Summary
Single cells in a population often respond differently to perturbations in the environment. Live-cell microscopy has enabled scientists to observe these differences at the single-cell level. Some advantages of live-cell imaging over population-based methods include better time resolution, higher sensitivity, automation, and richer datasets. One specific area where live-cell microscopy has made a significant impact is the field of NF-κB signaling dynamics, and recent efforts have focused on making live-cell imaging of these dynamics more high throughput. We highlight the major aspects of increasing throughput and describe a current system that can monitor, image and analyze the NF-κB activation of thousands of single cells in parallel.
Genetically identical, individual cells in a common environment often vary significantly in their response to perturbations. The behavioral variation between single cells can be observed experimentally using live-cell microscopy, as has been demonstrated in bacterial [1, 2], yeast [3], and mammalian systems [4–6]. One common feature of these studies is that measuring the dynamics of single cells yields new and exciting insights about phenotypic heterogeneity in a population.
NF-κB is a family of transcription factors that control the expression of hundreds of genes [7]. Beyond NF-κB’s importance as a key player in innate immunity [8], study of NF-κB activation as a case study in the field of systems biology has been very productive in recent years [9–15]. The initial computational models of NF-κB signaling demonstrated the power of combined computational and experimental analyses in understanding the NF-κB response to TNF-α [16]. Further studies continue to reveal the role of additional components [14, 17], the response to other ligands [11, 13], and the behavior of NF-κB in single cells [9, 12, 18].
Live-cell microscopy has made a substantial impact on the field of NF-κB dynamics, illuminating how population responses can be interpreted as the coordinated action of single cells. These microscopy-based studies have been limited to the analysis of relatively few cells, but recent developments have increased this throughput dramatically. The purpose of this short review is to highlight some important reasons why live-cell imaging has been useful in studying NF-κB, and then to summarize the workflow, methods and challenges of NF-κB imaging with a focus on recent advances that have made this technology high-throughput.
Why bother with single cell imaging?
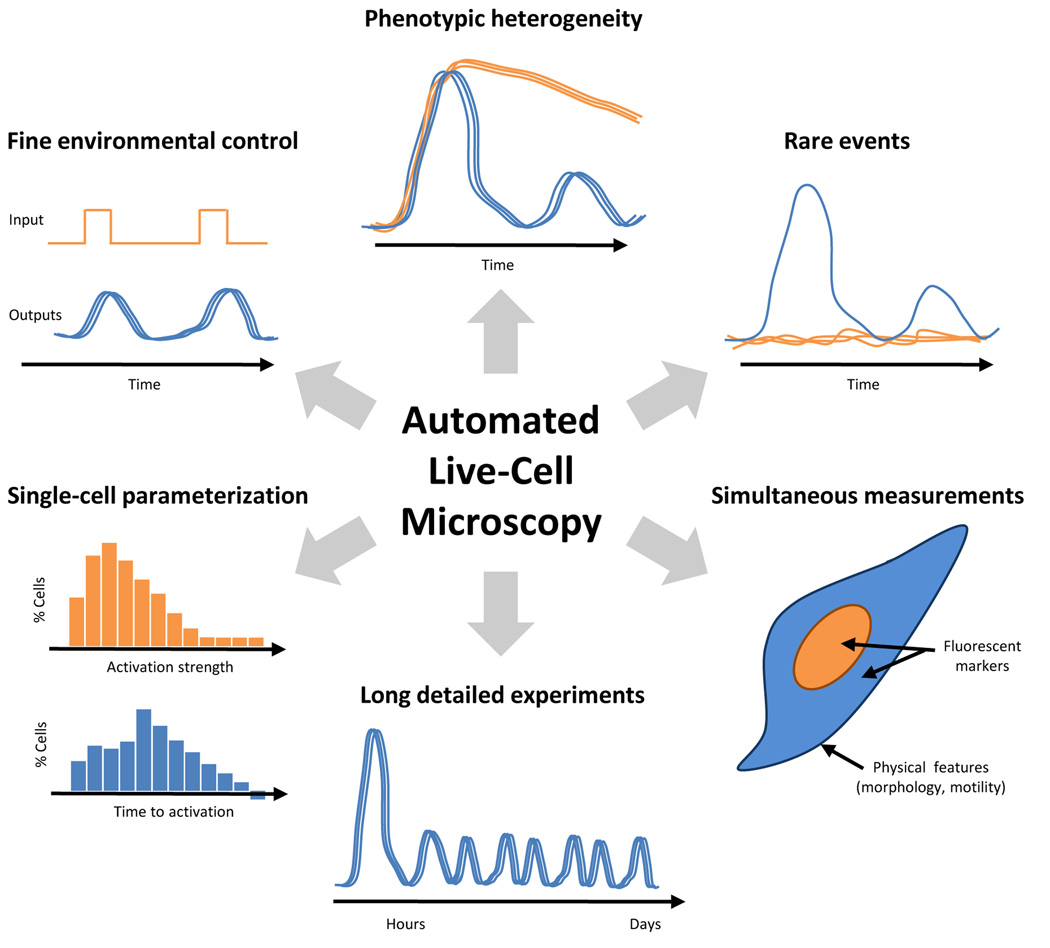
Live-cell imaging is relatively expensive to set up and maintain, so we begin by outlining several ways in which the data sets generated using live-cell microscopy can be much richer than those enabled by current population-based methods, several of which are shown schematically in Figure 1. Traditionally, NF-κB activation is measured by an electrophoretic mobility shift assay (EMSA) that detects the interaction of activated nuclear NF-κB with a labeled DNA probe [19]. EMSAs are not optimal for measuring the dynamics of NF-κB activation. For example, NF-κB activation dynamics occur with a time resolution of minutes and can persist for hours. The time resolution that can be obtained using the EMSA is typically limited by the length of the experiment, the number of wells in the gel comb, and the efficiency of the scientist. In a four-hour experiment with a 20-well gel, the time resolution will be about fifteen minutes. The maximum reasonable time resolution (based on the scientist’s efficiency) is probably on the order of five minutes and not sustainable for long time-courses. In contrast, live-cell imaging allows for the near-continuous measurement of NF-κB localization over longer time periods. An important detail is that while EMSAs measure only active nuclear NF-κB, fluorescence microscopy measures total nuclear NF-κB, active and inactive, and should be considered in building models from microscopy data. Although EMSA gels can quantified using a gel imager, live-cell microscopy has better dynamic range in detecting and quantifying NF-κB activation, as has now been well demonstrated. Finally, live-cell imaging requires far less sample than traditional EMSAs, Western blots or kinase assays.
Figure 1. The advantages of using automated, live-cell microscopy to study NF-κB dynamics.
Some advantages are biological, such as the ability to detect variation in cell behavior, from larger-scale phenotypic heterogeneity to rare events involving as little as a single cell, and deriving biological parameters based on single-cell measurements. Others are technical, including the ability to perform long experiments at fine time resolution, making several measurements simultaneously and having precise, dynamic environmental control.
Live-cell imaging is also generally automated, and it is possible to perform many detailed time- course experiments at once. In contrast, a well-trained experimentalist might be able to perform three or four EMSAs at a time. This advantage is particularly important, because NF-κB responds to hundreds of different ligands, cytokines, and chemicals [20] – each of which can presumably lead to different activation dynamics. To understand NF-κB signaling in the context of complex cellular environments it will be necessary to measure NF-κB dynamics under a wide variety of stimulation conditions, including activators [20] and inhibitors [21]. Most studies use a constant, saturating concentration of TNF-α, which probably does not reflect the physiological condition. Instead, researchers have now begun to study the NF-κB response to different TNF-α concentrations [10] and time-varying TNF-α stimulation [9, 14, 22]. Recent interests also include the NF-κB activation dynamics under different ligands [13, 18, 23], upon drug perturbations [24, 25], and in response to live or dead bacteria [26–29]. Furthermore, multiple outputs can be observed in the same cell through time, for example the dynamics of NF-κB localization and IκBα expression [12, 30].
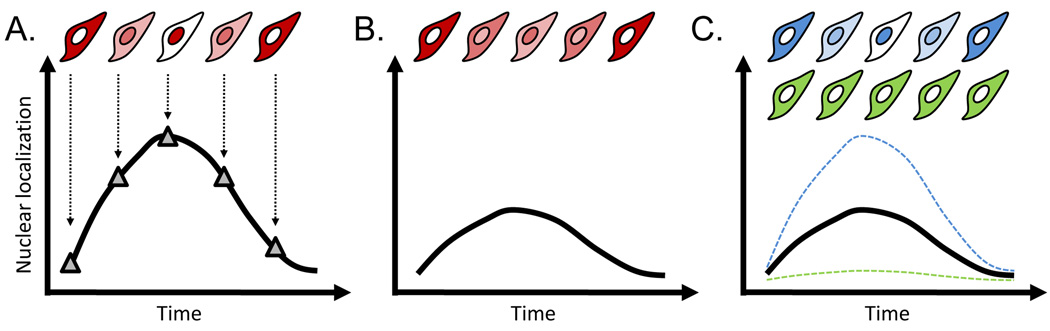
The most important argument for live-cell imaging, however, is that single-cell resolution adds a critical new dimension to the data. A typical EMSA measurement represents the collective NF-κB nuclear localization of around 106 cells. With the behavior of so many cells pooled together, important phenotypes exhibited by individual cells will likely be obscured. An example of the importance of single-cell resolution to the NF-κB dynamics is the response to varying concentrations of tumor necrosis factor-alpha (TNF-α). Earlier work at the population level showed that the overall localization of NF-κB to the nucleus was reduced as the TNF-α concentration was lowered [10]. Two models of the single-cell response to reduced TNF-α concentration can explain this observation equally well, as shown in Figure 2. First, all cells could exhibit similar dynamics but with a lowered peak activation. Alternatively, individual cells may have an all-or-none response, where a smaller number of cells respond to the signal. We have recently found experimental evidence supporting a combination of these two models [15].
Figure 2. Population measurements can obscure heterogeneous single-cell behaviors.
The NF-κB response of single cells to a high concentration of TNF-α, in terms of single-cell (top) and population (bottom) level behavior, is shown schematically in (A). At lower concentrations, the response is less pronounced, which can be explained by either of two models. (B) The first model is that all of the cells behave roughly identically and with a lower intensity response. (C) The second model is that some of the cells respond to the stimulus with a normal response (blue), while others do not respond at all (green). These two models are indistinguishable at the population level.
Another example of cellular phenotypic heterogeneity concerns the NF-κB response to the bacterial cell wall product lipopolysaccharide (LPS). LPS causes a stable nuclear localization for ~3 hours when measured at the population level [11]. Single-cell measurements of NF-κB localization under LPS stimulation conditions verified this population response, but revealed complex behaviors within the population. Specifically, cells exhibit one of two qualitatively different behaviors: in some, NF-κB activation is only transient, while in others, NF-κB persists in the nucleus for hours [18]. This result suggests that major qualitative behaviors exhibited by a significant percentage (roughly 45% in this case) of cells can be completely missed by population-based assays.
Other technologies, such as immunocytochemistry [31] or flow cytometry [32], can also achieve single-cell resolution. Many cells can be analyzed with flow cytometry and the throughput of immunocytochemistry has been recently improved with high-content cell screening strategies [22, 33]. In addition, they can detect functional modifications, the most important being phosphorylation. However, unlike live-cell imaging, they are unable to track the same cell through time and usually involve fixation of the cells. Therefore, live-cell imaging can reveal details about dynamic processes, but should also be combined with other functional assays.
Workflow, methods and challenges
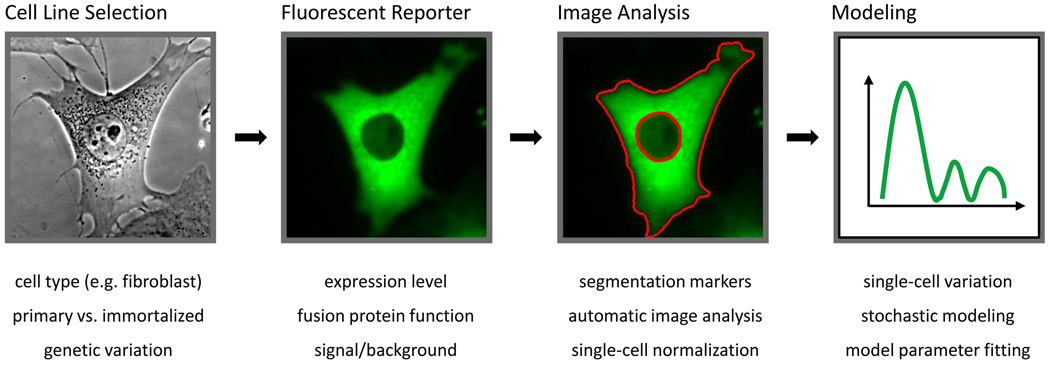
Based on these advantages, we anticipate a rise in the use of live-cell imaging to study NF-κB dynamics and discuss some of the major steps and decision points involved here (Figure 3). The general pipeline for live-cell imaging studies in NF-κB involves the selection of an appropriate cell line for study, fluorescence labeling to facilitate imaging, image analysis and data interpretation via modeling, as described below.
Figure 3. Workflow diagram for high-throughput single-cell imaging of NF-κB dynamics.
The steps involved in a live-cell microscopy study of NF-κB are outlined and corresponding difficulties and issues are listed below. Each box is described in more detail in the main text.
Cell line selection
The first step in the pipeline is to choose cells that will be suitable for imaging and appropriate for studying NF-κB dynamics. The cell types used for imaging NF-κB activation have been primary mouse embryonic fibroblasts (MEFs) [9], as well as MEFs immortalized through the 3T3 protocol [18], SK-N-AS neuroblastoma cells [12] or HeLa cells [12]. In general, the appropriateness of a cell line in any study must be carefully evaluated. For example, cell lines can display highly aberrant behavior after transformation, which may impact NF-κB expression or activity [34]. On the other hand, primary cells may be more difficult to label with a fluorescent reporter and to subpassage efficiently. Furthermore, in some cases it may be desirable to image a genetically identical population of cells, which requires the selection of a clonal line.
Surprisingly, although NF-κB activation is typically associated with innate immune signaling, relatively few studies have used relevant immune cell types. In the future, we expect that more researchers will turn to classic immune cell types or cell lines that mimic the properties of immune cells, such as the macrophage like RAW 264.7 line [26].
Fluorescent reporters
The next step is to visualize NF-κB activation, typically using a fluorescently labeled p65 subunit of NF-κB that is introduced through transfection or viral transduction. Typically, this process involves the overexpression of the fusion protein to increase brightness and improve image quality. However, variation in the expression level of NF-κB can affect the dynamics of activation [35]. It is therefore often necessary to achieve approximately endogenous levels of p65 expression. Expression with transfected plasmids can achieve endogenous expression through screening of clonal cells lines by transfection [9], or by tuning expression using inducible promoters or degradation tags [36]. Viral-based transduction can be used for a more careful control of gene copy number and may be combined with an endogenous promoter to mimic normal expression control [18]. It is preferable to introduce these constructs into a background that does not contain wild-type NF-κB since the unlabeled protein is not detected in imaging and the gene dosage will be inherently higher.
A new development is the generation of an EGFP-NF-κB knock-in mouse [37] that circumvents many of these concerns and allows for experimentation on genetically identical primary cells. The knock-in mouse is viable, but the functional properties of the fusion protein are slightly different, evidenced by lowered NF-κB dependent induction of IκBα, which is similar to virally transduced EGFP-NF-κB [18].
Expression of a fluorescent protein at the level of endogenous p65 is significantly lower than expression from a strong constitutive promoter (Covert lab, unpublished data). Typically, enough emission light from fluorescently labeled NF-κB can be collected to image every tenth of a second on a wide-field microscope. Exposure to fluorescence excitation light can lead to photobleaching and phototoxicity that can affect image quality and cell health. Imaging cells every few minutes avoids these concerns while still capturing all relevant NF-κB dynamics, although imaging frequency can be increased by lowering light source intensity and improving detection sensitivity with more efficient cameras and filters. In addition, media formulations with low background fluorescence can play a large role in improving image quality [38].
Single-cell transcriptional activity in living cells can be measured by driving the expression of a reporter with a promoter of interest. Common reporters include fluorescent proteins or bioluminescence from luciferase-based systems [12]. Fluorescent proteins require a maturation time before becoming active which lead to a delay before signal can be recorded, and it is best to choose fast maturing and destabilized variants. Bioluminescence has the advantage of very low background signal, but requires the addition of the substrate luciferin and long exposure times, although this is generally less of an issue with new brighter reporters [39].
Image analysis
Image analysis is likely the major bottleneck in high-throughput imaging studies, even with substantial efforts in software-based analysis tools [40]. Labeled cells are imaged using a standard fluorescence microscope under temperature and carbon dioxide control to maintain cell viability. The intensity of labeled p65 in the nucleus of cells over time is measured from the images to generate dynamic profiles of NF-κB activation. Image segmentation is required to determine the relative fluorescence intensity (correlating to the presence of p65) in the cellular nucleus and/or cytoplasm. At times when the intensity of fluorescence in the nucleus and cytoplasm are roughly equal, it is virtually impossible, by computer or by eye, to define the nucleus. Because of the need to define these nuclear regions, it is highly recommended that a nuclear marker also be introduced into the cells. Fluorescent proteins designed to localize to the nucleus, such as those linked to the nuclear protein H2B, are preferred for longer time-lapse experiments. With the nucleus clearly marked in a separate fluorescent channel, image segmentation is more efficient and the location of the nucleus can be used as an excellent prior for further cellular segmentation. In addition, it is often useful to normalize for differences in p65 expression, which occurs even in genetically identical cells. This requires segmentation of the cytoplasm, at least at one and possibly all time points.
Data interpretation via modeling
As stated above, computational modeling of NF-κB activation is well established and useful in interpreting dynamic data. The original modelwas fit to population data (EMSA and Western blot) [16], and modeling single cells presents a new challenge. First, it has now been shown that several of these parameter estimations made at the population level are significantly different from estimations made based on single-cell data [18]. The majority of the parameters fit to single-cell data involve the processes of translation and transcription or initial protein concentrations which are noisy in single cells. Second, modeling approaches are required which can capture single-cell variability as well as population-level responses. In such models, stochasticity may be encoded either at the cellular [18] or molecular [15, 41, 42] level. Additionally, none of the models capture all experimentally measured aspects of NF-κB activation. Large imaging datasets reveal distributions of many single-cell behaviors, and it is especially difficult to reproduce all observed behaviors in a single model. It will therefore become increasingly important to understand which dynamic behaviors are relevant biologically, as has been done with models of circadian rhythm [43], and fit models accordingly.
Towards high-throughput imaging
Taking these issues into account, we recently developed a platform that can culture, image and determine the NF-κB activation of thousands of single cells – all in a matter of days. To accomplish this, we used microfluidic cell culture [44] of a clonal 3T3 cell line expressing a p65-DsRed fluorescent fusion protein [12] and an H2B-GFP nuclear marker [45], created in a p65 knockout background [46]. The result was a near-endogenous level of p65 expression [18]. The images were obtained using an incubated and motorized epifluorescence microscope and were automatically processed to extract NF-κB localization dynamics (normalized by total cellular NF-κB at the first time point). We used this platform to study the effect of TNF-α concentration on NF-κB activation, and found that single cells respond heterogeneously to changing concentrations, both in terms of an all-or-none response (as depicted in Figure 2) as well as more graded responses [15]. A reduced percentage of cells respond to lowered stimulus concentrations, as has been observed in other mammalian systems, such as T-cell activation [47]. Live-cell microscopy makes it possible to follow NF-κB dynamics after the decision to activate and quantify graded concentration dependent behaviors such as oscillation timing. To model the variation in the response, we extended a stochastic model of NF-κB activation [42] that reproduces most of the observed behaviors.
Conclusion
In summary, single-cell microscopy has necessitated the reinterpretation of results from population-level studies due to the variation of single cells. Although variation in single-cell NF-κB dynamics is now a growing area of study, it remains to be determined if such variation is biologically relevant. In any situation where heterogeneity arises between cells, one also needs to consider whether or not the differences will lead to a phenotypic outcome [48], as has been shown in apoptosis [49, 50]. In addition, further work is needed to link single-cell transcriptional activity with single-cell transcription factor dynamics, likely through transcriptional reporters as described above. Such information would place NF-κB dynamics in the context of larger transcriptional regulatory networks [51].
With these techniques for high-throughput imaging, it is now possible to examine NF-κB signaling for hundreds of conditions that would have been infeasible with traditional assays. For example, whereas most NF-κB studies have focused on TNF-α stimulation, NF-κB activates in response to hundreds of different ligands, cytokines, and chemicals. The effects of these stimuli can now be examined in single-cell, high time resolution detail in about the same time as performing an EMSA. Additionally, given that many pathways converge on NF-κB, another important area of study is how multiple stimuli are integrated into a single NF-κB dynamic profile [52, 53]. Study of these combinatorial effects would be extremely difficult by traditional methods. Finally, as evidenced by several large imaging based siRNA screens [54] and dynamic proteomic studies [50, 55], live-cell microscopy assays can be used for high-throughput screening, significantly increasing the discovery process.
Acknowledgments
The authors would like to thank J. Hughey for assistance with the figures, and support from a Stanford Graduate Fellowship (to TKL) and an NIH/NCI Pathway to Independence Award (CA125994).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Timothy K. Lee, Email: tklee@stanford.edu.
Markus W. Covert, Email: mcovert@stanford.edu.
References
- 1.Balaban NQ, et al. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 2.Chabot JR, et al. Stochastic gene expression out-of-steady-state in the cyanobacterial circadian clock. Nature. 2007;450(7173):1249–1252. doi: 10.1038/nature06395. [DOI] [PubMed] [Google Scholar]
- 3.Skotheim JM, et al. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 2008;454(7202):291–296. doi: 10.1038/nature07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geva-Zatorsky N, et al. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2:2006.0033. doi: 10.1038/msb4100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sigal A, et al. Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat Methods. 2006;3(7):525–531. doi: 10.1038/nmeth892. [DOI] [PubMed] [Google Scholar]
- 6.Sprinzak D, et al. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465(7294):86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 8.Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25(51):6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 9. Ashall L, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324(5924):242–246. doi: 10.1126/science.1164860. The NF-κB response to repeated short pulses of TNF-α was examined using live-cell imaging. The frequency of these pulses changes the NF-κB dependent gene expression, suggesting that pulse frequency can specify transcriptional programs.
- 10.Cheong R, et al. Transient IkappaB kinase activity mediates temporal NFkappaB dynamics in response to a wide range of tumor necrosis factor-alpha doses. J Biol Chem. 2006;281(5):2945–2950. doi: 10.1074/jbc.M510085200. [DOI] [PubMed] [Google Scholar]
- 11.Covert MW, et al. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science. 2005;309(5742):1854–1857. doi: 10.1126/science.1112304. [DOI] [PubMed] [Google Scholar]
- 12.Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306(5696):704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 13.Werner S, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005 doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 14.Werner SL, et al. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22(15):2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tay S, et al. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466(7303):267–271. doi: 10.1038/nature09145. NF-κB dynamics were measured in high-throughput with a microfluidic cell culture system and live-cell imaging. The response of thousands of single cells was measured for a wide range of TNF-α concentrations. NF-κB dependent gene expression was also measured and a model was presented to explain these findings.
- 16.Hoffmann A, et al. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298(5596):1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 17.Basak S, et al. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128(2):369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee TK, et al. A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide. Science signaling. 2009;2(93):ra65. doi: 10.1126/scisignal.2000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellman LM, Fried MG. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat Protoc. 2007;2(8):1849–1861. doi: 10.1038/nprot.2007.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pahl HL. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene. 1999;18(49):6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 21.Gilmore TD, Herscovitch M. Inhibitors of NF-kappaB signaling: 785 and counting. Oncogene. 2006;25(51):6887–6899. doi: 10.1038/sj.onc.1209982. [DOI] [PubMed] [Google Scholar]
- 22. Cheong R, Wang CJ, Levchenko A. High content cell screening in a microfluidic device. Mol Cell Proteomics. 2009;8(3):433–442. doi: 10.1074/mcp.M800291-MCP200. A microfluidic culture system is used to stimulate, fix and immunostain cells in high-throughput under precisely controlled conditions. In addition, immunostaining allows for the readout of functional outputs such as phosphorylation state.
- 23.Ihekwaba AEC, et al. Bridging the gap between in silico and cell-based analysis of the nuclear factor-kappaB signaling pathway by in vitro studies of IKK2. FEBS J. 2007;274(7):1678–1690. doi: 10.1111/j.1742-4658.2007.05713.x. [DOI] [PubMed] [Google Scholar]
- 24.Sung M-H, et al. Dynamic effect of bortezomib on nuclear factor-kappaB activity and gene expression in tumor cells. Mol Pharmacol. 2008;74(5):1215–1222. doi: 10.1124/mol.108.049114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung M-H, Simon R. In silico simulation of inhibitor drug effects on nuclear factor-kappaB pathway dynamics. Mol Pharmacol. 2004;66(1):70–75. doi: 10.1124/mol.66.1.70. [DOI] [PubMed] [Google Scholar]
- 26.James CD, et al. Nuclear translocation kinetics of NF-kappaB in macrophages challenged with pathogens in a microfluidic platform. Biomed Microdevices. 2009;11(3):693–700. doi: 10.1007/s10544-008-9281-5. [DOI] [PubMed] [Google Scholar]
- 27.Bartfeld S, et al. High-throughput and single-cell imaging of NF-kappaB oscillations using monoclonal cell lines. BMC Cell Biol. 2010;11:21. doi: 10.1186/1471-2121-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadler C, et al. The type III secretion effector NleE inhibits NF-kappaB activation. PLoS Pathog. 2010;6(1):e1000743. doi: 10.1371/journal.ppat.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartfeld S, et al. Temporal resolution of two-tracked NF-kappaB activation by Legionella pneumophila. Cell Microbiol. 2009;11(11):1638–1651. doi: 10.1111/j.1462-5822.2009.01354.x. [DOI] [PubMed] [Google Scholar]
- 30.Nelson G, et al. Multi-parameter analysis of the kinetics of NF-kappaB signalling and transcription in single living cells. J Cell Sci. 2002;115(Pt 6):1137–1148. doi: 10.1242/jcs.115.6.1137. [DOI] [PubMed] [Google Scholar]
- 31.Soen Y, et al. Exploring the regulation of human neural precursor cell differentiation using arrays of signaling microenvironments. Mol Syst Biol. 2006;2:37. doi: 10.1038/msb4100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez OD, Nolan GP. Simultaneous measurement of multiple active kinase states using polychromatic flow cytometry. Nat Biotechnol. 2002;20(2):155–162. doi: 10.1038/nbt0202-155. [DOI] [PubMed] [Google Scholar]
- 33.Perlman ZE, et al. Multidimensional drug profiling by automated microscopy. Science. 2004;306(5699):1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 34.Gapuzan M-ER, et al. Immortalized fibroblasts from NF-kappaB RelA knockout mice show phenotypic heterogeneity and maintain increased sensitivity to tumor necrosis factor alpha after transformation by v-Ras. Oncogene. 2005;24(43):6574–6583. doi: 10.1038/sj.onc.1208809. [DOI] [PubMed] [Google Scholar]
- 35.Nelson G, et al. Multi-parameter analysis of the kinetics of NF-kappaB signalling and transcription in single living cells. J Cell Sci. 2002;115(Pt 6):1137–1148. doi: 10.1242/jcs.115.6.1137. [DOI] [PubMed] [Google Scholar]
- 36.Banaszynski LA, et al. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126(5):995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. De Lorenzi R, et al. GFP-p65 knock-in mice as a tool to study NF-kappaB dynamics in vivo. Genesis. 2009;47(5):323–329. doi: 10.1002/dvg.20468. Homologous recombination was used to introduce the sequence encoding EGFP-p65 in place of the rela gene. Cells derived from these mice will alleviate many of the concerns with live-cell imaging of fluorescent protein fusions.
- 38.Bogdanov AM, et al. Cell culture medium affects GFP photostability: a solution. Nat Methods. 2009;6(12):859–860. doi: 10.1038/nmeth1209-859. [DOI] [PubMed] [Google Scholar]
- 39.Hoshino H, Nakajima Y, Ohmiya Y. Luciferase-YFP fusion tag with enhanced emission for single-cell luminescence imaging. Nat Methods. 2007;4(8):637–639. doi: 10.1038/nmeth1069. [DOI] [PubMed] [Google Scholar]
- 40.Walter T, et al. Visualization of image data from cells to organisms. Nat Methods. 2010;7(3 Suppl):S26–S41. doi: 10.1038/nmeth.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipniacki T, Kimmel M. Deterministic and stochastic models of NFkappaB pathway. Cardiovasc Toxicol. 2007;7(4):215–234. doi: 10.1007/s12012-007-9003-x. [DOI] [PubMed] [Google Scholar]
- 42.Lipniacki T, et al. Single TNFalpha trimers mediating NF-kappaB activation: stochastic robustness of NF-kappaB signaling. BMC Bioinformatics. 2007;8:376. doi: 10.1186/1471-2105-8-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirsky HP, et al. A model of the cell-autonomous mammalian circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(27):11107–11112. doi: 10.1073/pnas.0904837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gómez-Sjöberg R, et al. Versatile, fully automated, microfluidic cell culture system. Anal Chem. 2007;79(22):8557–8563. doi: 10.1021/ac071311w. [DOI] [PubMed] [Google Scholar]
- 45.Lois C, et al. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295(5556):868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 46.Beg AA, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274(5288):782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 47.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005;3(11):e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;141(4):559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spencer SL, et al. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459(7245):428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen AA, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322(5907):1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 51.Seok J, et al. A dynamic network of transcription in LPS-treated human subjects. BMC Syst Biol. 2009;3:78. doi: 10.1186/1752-0509-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsueh RC, et al. Deciphering signaling outcomes from a system of complex networks. Science signaling. 2009;2(71):ra22. doi: 10.1126/scisignal.2000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Natarajan M, et al. A global analysis of cross-talk in a mammalian cellular signalling network. Nat Cell Biol. 2006;8(6):571–580. doi: 10.1038/ncb1418. [DOI] [PubMed] [Google Scholar]
- 54.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464(7289):721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geva-Zatorsky N, et al. Protein Dynamics in Drug Combinations: a Linear Superposition of Individual-Drug Responses. Cell. 2010;140(5):643–651. doi: 10.1016/j.cell.2010.02.011. [DOI] [PubMed] [Google Scholar]