminus (Figure 1B). The CTDs of the two TgMIC8 homologues possess a penultimate tryptophan residue but are not of acidic nature. Conversely, TgAMA1 contains the C-terminal acidic residues, but the most C-terminal tryptophan is 21 residues in from the C-terminus (W520). This residue lies within a FW motif that is highly conserved in the AMA1 homologues of different apicomplexans (Hehl et al., 2000, Donahue et al., 2000) and is known to be essential for invasion in P. falciparum (Treeck et al., 2009). The short TgMIC16 CTD does not exhibit any feature of the aldolase-binding motifs.
TgAMA1, TgMIC2, TgMIC6 and TgMIC12 possess a rhomboid cleavage site, IAGG or IAGL, at a conserved position within the TMD, and were previously shown to be cleaved in the parasite and in in vitro cleavage assays (Opitz et al., 2002, Urban & Freeman, 2003, Dowse et al., 2005, Brossier et al., 2005, Howell et al., 2005, Buguliskis et al.). A very similar motif is found at the corresponding position within the TMD of TgMIC16 and in a different position within the TMD of TgMIC8. No rhomboid cleavage motif could be identified in the TMD of TgMIC8.2.
From the two motifs shown to be essential for TgMIC2 targeting to the micronemes (Di Cristina et al., 2000), the sequence SYHYY is not conserved in any of the CTDs of the TM-MICs analyzed, whereas the motif EIEYE is strictly conserved in the tail of TgMIC6. A sequence resembling EIEYE is similarly positioned in the TgMIC12 and TgAMA1 CTDs but the critical last glutamine residue is only present in TgMIC12. No sequence reminiscent of such a targeting motif could be identified in the CTDs of the TgMIC8 family members or TgMIC16.
Several TM-MICs lack trafficking signals in their CTDs
The TM-CTDs of TgMIC2, TgMIC6 and TgMIC12 were shown to be able to target to the micronemes the surface antigen 1 protein (TgSAG1), lacking its GPI anchor signal (Di Cristina et al., 2000, Opitz et al., 2002, Reiss et al., 2001). From these studies it was concluded that these three TM-MICs act as escorters, bringing the soluble components of their respective complexes to the organelle. TgMIC8 was initially suspected to act as an escorter based on the ability of a GPI anchored TgMIC8 construct to bring TgMIC3 to the plasma membrane (Meissner et al., 2002). However this experiment only demonstrated that TgMIC3 and TgMIC8 were part of the same complex and the escorter hypothesis had to be revisited in the light of a recent study, which established that the soluble partner TgMIC3 was correctly targeted to the micronemes, even in the absence of TgMIC8 (Kessler, 2008).
To assess if TgMIC8, TgMIC8.2 and TgMIC16 contain trafficking information, their TM-CTDs were C-terminally fused to the SAG1 coding sequence, lacking its GPI anchoring signal, and to a Ty-1 tag epitope, and expressed under the control of the TgMIC2 promoter (the constructs are depicted in Figure S2). Depending on the information carried by the CTDs, these chimeras were expected to travel through the secretory pathway and to be secreted onto the parasite surface, either via the micronemes by extracellular parasites at the time of invasion, or via the dense granules (DGs) in a constitutive fashion.
In parasites expressing pMS1tyMIC16TM-CTD or pMS1tyMIC8TM-CTD, the fusion proteins accumulated in the DGs and were delivered to the parasitophorous vacuole (PV), as shown by co-localization with the DG marker GRA3 (Figure 3). An identical SAG1 fusion with the TM-CTD of TgMIC8.2 (pMS1tyMIC8.2TM-CTD) accumulated in the trans-Golgi, since it accumulated in a compartment in the proximity of cis-Golgi as shown by staining with the marker GRASP-YFP (Pelletier et al., 2002) (Figure 3). Due to the presence of a TMD, the chimeric proteins were expected to accumulate at the plasma membrane (PM). The absence of PM staining suggests that these proteins are cleaved once delivered to the plasma membrane, and what is being detected is the processed form.
Figure 3.
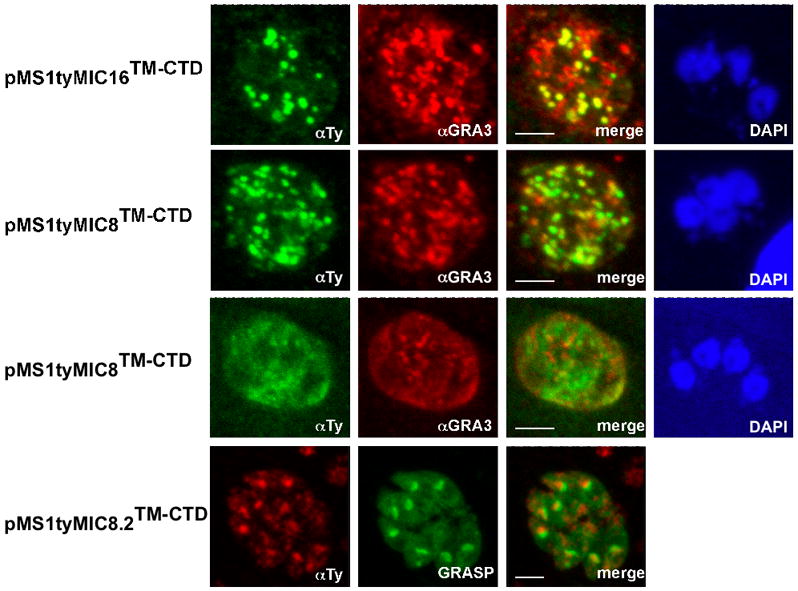
Localization of several stably transfected chimeras in the parasite using double immunofluorescence analysis and confocal microscopy. Anti-GRA3 or GRASP-YFP were used as dense granules and Golgi markers, respectively. Scale bars indicate 5μm. pMS1tyMIC16TM-CTD and pMS1tyMIC8TM-CTD expressing parasites were stained with anti-Ty-1 (in green) and anti-GRA3 (in red). pMS1tyMIC8.2TM-CTD was stained with anti-Ty-1 (in red) and co-localized with expression of GRASP-YFP (in green). The nucleus was stained with DAPI (in blue).
Insight into the trafficking of TgAMA1 was performed by expressing its TM-CTD C-terminally fused to the SAG1 and Ty-1 tag epitope, under the control of the endogenous promoter. The fusion pAS1tyAMA1TM-CTD localized to the DGs, and not to the PM, as previously shown for pMS1tyMIC16TM-CTD and pMS1tyMIC8TM-CTD, suggesting that this protein also undergoes proteolysis (Figure 4). A series of truncated variants of TgAMA1 fused N-terminally to a His tag, under the control of the endogenous promoter, were also expressed. Unlike pAS1tyAMA1TM-CTD, pAhisAMA1ΔTM-CTD, encoding the ectodomain only, or pAhisAMA1ΔCTD, encoding the ectodomain and TMD, were predominantly targeted to the micronemes, as shown by co-localization with TgMIC4. The same localization was obtained for the full-length protein, pAhisAMA1 (Figure 4). These data suggest that the ectodomain, but not the CTD, of TgAMA1 assures correct trafficking to the micronemes potentially via interaction with a yet unidentified protein. This is in accordance with the observations made on the Plasmodium AMA1 and other micronemal proteins (Treeck et al., 2009, Treeck et al., 2006, Healer et al., 2002). These results indicate that unlike TgMIC2, TgMIC6 and TgMIC12, none of the other TM-MICs analyzed here carry the necessary signal in their CTD to travel to the micronemes.
Figure 4.
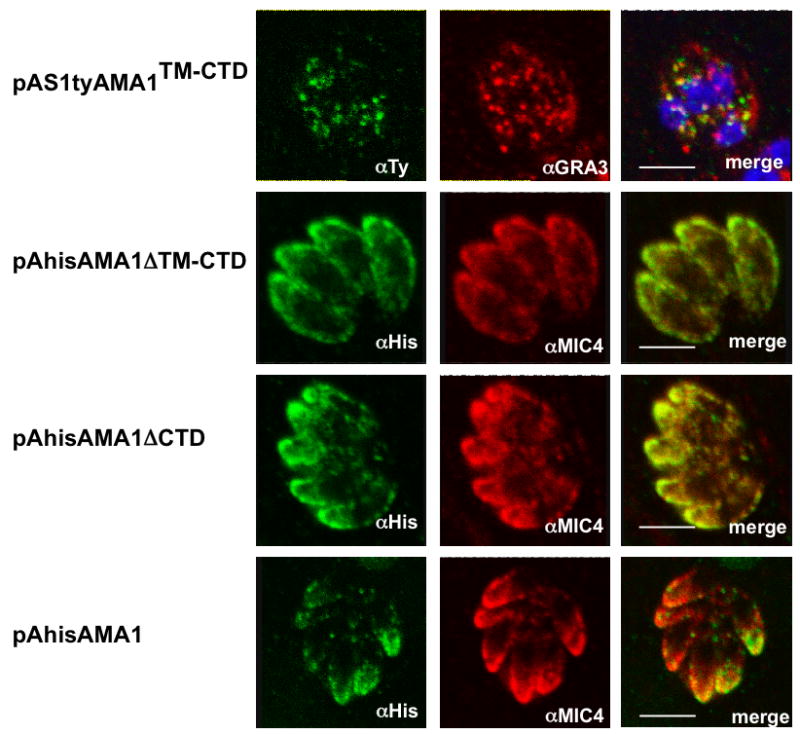
Localization of several stably transfected chimeras in the parasite using double immunofluorescence analysis and confocal microscopy and schemes of the different constructs. Anti-MIC4 was used as micronemal marker. Scale bars indicate 5μm. pAS1tyAMA1TM-CTD expressing parasites were stained with anti-Ty-1 (in green) and co-localized with anti-GRA3 (in red). pAhisAMA1ΔTM-CTD, pAhisAMA1ΔCTD and pAhisAMA1 expressing parasites were stained with anti-his (in green) and co-localized with anti-MIC4 (in red).
Several TM-MICs are susceptible to cleavage within the membrane-spanning domain
To determine if the SAG1-ty-TM-CTD chimeras were serving as substrates for intramembrane cleavage we generated parasites expressing SAG1 fusion constructs mutated in the predicted rhomboid cleavage sites (the mutated residues are boxed in the schemes depicted in Figures 5B–5E). Analysis by IFA of the corresponding transgenic parasite lines revealed that there was a dramatic change on the subcellular localization when compared to the wild type chimeras (Figure 3). The mutant fusion proteins accumulated at the PM and residually at the DGs (Figure 5A), suggesting that they were indeed subject to intramembrane proteolysis and introduction of the mutations conferred resistance to cleavage and accumulation at the parasite surface.
Figure 5.
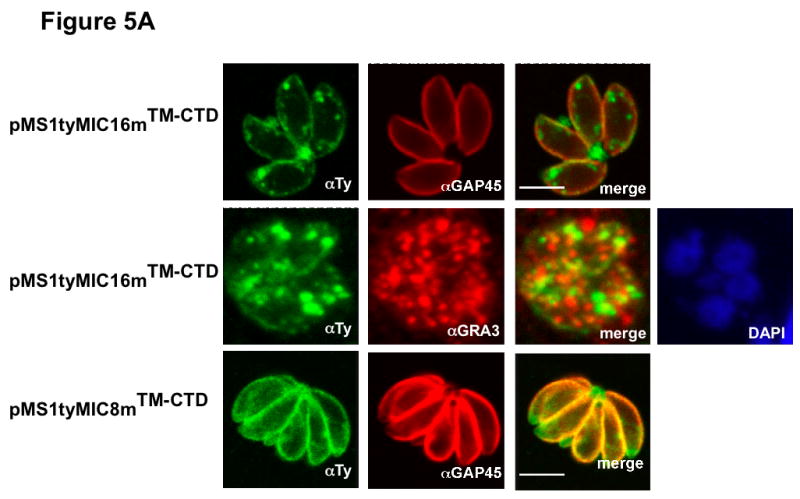
A Subcellular distribution of the SAG1-TM-CTD mutant chimeras by IFA and documented by confocal microscopy. Anti-GAP45 antibodies (in red), Anti-GRA3 (in red) and GRASP-YFP (in green) were used as IMC, DGs and Golgi markers, respectively. The nucleus was stained with DAPI (blue). pMS1tyMIC16mTM-CTD, pMS1tyMIC8mTM-CTD (in green) andpAS1tyAMA1mTM-CTD (in red) expressing parasites were stained with anti-Ty-1. Scale bars indicate 5μm.
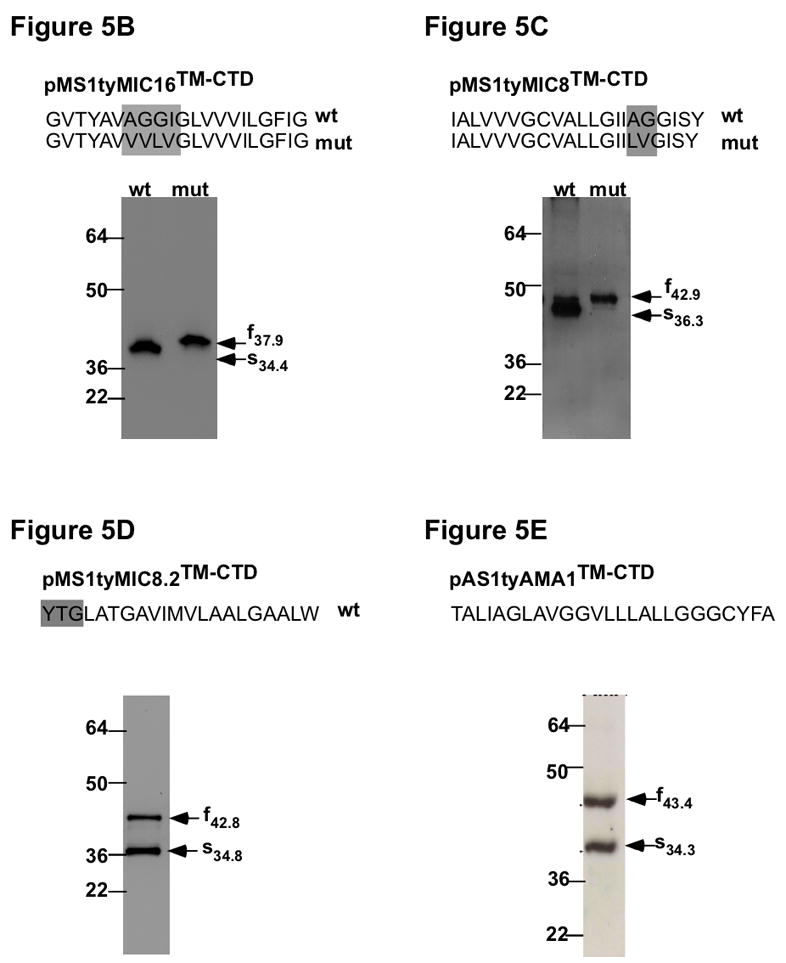
B–E Analysis of the cleavage events in the different chimeras. On the top, is shown the TMD of each TM-MIC in the original (wt) and mutated chimera (mut) and the residues mutated in the rhomboid cleavage motif are boxed. Below are western-blot analysis of lysates from parasites stably expressing the different pMS1tyMICTM-CTD fusion constructs. Indicated by arrows are the migration of the full (f) and shed (s) forms with indication of the molecular weight. B Migration of pMS1tyMIC16TM-CTD (wt) and pMS1tyMIC16mTM-CTD (mut). C Migration of pMS1tyMIC8TM-CTD (wt) and pMS1tyMIC8mTM-CTD (mut). D Migration of pMS1tyMIC8.2TM-CTD. E Migration of pAS1tyAMA1TM-CTD. The proteins were detected with anti-Ty-1. Molecular weight markers are indicated in kDa.
To confirm that the changes in localization coincided with abrogation of cleavage, western blot analyses were performed on total lysates from transgenic parasites expressing wild type or mutated SAG1-ty-TM-CTD chimeras (an additional blot can be seen on Figure S2). pMS1tyMIC16TM-CTD is detectable as a processed form that is no longer detected in the mutant chimera, pMS1tyMIC16mTM-CTD, when the putative rhomboid cleavage site AGGI was mutated to VVLV. The size difference between the processed and non-processed forms suggests that this cleavage is occurring downstream of the Ty-1 epitope within the TMD (Figure 5B). Similarly, when the motif IAGG in pMS1tyMIC8TM-CTD was mutated to IILV in pMS1tyMIC8mTM-CTD, there was a change in the migration pattern, compatible with the occurrence of a proteolytic cleavage downstream of the Ty-1 epitope, within the putative cleavage motif (Figure 5C). In sharp contrast to all the other rhomboid cleavage sites identified to date in apicomplexan substrates, this potential cleavage motif lies close to the cytoplasmic side of the TgMIC8 TMD (Figure 1A).
Expression of pMS1tyMIC8.2TM-CTD led to the generation of two products suggesting that the protein undergoes proteolytic maturation (Figure 5D). The smaller product shows the same migration behaviour on SDS page as the intramembrane cleavage product observed for pMS1tyMIC16TM-CTD and other fusions (Figure S2), suggesting that the cleavage occurs within or close to the TMD but there is no recognizable rhomboid cleavage site in the TMD of TgMIC8.2 and therefore we could not test for rhomboid cleavage. pAS1ty1AMA1TM-CTD was mainly detected in the DGs and was subject to proteolysis at a site compatible with intramembrane cleavage as determined by western-blot (Figures 4 and 5E). In fact shedding of TgAMA1 from the parasite surface, during invasion, was previously reported to occur by proteolytic cleavage at a precise site within the TMD (Howell et al., 2005, Buguliskis et al.).
These results indicate that pAS1tyAMA1TM-CTD, pMS1tyMIC8TM-CTD and pMS1tyMIC16TM-CTD, are cleaved likely by a rhomboid protease at the plasma membrane.
Several TM-MICs bind to aldolase
To determine whether other TM-MICs besides TgMIC2 can interact with aldolase, we examined the ability of bacterially expressed GST-CTD fusions to bind to aldolase by in vitro pull-down assays. Purified recombinant rabbit aldolase was used as source of aldolase and GST-MIC2CTD and GST alone served as positive and negative controls, respectively. The sequences of the TM-MICs used to generate the GST-fusions are listed in Figure S3. The experiment was repeated several times using independent purifications of each GST fusion and reproducibly showed that GST-MIC8.2CTD and GST-MIC16CTD were unable to bind to aldolase. In contrast, significant binding was monitored with GST-MIC6CTD, GST-MIC12CTD and GST-AMA1CTD (Figure 6A), confirming previous results with TgMIC6 (Zheng et al., 2009). In the case of GST-MIC8CTD, no conclusions could be taken regarding binding, due to aberrant migration of the protein on the gel, possibly result of protein un-stability.
Figure 6. In vitro aldolase binding assay.
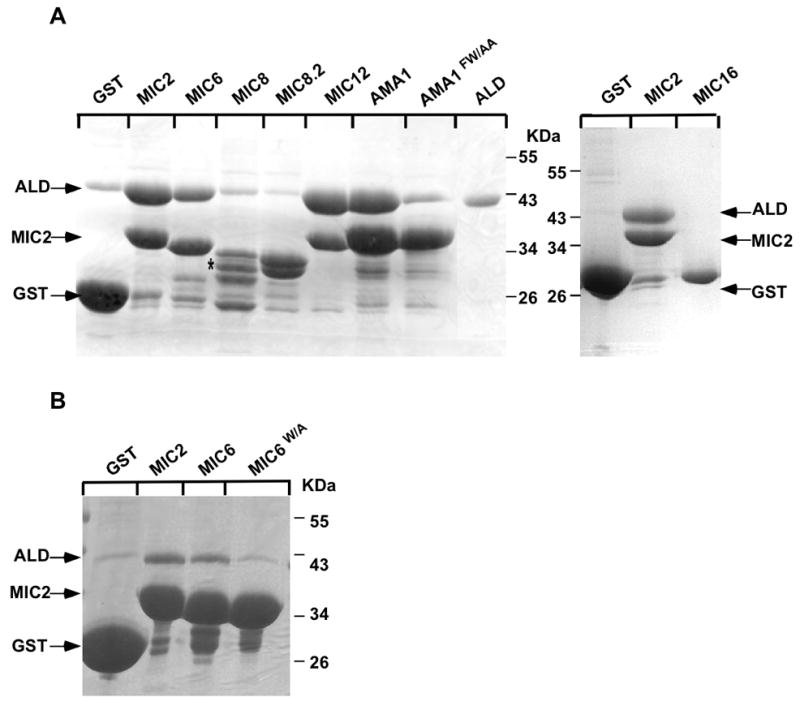
A. GST-pull-down assays were performed with GST alone or with the GST fusion constructs GST-MIC2CTD, GST-MIC6CTD, GST-MIC8CTD, GST-MIC8.2CTD, GST-MIC12CTD, GST-AMA1CTD and aldolase alone. GST-MIC16CTD and GST-MIC6W/ACTD were tested separately with GST-MIC2CTD. Note that GST-MIC8CTD migration is aberrant, as indicated by an asterisk.
B. GST-pull-down assays were performed with GST alone or with the GST fusion constructs GST-MIC2CTD, GST-MIC6CTD, GST-MIC6W/ACTD and aldolase alone. SDS-page gel were stained with coomassie-blue. The migration of aldolase, GST and GST-MIC2CTD is indicated by arrows.
It is known that mutation of the conserved tryptophan residue at the C-terminus of PbTRAP, PfTRAP, PfTLP and TgMIC2 abrogates interaction with aldolase (Buscaglia et al., 2003, Heiss et al., 2008, Jewett & Sibley, 2003). TgMIC6 possesses a tryptophan residue in the same position as the one in TgMIC2 (Figure 1B), suggesting that this is the residue responsible for binding to aldolase. Indeed a GST-MIC6mCTD, in which F349 was replaced by an alanine residue (MIC6W/A) (Figure S3), showed a significant reduction in binding to aldolase (Figure 6B). Although there is not a tryptophan residue at the extreme C-terminus of TgAMA1, site-directed mutagenesis was performed to mutate F519W520, which is more distal to the C-terminus but represents a highly conserved motif in all apicomplexan AMA1 proteins and precedes a stretch of acidic residues in the TgAMA1 CTD (Figure S3). Intriguingly, the replacement of F519W520 by AA (AMA1FW/AA) led to a significant reduction in the binding of GST-AMA1mCTD to aldolase (Figure 6A).
Mutations in the CTD of TgAMA1 that block binding to aldolase inhibit invasion
The availability of mutant parasite strains in which the TgMIC6 and TgAMA1 genes have been disrupted by double homologous recombination offered the opportunity to examine the importance of the tryptophan residue in TgMIC6 and TgAMA1 for invasion (Reiss et al., 2001, Mital et al., 2005).
A mutant of TgMIC6, TgMIC6W/A-Ty, was generated in which the residue W348, lying in a similar position as the tryptophan residue involved in TgMIC2 binding to aldolase, was converted to an alanine residue (Figure 1B). TgMIC6-Ty and TgMIC6W/A-Ty expressing vectors were used to complement the mic6ko strain and the resulting proteins were shown to localize to the micronemes (Figure 7A). Given that TgMIC6 is acting as escorter, in the absence of the protein, the soluble partners of its adhesive complex, TgMIC1 and TgMIC4, are mistargeted to the DGs and hence unable to participate in the invasion process (Reiss et al., 2001). In consequence, the mic6ko mutant is virtually comparable to a triple-knockout of TgMIC6, TgMIC1 and TgMIC4 (Reiss et al., 2001), in a situation parallel to the mic1ko strain, where TgMIC4 and TgMIC6 fail to traffic to the micronemes. Consistent with the invasiveness of mic1ko (Cerede et al., 2005), mic6ko shows about a 50% reduction of invasion efficiency compared to the RH-2YFP strain, which was used as an internal standard for parasite fitness (Figure 7B). Complementation of mic6ko with either MIC6Ty or MIC6W/ATy restored the invasion phenotype to a level comparable to wild type level. Gliding assays showed that mic6ko parasites are not defective in gliding and, as expected, the MIC6W/ATy complemented parasites also glide normally (Figure 7C). These results suggest that the residue W348, and therefore aldolase binding, is not critical for the function of the TgMIC4-MIC1-MIC6 complex during invasion.
Figure 7. Invasion and gliding assays of mic6ko and complemented strains. The penultimate tryptophan residue in the CTD of TgMIC6 is not critical for productive invasion.

A. IFA documented by confocal microscopy of intracellular parasites deficient in TgMIC6 (mic6ko) and complemented with TgMIC6-Ty (mic6ko/MIC6Ty) or MIC6W/ATy (mic6ko/MIC6W/ATy). Co-localization with the micronemal marker TgMIC4 was assessed using antibodies anti-TgMIC4 (red) and anti-Ty-1 (green). Scale bars indicate 5μm.
B. Quantification of the relative invasion efficiency of the four parasite strains as determined by a cell invasion assay normalized to RH-YFP strain co-cultivated and used as internal standard for parasite fitness. Error bars indicate standard deviations.
C. Gliding assays of mic6ko and mic6ko/MIC6W/ATy parasites. The trails were stained with anti-SAG1. The arrow indicates a trail.
To study whether the residues F519W520 contribute to the function of the TgAMA1 during invasion we used a previously reported TgAMA1 conditional knockout parasite (ama1koi; Mital et. al., 2005), in which the expression of wild type (myc-tagged) TgAMA1 can be controlled by the addition of anhydrotetracycline (ATc). In the absence of ATc, AMA1myc is expressed in these parasites and they are fully invasive; in the presence of ATc, AMA1myc expression is repressed and the parasites are severely defective in invasion (Mital et al., 2005). The ama1koi parasites were transfected with plasmids encoding Flag-tagged wild type or mutant TgAMA1 (AMA1WTFlag and AMA1FW/AAFlag, respectively), and independent clones expressing similar levels of AMA1WTFlag and AMA1FW/AAFlag in the presence of ATc were isolated. Both the wild type and mutant proteins localized to the apical end of the parasite, as shown by co-localization with M2AP, indicating proper localization (Figure 8A and data not shown). While AMA1WTFlag was able to complement the ATc-induced invasion defect in the ama1koi parasites, AMA1FW/AAFlag was not (Figure 8B). These data demonstrate that the hydrophobic residues F519W520 within the CTD of TgAMA1 are essential for both aldolase binding and host cell invasion.
Figure 8. Invasion assay of TgAMA1 depleted parasites and complemented strains. Residues F519W520 within the CTD of TgAMA1 are critical for productive invasion.

A. IFA analysis of AMA1FW/AAFlag expressed in the AMA1 conditional knockout (ama1koi) parasites shows proper colocalization of the mutant protein with the microneme marker protein, M2AP. Two independent AMA1FW/AAFlag-expressing clones are shown (AMA1FW/AAFlag-1 and -2), as are the corresponding DIC images (left panels). Scale bars indicate 5 μm.
B. Invasion assay. Host cell invasion by AMA1Flag, AMA1FW/AAFlag-1, AMA1FW/AAFlag-2, and ama1koi parasites, each grown in the presence of ATc for 36 h, was measured 1 h postinfection using the laser scanning cytometer-based assay. The average number of intracellular parasites per scan area is presented (two independent experiments, two replicates within each experiment), with error bars representing the SD of the mean between experiments. The invasion levels for each parasite population (expressed relative to the AMA1Flag parasites) are shown. The total number of intracellular parasites counted is also listed below each sample. * p < 0.05, relative to AMA1Flag.
Discussion
MIC complexes serve essential roles during host cell invasion, by mediating parasite attachment, MJ formation and bridging of the host cell receptors to the actomyosin system, hence promoting gliding and invasion. The smooth transition through the various steps of the invasion process requires a high level of coordination not only between the different MIC complexes but also between each component of a given complex. The TM-MICs, in particular are multitasks and execute distinct functions that are specified by their modular design. The ectodomains, on one hand, recruit microneme or rhoptry proteins to the complex and, in several instances also interact directly with host cell receptors; and the TM-CTDs, on the other hand, contribute to targeting, proteolytic shedding and connection to the actomyosin system of the parasite. TgMIC2 and other members of the TRAP family are suited to carry out these multiple tasks (Morahan et al., 2009).
In this study, we have investigated and compared to TgMIC2, the biological properties of the TM-MICs associated with the three other major MIC complexes known to be involved in invasion, as well as of TgMIC12, TgMIC8.2 and TgMIC16, whose functions remain to be established.
Targeting to the micronemes, as demonstrated for the rhoptries (Ngo et al., 2003, Richard et al., 2009), resembles post-TGN targeting in other eukaryotes (Sheiner & Soldati-Favre, 2008). Complexes of soluble and TM-MICs are formed in the ER and travel through the secretory pathway until they are finally secreted (Huynh et al., 2003, Reiss et al., 2001). Some TM-MICs have been shown to act as escorters, implying that their CTDs are recognized by components of the vesicular sorting machinery (Meissner et al., 2002). Consistent with this idea, two micronemal targeting motifs, SYHYY and EIEYE, were identified in TgMIC2 (Di Cristina et al., 2000). The apparent absence of such motifs in the CTDs of TgAMA1, TgMIC16 and the TgMIC8 family members is in agreement with the findings here that the respective SAG1-ty-TM-CTD chimeras fail to traffic to the micronemes. Consequently, these TM-MICs do not function as escorters and are likely to interact, via their ectodomains, with other proteins that carry a determinant for micronemal targeting. Consistent with this hypothesis, the chimera the chimera MIC8 fused to the CTD of P. berghei TRAP localizes to the micronemes (Kessler, 2008) although the PbTRAP TM-CTD does not confer trafficking to micronemes in T. gondii (Di Cristina et al., 2000). Similarly, the refined analysis of TgAMA1 clearly established that it is the ectodomain of the protein that carries the necessary traffic information to the micronemes. Studies on AMA1 in P. falciparum led to the same conclusion (Treeck et al., 2009, Treeck et al., 2006, Healer et al., 2002). These observations imply that TgAMA1, TgMIC8, TgMIC8.2 and TgMIC16 may belong to complexes that are composed of more than one type of TM-MIC.
These observations imply that TgAMA1, TgMIC8, TgMIC8.2 and TgMIC16 may belong to complexes that are composed of more than one type of TM-MIC.
The SAG1-ty-TM-CTDs chimeras localized either to the DGs and PV (TgMIC8, TgMIC16 and TgAMA1), or were retained in the Golgi (TgMIC8.2). An alignment of the TM-CTDs of the selected TM-MICs predicted the presence of a rhomboid cleavage motif similar to IAGG in the TMDs of TgMIC2, TgMIC6, TgMIC12, TgAMA1 and TgMIC16. An IAGG motif is also present within the TMD of TgMIC8, but it is significantly shifted within the TMD, closer to the CTD. No apparent rhomboid cleavage site signature could be identified in the TMD of TgMIC8.2. Consistent with cleavage at the rhomboid cleavage motif, proteolytic processing at the expected position was observed for pMS1tyMIC8TM-CTD, pMS1tyMIC16TM-CTD and pAS1tyAMA1TM-CTD chimeric proteins. To provide further evidence for intramembrane processing by a rhomboid, point mutations were introduced in the identified cleavage motifs. The majority of the mutations introduced abrogated processing and thus provided strong evidence that the chimeras are cleaved within their TMD by a rhomboid-like protease. Interestingly, while TgMIC8 appears to be cleaved by a rhomboid, this proteolysis occurs much closer to the cytoplasmic region of the TMD than in all the other TM-MICs. This may allow the direct release of the CTD into the cytoplasm, where it can initiate a signaling cascade, as previously proposed (Kessler, 2008). Given the absence of a recognizable rhomboid cleavage motif in the TMD of TgMIC8.2, we could not assess the nature of the processing event. However, the cleavage product runs at a size compatible with intramembrane processing and TgMIC8.2-CTD can replace that of TgMIC8 (Kessler, 2008), which is susceptible to intramembrane cleavage. It is consequently still plausible that the TgMIC8.2-chimera is as well processed within the TMD.
Preventing the proteolytic cleavage by mutagenesis had an anticipated impact on the localization of the SAG1-ty-TM-CTD chimeras. All the mutant chimeras showed a dramatic change in subcellular localization, accumulating at the parasite’s surface, indicative of cleavage abrogation. In a prior study, a similar accumulation at the parasite plasma membrane was observed for the uncleaved SAG1TgMIC12TM-CTD mutant, which was mistargeted to the DGs (Opitz et al., 2002). This observation suggested that MPP1 is a constitutively active rhomboid-like protease at the plasma membrane of the parasite. We cannot discriminate between cleavage of the SAG1-ty-TM-CTD constructs at the plasma membrane or inside the parasite, but it is likely that TgMIC16 and TgMIC8 fusion constructs are cleaved by MPP1, because the corresponding non-cleaved mutants accumulate at the parasite’s surface. The unambiguous assignment of each of the TM-MIC substrates to a given protease awaits further investigations.
Several Plasmodium TM-MIC proteins have been reported to interact with the F-actin binding protein aldolase and this way bridge the host cell surface with the actomyosin motor of the parasite. These proteins share the structural features characteristic of TRAP, namely an N-terminal secretion signal, a van Willebrand A-domain, one or more TSR domains, a TMD with a rhomboid-cleavage motif and an acidic CTD with a unique tryptophan residue close to the C-terminus (Morahan et al., 2009). In T. gondii, TgMIC6 and TgMIC2 are the only TM-MICs shown to bind to aldolase (Jewett & Sibley, 2003, Starnes et al., 2006, Zheng et al., 2009), in a model compatible TgMIC2 redistribution along the parasite’s surface upon invasion (Carruthers & Sibley, 1999) and demonstrated role in motility and invasion (Huynh & Carruthers, 2006). The patches of acidic amino acids constituting the aldolase binding site within the TgMIC2 CTD (Starnes et al., 2006) are not strictly conserved in the TgMIC6, TgMIC12 and TgAMA1 CTDs (Figure S4) but as shown in this study, these proteins are able to bind to aldolase in an in vitro pull down assay. This suggests that the composition in acidic amino acids and their precise location within the CTD can accomodate a level of variation. The second prominent feature of aldolase binding is the presence of a conserved tryptophan residue at the extreme C-terminus of the CTD. All the TM-MICs studied here possess this residue except TgAMA1 and TgMIC16, and as shown by mutation of the residue in TgMIC6, the residue mediates binding to aldolase. Although the TgMIC8 family members possess a tryptophan residue at the extreme C-terminus, the acidic patch is absent and none of these CTDs bind to aldolase in the in vitro assay. This is in accordance with functional analysis showing no motility defect in TgMIC8 depleted parasites (Kessler, 2008). In contrast, TgAMA1 is able to bind to aldolase without an extreme C-terminal tryptophan, although TgAMA1 does contain a tryptophan just N-terminal to a patch of acidic residues (W520; Figure S4) within an FW motif that is well conserved among AMA1 homologues in other Apicomplexans (Hehl et al., 2000). Mutation of this FW motif in TgAMA1 disrupted both aldolase binding and invasion. This result suggests that TgAMA1 serves as a bridging protein that physically connects the glideosome (via its CTD) to other components of the MJ complex (via its ectodomain) and thus plays a critical role in the posterior translocation of the MJ complex during invasion. However, during invasion the majority of the TgAMA1 is not restricted to the MJ but is found over the parasite’s surface (Alexander et al., 2005, Howell et al., 2005). This suggests that there may be two pools of TgAMA1 at the parasite surface, one of which is bound to aldolase and is responsible for anchoring the MJ complex to the actomyosin motor. Whether the remaining fraction of TgAMA1 serves a distinct function or is simply available to be recruited to the MJ is unknown, but is it possible that the two pools of TgAMA1 are distinguished by different post-translational modifications of their CTDs, such as phosphorylation (Treeck et al., 2009). Disappointingly, our repeated efforts to monitor TgAMA1, or even TgMIC2, interaction with aldolase in the parasite by co-immunoprecipitation were unfruitful.
The extreme C-terminus of TgMIC12 shares a nearly strictly conserved amino acid sequence with TgMIC2, and this reflects its comparable propensity to bind to aldolase in pull down experiments. Given the fact that no functional data are available to date on TgMIC12, the physiological relevance of these observations is not known.
TgMIC6 binds less efficiently to aldolase compared to the CTDs of TgMIC2 or TgMIC12, and this can simply reflect the fact that fewer acidic residues are present at its extreme C-terminus. TgMIC6 wild type or TgMIC6 carrying a W348/A mutation are both able to functionally complement the invasion defect in parasites depleted of TgMIC6, suggesting that the tryptophan residue may not be crucial for the function of the TgMIC1-MIC4-MIC6 complex. Consistent with these findings mic6ko showed not defect in gliding motility, suggesting that the function of TgMIC1-MIC4-MIC6 complex in invasion might be assisted via the formation of a macrocomplex by another TM-MIC that connects to the actomyosin system.
It remains to be determined for TgMIC12, if aldolase plays a role in its functions in vivo and if it reflects a direct interaction with the actomyosin motor or another biological role. Taken together, there is an excellent correlation between the predictions made from sequence analysis and the three biological properties examined experimentally: presence of trafficking determinants, susceptibility to rhomboid protease cleavage and binding to aldolase. Moreover the assessed properties of the TM-MICs are in good accordance with their functional contribution to the individual steps of the invasion process (Table 1).
Table 1.
| TM-MIC | MIC2 | MIC12 | MIC6 | MIC8 | MIC8.2 | AMA1 | MIC16 |
|---|---|---|---|---|---|---|---|
| Role in motility and invasion | Motility, Host cell binding | ND | Host cell binding | Rhoptry secretion | CTD complements MIC8 | MJ formation, Rhoptry secretion | ND |
| Trafficking determinant | ++ (Di Cristina et al., 2000) |
+ (Opitz et al., 2002) |
+ (Reiss et al., 2001) |
- TS |
- TS |
- TS |
- TS |
| Binding to aldolase | + (Buscaglia et al., 2003, Jewett & Sibley, 2003, Heiss et al., 2008) |
+ TS |
+ TS |
- TS |
- TS |
+ TS |
- TS |
| Substrate for rhomboid cleavage | + (Opitz et al., 2002, Brossier et al., 2003, Urban & Freeman, 2003, Brossier et al., 2005, Dowse et al., 2005) |
+ (Opitz et al., 2002, Brossier et al., 2003, Urban & Freeman, 2003, Brossier et al., 2005, Dowse et al., 2005) |
+ (Opitz et al., 2002, Brossier et al., 2003, Urban & Freeman, 2003, Brossier et al., 2005, Dowse et al., 2005) |
+ TS |
- TS |
+ (Howell et al., 2005) |
+ TS |
TS – this study; ND - not determined.
Material and Methods
Reagents and parasite culture
Restriction enzymes were purchased from New England Biolabs and secondary antibodies for western blots and IFA from Molecular Probes. T. gondii tachyzoites (RH strain wild-type and RHhxgprt−) were grown in human foreskin fibroblasts (HFF) or Vero cells in Dulbecco’s Modified Eagle’s Medium (DMEM, GIBCO) supplemented with 5% fetal calf serum (FCS), 2mM glutamine and 25 μg/ml gentamicin. ama1koi parasites were cultured in DMEM supplemented with 1% fetal bovine serum, 25μg/ml mycophenolic acid, 50μg/ml xanthine, and 6.8μg/ml chloramphenicol (Mital et. al., 2005). Downregulation of AMA1-myc expression in intracellular ama1koi parasites was achieved by incubation of infected cells for 36 hr in medium containing 1,5μg/ml anhydrotetracycline (ATc, Clontech).
Cloning of DNA constructs
For determination of MIC16 localization in T. gondii, the full-length gene was amplified from tachyzoite cDNA by PCR using the primers 1969 and 1971 (Supplementary table 1). The PCR product was purified, digested with EcoR1 and NsiI, and cloned into the corresponding sites in pTUB8Ty (Meissner et al., 2002).
For expression in T. gondii as N-terminal SAG1-Ty fusions, DNA fragments coding for the TMD and CTD of MIC8, MIC8.2 and MIC16 were amplified from tachyzoite cDNA by PCR using the primers 1887 and 1888, 1889 and 1890, 1970 and 1972 (supplementary table 1). MIC8TMCTD, MIC8.2TMCTD and MIC16TMCTD were digested with SalI and PacI and each fragment was cloned into the corresponding sites in pMSAG1Ty vector, which drives expression under control of the TgMIC2 promoter, originating pMSAG1tyMIC8TMCTD, pMSAG1tyMIC8.2TMCTD and pMSAG1tyMIC16TMCTD (Di Cristina et al., 2000).
For expression of the different AMA1 constructs under control of its endogenous promoter in RH strain T. gondii, DNA fragments coding for TMD and CTD, the full length protein, only the ectodomain (aminoacids 1-456) or the ectodomain and TMD (aminoacids 1-479) were amplified from tachyzoite cDNA by PCR using the primers 1079 and 1080, 2225 and 1080, 2247 and 2249 and 2247 and 2248, respectively (Supplementary table 1). The three last pair of primers added a 8His tag immediately after amino acid 25. AMA1TMCTD was digested with XhoI and PacI and cloned in pMSAG1Ty vector, originating pMSAG1tyAMA1TMCTD, and the other PCR products were digested with Nsi1 and Pac1 and cloned into the corresponding sites in pROP1 vector, originating pROPhisAMA1, pROPhisAMA1ΔTM-CTD and pROPhisAMA1ΔCTD (Soldati et al., 1998). The AMA1 promoter was amplified with primers 2462 and 2463 and cloned between Kpn1 and Nsi sites in the pROP1 vectors, originating pAhisAMA1, pAhisAMA1ΔTM-CTD and pAhisAMA1ΔCTD, or between Kpn1 and Nsi1 sites in the pMSAG1Ty vector expressing AMA1 TM and CTD, originating pAS1tyAMA1TM-CTD. Generation of plasmid pSK+A/AMA1-Flag for expression of Flag-tagged TgAMA1 in the ama1koi parasites has been described elsewhere (Parussini et al, submitted).
For bacterial expression, DNA fragments corresponding to TM-CTDs of TgMIC2, TgMIC12, TgMIC8, TgMIC8.2, TgMIC6, TgMIC6W/A, TgAMA1 and TgMIC16 were amplified from tachyzoite cDNA by PCR using the primers 176 and 177, 1599 and 710, 325 and 326, 1832 and 1833, 211 and 212, 211 and 3100, 1948 and 1949 and 2001 and 2002, respectively (supplementary table 1). PCR products were purified using the Easy Pure-DNA Purification Kit (Biozym). MIC2TMCTD, MIC6TMCTD and MIC6W/ATMCTD were digested with EcoRI and SalI, MIC12 TMCTD and AMA1 TMCTD with EcoRI and XhoI, MIC8TMCTD with BamHI and MIC8.2TMCTD and MIC16TMCTD with BamHI and XhoI. Each fragment was cloned into the corresponding sites in the pGEX4T1 vector to generate N-terminal GST fusions.
Mutated constructs
To mutate the motif FW to AA on the AMA1CTD of the plasmid pGEX4T1-AMA1TMCTD the primers 1830 and 1831 were used in a site-directed mutagenesis reaction using the commercial QuikChange II Site-Directed Mutagenesis Kit (Stratagen) according to the manufacturer’s instructions. Similarly, the residues AGG, YTG, AG or AGGI were mutated to ILV, VL or VVLV in the TMDs of MIC8, MIC8.2 and MIC16 in the plasmids pMSAG1tyMIC8TMCTD, pMSAG1tyMIC8.2TMCTD and pMSAG1tyMIC16TMCTD, respectively, using the primers 2003 and 2004, 2005 and 2006 or 2226 and 2227, respectively. To mutate F519W520 to AA on the AMA1 cytosolic tail in pSK+A/AMA1-Flag, primers TgAMA1FW/AA.f and TgAMA1FW/AA.r (Supplementary Table 1) were used for site directed mutagenesis as described above, generating the vector pSK+A/AMA1FW/AAFlag.
Protein expression and purification
pGEX-4T1 vectors encoding the MIC2, MIC12, MIC6, MIC8, MIC8.2, MIC16 or AMA1 TMCTD GST fusion proteins were transformed into the E. coli BL21 strain (Novagen, Madison, WI). Protein expression was induced using 1mM isopropyl-beta-d-thiogalactopyranoside (IPTG) for four hours at 37 °C. Bacterial pellets were resuspended in 1X PBS supplemented with 1mg/ml of Lysozym, 10μg/ml DNAse, 20μg/ml RNAse and 1mM PMSF, and were allowed to homogenize for 30 minutes at 4 °C, following which, cells were disrupted by 5 consecutive cycles freeze/thaw. After centrifugation (30 minutes, 30 000RPM), the supernatant containing the soluble GST-fusions was collected, and purified using GSH-beads (Glutathione Sepharose 4 Fast Flow, Amersham) according to the manufactures advice.
GST-Fusion protein pull-down experiment
Glutathione-sepharose beads were incubated with 0.5mg of GST fusion proteins or GST for 1 hr at 4 °C. Beads were washed twice with PBS and once with buffer XB (50mM KCL, 20mM Hepes, 2mM MgCL2, 0.2mM EDTA, 0.2% Tween2, pH7.7), prior to the addition of approximately 400μg of recombinant aldolase (Sigma). Aldolase was incubated with the GST-fusion purified proteins for 4 hours at 4 °C, washed five times with buffer XB, and bound proteins were eluted using SDS–PAGE-loading buffer supplemented with 100mM DTT.
Parasite transfection and selection of clonal stable lines
Parasites transfection was performed by electroporation as previously described (Soldati & Boothroyd, 1993). The HXGPRT gene was used as a positive selectable marker in the presence of mycophenolic acid (25 mg/ml) and xanthine (50 mg/ml) as described previously (Donald et al., 1996). Briefly, freshly released parasites (5×107) of the RHhxgprt strain were resuspended in cytomix buffer in the presence of 50–80μg of linearized plasmid carrying the selectable marker gene and the expression cassette containing the DNA sequences. Transfection of ama1koi conditional knockout parasites was carried out by electroporation using 2.6×107 freshly released parasites, 100 μg of pSK+A/AMA1-Flag or pSK+A/AMA1FW/AAFlag, and 6 μg of pDHFR*.Tsc3ABP (Roos et. al., 1997) plasmids. Parasites were electroporated at 2 kV, 25 mF, 48 V using a BTX electroporator (Harvard biosciences, Holliston, MA, USA) before being added to a monolayer of HFF cells in the presence of mycophenolic acid/xanthine. Selection with 1μM pyrimethamine was initiated 24 hr later and continued for 7–10 days, after which resistant clones were isolated by limiting dilution.
Western blotting
For AMA1 Western blots, 108 parasites were harvested after complete lysis of the host cell monolayer and extracted for 30 min at 4 °C in 1 ml of RIPA buffer (50 mM Tris-HCl, pH 7.5; 1 % (vol/vol) Triton X-100; 0.5 % (vol/vol) sodium deoxycholate; 0.2 % (wt/vol) SDS; 100 mM NaCl2; 5 mM EDTA) containing protease inhibitors (Sigma P8340), added directly to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer and boiled for 10 min. For all other samples, extracts from 2 × 107 parasites were prepared in 1x PBS by five consecutive freeze/thaw cycles with intermediate homogenization, following two consecutive sonications, and the suspension was boiled in SDS–PAGE loading buffer containing 100mM DTT. SDS-PAGE was performed using standard methods. Separated proteins were transferred to a nitrocellulose membrane using a semidry electroblotter. Western blots were performed using anti-Ty1 mAb (Bastin et al., 1996), anti-AMA1 mAb (B3.90, Donahue et. al., 2000), anti-myc mAb (Mital et al, 2005) and anti-Flag mAb (Sigma F3165) in 5% non-fat milk powder in 1X PBS. As secondary antibody, a peroxidase-conjugated goat anti-mouse or anti-rabbit antibody was used (Molecular Probes, Paisley, UK). Bound antibodies were visualized using either the ECL system (Amersham Corp) or with SuperSignal™ West Pico chemiluminescent substrates (Pierce).
IFA and confocal microscopy
All manipulations were carried out at room temperature. Intracellular parasites grown in HFF seeded on glass coverslips were fixed with 4% paraformaldehyde (PFA) for 10 minutes or 4% PFA-0.002% Glutaraldehyde for 10 minutes. Following fixation, slides were quenched in 1X PBS-0.1M glycine. Cells were then permeabilized in 1X PBS-0.2 % Triton-X-100 (PBS/Triton) for 20 minutes and blocked in the same buffer supplemented with 2% FCS (PBS/Triton/FCS). Slides were incubated for 60 minutes with the primary antibodies anti-GAP45, anti-MIC4 (Brecht et al., 2001) anti-Myc, anti-GRA3 (kindly provided by JF Dubremetz) or anti-Ty1 diluted in PBS/Triton/FCS, washed and incubated for 40 minutes with Alexa488- or Alexa594-conjugated goat anti-mouse or goat anti-rabbit IgGs diluted in PBS/Triton/FCS. DAPI staining was performed with a concentration of 0.1mg-DAPI/mL 1XPBS for 5 minutes incubation before slides were mounted in Fluoromount G (Southern Biotech) and stored at 4°C in the dark. Micrographs were obtained on a Zeiss Axioskop 2 equiped with an Axiocam color CCD camera. Images where recorded and treated on computer through the AxioVision™ software. Confocal images were collected with a Leica laser scanning confocal microscope (TCS-NTDM/IRB) using a 63 Plan-Apo objective with NA 1.40. Optical sections were recorded at 250 nm per vertical step and four times averaging.
Cell invasion assays
Comparison of different T. gondii strains for invasion efficiency was done using an RH-2YFP strain as internal standard. A confluent 60mm-dish of human foreskin fibroblasts was heavily infected with a mixture of the strain of interest and RH-2YFP parasites. Some hours later the dish was washed to remove any non-invaded parasites. Two days later parasites egressed from their host cells and were collected by centrifugation at 240g, RT for 10 minutes and resuspended in 5ml culture medium (DMEM complemented with 2mM L-Glutamine, 5% FCS and 25μg/ml Gentamicin) preheated to 37°C. From this suspension a 1:10 dilution was made in preheated medium, the ratio of non-YFP to YFP parasites was determined in a Neubauer chamber (ratio between 0.8 and 7) and 500μl were inoculated into a well on a 24-well IFA plate. Invasion was allowed to take place for 1h at 37°C, then the wells were washed two times in CM-PBS (1 mM CaCl2, 0.5 mM MgCl2 in PBS) and refilled with fresh medium. The plate was incubated for another 24–32 hours in order for the parasites to devide. Afterwards cells were fixed with 4% paraformaldehyde for 20 min, followed by 3 min incubation with 0.1 M glycine in PBS to quench the reaction and subjected to an indirect immunofluorescence assay (IFA). Fixed cells were permeabilised with 0.2% Triton-X100 in PBS for 20 min and blocked in 2% bovine serum albumin, 0.2% Triton-X100 in PBS for 20 min. The cells were then stained with rabbit anti-TgMLC antibodies followed by Alexa 594 goat anti-rabbit antibodies (Molecular Probes). Total number of parasite vacuoles and RH-2YFP parasite vacuoles were counted on 20 microscopic fields on each IFA-slide with a minimum of 750 vacuoles in total per slide. Only vacuoles containing at least two parasites were counted. The ratio of YFP to non-YFP vacuoles was calculated and compared to the ratio obtained from live parasites at the beginning of the experiment. Each experiment has been repeated six times.
Alternatively, host cell invasion was measured using a laser scanning cytometer-based assay (Mital et al., 2006). Briefly, parasites grown for 36 hr in the presence of ATc were harvested, added to HFF monolayers and incubated at 37°C. One hr post-infection, the coverslips were fixed, blocked, and labeled with an anti-SAG1 antibody (mAb GII-9; Argene, North Massapequa USA) followed by an R-phycoerythrin-conjugated secondary antibody (“orange,” DAKO, Carpenteria USA). Samples were then permeabilized, blocked, and labeled with anti-SAG1 followed by an Alexa647-conjugated secondary antibody (“red,” Molecular Probes). Samples were analyzed on a CompuCyte Laser Scanning Cytometer equipped with a BX50 upright fluorescence microscope (Olympus America, Melville USA), 20X objective (N.A. 0.5), argon ion (488 nm) and helium/neon (633 nm) lasers, and three filter blocks/photomultiplier tubes (530–555 nm [green], 600–640 nm [orange], and 650 nm [long-red]). Data were acquired and analyzed using Wincyte 3.4 Software (CompuCyte, Cambridge USA). Red parasites were counted to determine the total number of parasites per field. The number of orange, extracellular parasites was counted and subtracted from the total to calculate the number of invaded parasites per field. One-way ANOVA and Dunnett’s Multiple Comparison post-test were used to determine the significance of differences between groups. P values of less than 0.05 were considered significant.
Gliding motility assay
Freshly egressed tachyzoites were filtered, pelleted, and resuspended in Calcium-Saline containing 1μM of ionomycin. The suspension was deposited on coverslips previously coated with Poly-L-Lysine (3 hr at RT). Parasites were fixed with PAF/GA and IFA using the α-SAG1 antibody was performed to visualize the trails.
Supplementary Material
Acknowledgments
We thank Anne Kelsen for providing expert technical support. This work is part of the activities of the BioMalPar European Network of Excellence supported by a European grant (LSHP-CT-2004-503578) from the Priority 1 “Life Sciences, Genomics and Biotechnology for Health” in the 6th Framework Program, from supports to DS by the Swiss National Foundation and the Howard Hughes Medical Institute. Additional funding was provided by USPHS grant AI063276 (GEW). JMS is a recipient of the EU-funded Marie Curie Action MalParTraining (MEST-CT-2005-020492), “The challenge of malaria in the post-genomic Era”.
Abbreviations
- IFA
Indirect immunofluorescence assay
- ROM
Rhomboid protease
- MIC
microneme protein
- TM-MIC
type-I single transmembrane MIC
- ADL
aldolase
- CTD
cytoplasmic C-terminal domain
- TMD
transmembrane domain
- DG
dense granules
- RON
rhoptry neck protein
- ROP
rhoptry bulb protein
- TSR
thrombospondin repeat
- TRAP
thrombospondin-related anonymous protein
- TGN
trans-Golgi network
References
- Alexander DL, Mital J, Ward GE, Bradley P, Boothroyd JC. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 2005;1:e17. doi: 10.1371/journal.ppat.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P, Bagherzadeh Z, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Molecular and biochemical parasitology. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- Besteiro S, Michelin A, Poncet J, Dubremetz JF, Lebrun M. Export of a Toxoplasma gondii rhoptry neck protein complex at the host cell membrane to form the moving junction during invasion. PLoS pathogens. 2009;5:e1000309. doi: 10.1371/journal.ppat.1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenschein TM, Friedrich N, Childs RA, Saouros S, Carpenter EP, Campanero-Rhodes MA, Simpson P, Chai W, Koutroukides T, Blackman MJ, Feizi T, Soldati-Favre D, Matthews S. Atomic resolution insight into host cell recognition by Toxoplasma gondii. EMBO J. 2007;26:2808–2820. doi: 10.1038/sj.emboj.7601704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht S, V, Carruthers B, Ferguson DJ, Giddings OK, Wang G, Jakle U, Harper JM, Sibley LD, Soldati D. The toxoplasma micronemal protein MIC4 is an adhesin composed of six conserved apple domains. The Journal of biological chemistry. 2001;276:4119–4127. doi: 10.1074/jbc.M008294200. [DOI] [PubMed] [Google Scholar]
- Brossier F, Jewett TJ, Lovett JL, Sibley LD. C-terminal processing of the toxoplasma protein MIC2 is essential for invasion into host cells. J Biol Chem. 2003;278:6229–6234. doi: 10.1074/jbc.M209837200. [DOI] [PubMed] [Google Scholar]
- Brossier F, Jewett TJ, Sibley LD, Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc Natl Acad Sci U S A. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brydges SD, Harper JM, Parussini F, Coppens I, Carruthers VB. A transient forward-targeting element for microneme-regulated secretion in Toxoplasma gondii. Biol Cell. 2008;100:253–264. doi: 10.1042/BC20070076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buguliskis JS, Brossier F, Shuman J, Sibley LD. Rhomboid 4 (ROM4) affects the processing of surface adhesins and facilitates host cell invasion by Toxoplasma gondii. PLoS Pathog. 6:e1000858. doi: 10.1371/journal.ppat.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia CA, Coppens I, Hol WG, Nussenzweig V. Sites of interaction between aldolase and thrombospondin-related anonymous protein in plasmodium. Mol Biol Cell. 2003;14:4947–4957. doi: 10.1091/mbc.E03-06-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers V, Boothroyd JC. Pulling together: an integrated model of Toxoplasma cell invasion. Curr Opin Microbiol. 2007;10:83–89. doi: 10.1016/j.mib.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Carruthers VB, Sherman GD, Sibley LD. The Toxoplasma adhesive protein MIC2 is proteolytically processed at multiple sites by two parasite-derived proteases. J Biol Chem. 2000;275:14346–14353. doi: 10.1074/jbc.275.19.14346. [DOI] [PubMed] [Google Scholar]
- Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur J Cell Biol. 1997;73:114–123. [PubMed] [Google Scholar]
- Carruthers VB, Sibley LD. Mobilization of intracellular calcium stimulates microneme discharge in Toxoplasma gondii. Mol Microbiol. 1999;31:421–428. doi: 10.1046/j.1365-2958.1999.01174.x. [DOI] [PubMed] [Google Scholar]
- Cerede O, Dubremetz JF, Soete M, Deslee D, Vial H, Bout D, Lebrun M. Synergistic role of micronemal proteins in Toxoplasma gondii virulence. J Exp Med. 2005;201:453–463. doi: 10.1084/jem.20041672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Harb OS, Roos DS. In silico identification of specialized secretory-organelle proteins in apicomplexan parasites and in vivo validation in Toxoplasma gondii. PLoS ONE. 2008;3:e3611. doi: 10.1371/journal.pone.0003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristina M, Spaccapelo R, Soldati D, Bistoni F, Crisanti A. Two conserved amino acid motifs mediate protein targeting to the micronemes of the apicomplexan parasite Toxoplasma gondii. Mol Cell Biol. 2000;20:7332–7341. doi: 10.1128/mcb.20.19.7332-7341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald RG, Carter D, Ullman B, Roos DS. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. The Journal of biological chemistry. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- Dowse TJ, Pascall JC, Brown KD, Soldati D. Apicomplexan rhomboids have a potential role in microneme protein cleavage during host cell invasion. Int J Parasitol. 2005;35:747–756. doi: 10.1016/j.ijpara.2005.04.001. [DOI] [PubMed] [Google Scholar]
- El Hajj H, Papoin J, Cerede O, Garcia-Reguet N, Soete M, Dubremetz JF, Lebrun M. Molecular signals in the trafficking of the Toxoplasma gondii protein MIC3 to the micronemes. Eukaryot Cell. 2008 doi: 10.1128/EC.00413-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JM, Huynh MH, Coppens I, Parussini F, Moreno S, Carruthers VB. A cleavable propeptide influences Toxoplasma infection by facilitating the trafficking and secretion of the TgMIC2-M2AP invasion complex. Mol Biol Cell. 2006;17:4551–4563. doi: 10.1091/mbc.E06-01-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehl AB, Lekutis C, Grigg ME, Bradley PJ, Dubremetz JF, Ortega-Barria E, Boothroyd JC. Toxoplasma gondii homologue of plasmodium apical membrane antigen 1 is involved in invasion of host cells. Infect Immun. 2000;68:7078–7086. doi: 10.1128/iai.68.12.7078-7086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss K, Nie H, Kumar S, Daly TM, Bergman LW, Matuschewski K. Functional characterization of a redundant Plasmodium TRAP-family invasin, TRAP-like protein (TLP), by aldolase binding and a genetic complementation test. Eukaryot Cell. 2008 doi: 10.1128/EC.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SA, Hackett F, Jongco AM, Withers-Martinez C, Kim K, Carruthers VB, Blackman MJ. Distinct mechanisms govern proteolytic shedding of a key invasion protein in apicomplexan pathogens. Mol Microbiol. 2005;57:1342–1356. doi: 10.1111/j.1365-2958.2005.04772.x. [DOI] [PubMed] [Google Scholar]
- Howell SA, Well I, Fleck SL, Kettleborough C, Collins CR, Blackman MJ. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J Biol Chem. 2003;278:23890–23898. doi: 10.1074/jbc.M302160200. [DOI] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Toxoplasma MIC2 is a major determinant of invasion and virulence. PLoS Pathog. 2006;2:e84. doi: 10.1371/journal.ppat.0020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh MH, Rabenau KE, Harper JM, Beatty WL, Sibley LD, Carruthers VB. Rapid invasion of host cells by Toxoplasma requires secretion of the MIC2-M2AP adhesive protein complex. Embo J. 2003;22:2082–2090. doi: 10.1093/emboj/cdg217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Sibley LD. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol Cell. 2003;11:885–894. doi: 10.1016/s1097-2765(03)00113-8. [DOI] [PubMed] [Google Scholar]
- Jewett TJ, Sibley LD. The toxoplasma proteins MIC2 and M2AP form a hexameric complex necessary for intracellular survival. J Biol Chem. 2004;279:9362–9369. doi: 10.1074/jbc.M312590200. [DOI] [PubMed] [Google Scholar]
- Kessler H, Herm-Götz A, Hegge S, Rauch M, Soldati-Favre D, Frischknecht F, Meissner M. Microneme protein 8 is a new essential invasion factor in Toxoplasma gondii. Journal of Cell Science. 2008;121(3) doi: 10.1242/jcs.022350. [DOI] [PubMed] [Google Scholar]
- Meissner M, Reiss M, Viebig N, Carruthers VB, Toursel C, Tomavo S, Ajioka JW, Soldati D. A family of transmembrane microneme proteins of Toxoplasma gondii contain EGF-like domains and function as escorters. J Cell Sci. 2002;115:563–574. doi: 10.1242/jcs.115.3.563. [DOI] [PubMed] [Google Scholar]
- Mital J, Meissner M, Soldati D, Ward GE. Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Mol biol cell. 2005;16:4341–4349. doi: 10.1091/mbc.E05-04-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mital J, Schwarz J, Taatjes DJ, Ward GE. Laser scanning cytometer-based assays for measuring host cell attachment and invasion by the human pathogen Toxoplasma gondii. Cytometry A. 2006;69:13–19. doi: 10.1002/cyto.a.20202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan BJ, Wang L, Coppel RL. No TRAP, no invasion. Trends in Parasitology. 2009;25:77–84. doi: 10.1016/j.pt.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Ngo HM, Yang M, Paprotka K, Pypaert M, Hoppe H, Joiner KA. AP-1 in Toxoplasma gondii mediates biogenesis of the rhoptry secretory organelle from a post-Golgi compartment. J Biol Chem. 2003;278:5343–5352. doi: 10.1074/jbc.M208291200. [DOI] [PubMed] [Google Scholar]
- Opitz C, Di Cristina M, Reiss M, Ruppert T, Crisanti A, Soldati D. Intramembrane cleavage of microneme proteins at the surface of the apicomplexan parasite Toxoplasma gondii. Embo J. 2002;21:1577–1585. doi: 10.1093/emboj/21.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, Stern CA, Pypaert M, Sheff D, Ngo HM, Roper N, He CY, Hu K, Toomre D, Coppens I, Roos DS, Joiner KA, Warren G. Golgi biogenesis in Toxoplasma gondii. Nature. 2002;418:548–552. doi: 10.1038/nature00946. [DOI] [PubMed] [Google Scholar]
- Periz J, Ryan R, Blake DP, Tomley FM. Eimeria tenella microneme protein EtMIC4: capture of the full-length transcribed sequence and comparison with other microneme proteins. Parasitology research. 2009;104:717–721. doi: 10.1007/s00436-008-1301-0. [DOI] [PubMed] [Google Scholar]
- Rabenau KE, Sohrabi A, Tripathy A, Reitter C, Ajioka JW, Tomley FM, Carruthers VB. TgM2AP participates in Toxoplasma gondii invasion of host cells and is tightly associated with the adhesive protein TgMIC2. Mol Microbiol. 2001;41:537–547. doi: 10.1046/j.1365-2958.2001.02513.x. [DOI] [PubMed] [Google Scholar]
- Reiss M, Viebig N, Brecht S, Fourmaux MN, Soete M, Di Cristina M, Dubremetz JF, Soldati D. Identification and characterization of an escorter for two secretory adhesins in Toxoplasma gondii. J Cell Biol. 2001;152:563–578. doi: 10.1083/jcb.152.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard D, Kats LM, Langer C, Black CG, Mitri K, Boddey JA, Cowman AF, Coppel RL. Identification of rhoptry trafficking determinants and evidence for a novel sorting mechanism in the malaria parasite Plasmodium falciparum. PLoS Pathog. 2009;5:e1000328. doi: 10.1371/journal.ppat.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saouros S, Edwards-Jones B, Reiss M, Sawmynaden K, Cota E, Simpson P, Dowse TJ, Jakle U, Ramboarina S, Shivarattan T, Matthews S, Soldati-Favre D. A novel galectin-like domain from Toxoplasma gondii micronemal protein 1 assists the folding, assembly, and transport of a cell adhesion complex. J Biol Chem. 2005;280:38583–38591. doi: 10.1074/jbc.C500365200. [DOI] [PubMed] [Google Scholar]
- Sawmynaden K, Saouros S, Friedrich N, Marchant J, Simpson P, Bleijlevens B, Blackman MJ, Soldati-Favre D, Matthews S. Structural insights into microneme protein assembly reveal a new mode of EGF domain recognition. EMBO Rep. 2008;9:1149–1155. doi: 10.1038/embor.2008.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheiner L, Soldati-Favre D. Protein trafficking inside Toxoplasma gondii. Traffic. 2008;9:636–646. doi: 10.1111/j.1600-0854.2008.00713.x. [DOI] [PubMed] [Google Scholar]
- Soldati D, Boothroyd JC. Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science (New York, NY) 1993;260:349–352. doi: 10.1126/science.8469986. [DOI] [PubMed] [Google Scholar]
- Starnes GL, Jewett TJ, Carruthers VB, Sibley LD. Two separate, conserved acidic amino acid domains within the Toxoplasma gondii MIC2 cytoplasmic tail are required for parasite survival. J Biol Chem. 2006;281:30745–30754. doi: 10.1074/jbc.M606523200. [DOI] [PubMed] [Google Scholar]
- Straub KW, Cheng SJ, Sohn CS, Bradley PJ. Novel components of the Apicomplexan moving junction reveal conserved and coccidia–restricted elements. Cell Microbiol. 2008;4:590–603. doi: 10.1111/j.1462-5822.2008.01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Duquette M, Liu JH, Lawler J, Wang JH. The crystal structure of the heparin-binding reelin-N domain of f-spondin. Journal of molecular biology. 2008;381:1213–1223. doi: 10.1016/j.jmb.2008.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tossavainen H, Pihlajamaa T, Huttunen TK, Raulo E, Rauvala H, Permi P, Kilpelainen I. The layered fold of the TSR domain of P. falciparum TRAP contains a heparin binding site. Protein science. 2006;15:1760–1768. doi: 10.1110/ps.052068506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treeck M, Struck NS, Haase S, Langer C, Herrmann S, Healer J, Cowman AF, Gilberger TW. A conserved region in the EBL proteins is implicated in microneme targeting of the malaria parasite Plasmodium falciparum. J Biol Chem. 2006;281:31995–32003. doi: 10.1074/jbc.M606717200. [DOI] [PubMed] [Google Scholar]
- Treeck M, Zacherl S, Herrmann S, Cabrera A, Kono M, Struck NS, Engelberg K, Haase S, Frischknecht F, Miura K, Spielmann T, Gilberger TW. Functional analysis of the leading malaria vaccine candidate AMA-1 reveals an essential role for the cytoplasmic domain in the invasion process. PLoS Pathog. 2009;5:e1000322. doi: 10.1371/journal.ppat.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- Witcombe DM, Ferguson DJ, Belli SI, Wallach MG, Smith NC. Eimeria maxima TRAP family protein EmTFP250: subcellular localisation and induction of immune responses by immunisation with a recombinant C-terminal derivative. Int J Parasitol. 2004;34:861–872. doi: 10.1016/j.ijpara.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Zheng B, He A, Gan M, Li Z, He H, Zhan X. MIC6 associates with aldolase in host cell invasion by Toxoplasma gondii. Parasitol Res. 2009;2:441–5. doi: 10.1007/s00436-009-1401-5. [DOI] [PubMed] [Google Scholar]
- Zhou XW, Blackman MJ, Howell SA, Carruthers VB. Proteomic analysis of cleavage events reveals a dynamic two-step mechanism for proteolysis of a key parasite adhesive complex. Mol Cell Proteomics. 2004;3:565–576. doi: 10.1074/mcp.M300123-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.