Abstract
ACTIVITY is a database on DNA/RNA site sequences with known activity magnitudes, measurement systems, sequence-activity relationships under fixed experimental conditions and procedures to adapt these relationships from one measurement system to another. This database deposits information on DNA/RNA affinities to proteins and cell nuclear extracts, cutting efficiencies, gene transcription activity, mRNA translation efficiencies, mutability and other biological activities of natural sites occurring within promoters, mRNA leaders, and other regulatory regions in pro- and eukaryotic genomes, their mutant forms and synthetic analogues. Since activity magnitudes are heavily system-dependent, the current version of ACTIVITY is supplemented by three novel sub-databases: (i) SYSTEM, measurement systems; (ii) KNOWLEDGE, sequence-activity relationships under fixed experimental conditions; and (iii) CROSS_TEST, procedures adapting a relationship from one measurement system to another. These databases are useful in molecular biology, pharmacogenetics, metabolic engineering, drug design and biotechnology. The databases can be queried using SRS and are available through the Web, http://wwwmgs.bionet.nsc.ru/systems/Activity/.
INTRODUCTION
As a part of many genome DNA sequencing and annotation efforts (1), many databases on DNA/RNA functional site locations have been developed, i.e., TRANSFAC (2), TRRD (3), SELEX_DB (4), etc. These information resources led to the design of a large body of web tools recognising these sites, in particular, TESS (5), the TRANSFAC-based expert system (2), MatInspector (6), MATRIX SEARCH (7), etc. However, new problems such as relating single nucleotide polymorphism (SNPs) analysis to known clinical phenotypes demand novel approaches for this mutation data analysis (8). This analysis is partially aimed at predicting regulatory DNA/RNA sites, the biological activity of which could be either increased, or decreased, or appeared de novo due to a point mutation. In this way, by using experimental data on the point mutations G663A and G666T in the #6 intron of the TDO2 gene, which causes mental disorders and reduces DNA–protein complex mobility in gel shift experiments, Ponomarenko and colleagues (9) predicted that the YY1-site was damaged by these mutations and later verified this fact by an anti-YY1 antibody test. This experiment was the first successful site prediction based on the alteration of DNA affinity in a nuclear extract, and demonstrates that data on site activity is useful for site recognition relevant to regulatory SNPs.
Experimental data on site activity are well represented in the literature. McClure and co-workers were the first to accumulate data on natural Escherichia coli promoter strength and to apply them to the prediction of the strength of some other natural promoters (10). Later, Jonsson et al. (11), on the basis of experimental data on natural E.coli promoter strengths, developed a method applicable to the analysis of artificial point mutations in natural E.coli promoters. Berg and von Hippel have collected data on the activities of prokaryotic activator and repressor binding sites, these data being the foundation for the commonly accepted statistical-mechanical theory of DNA–protein interactions (12,13). Stormo and co-workers (14,15) were the first to apply a wide spectrum of mutational events, mutagen-dependent hot spots, nonsense codon suppression and ribosome binding sites to sequence-activity analysis. Kraus et al. (16) have initiated investigations in eukaryotes: they studied transcription initiator (Inr-element) and TATA-box activities and predicted them successfully. Recently, Ponomarenko et al. (17) developed a database on transcription factor binding site activities, conformational and physicochemical DNA properties correlating to site activities and web tools predicting the activity of these sites in an arbitrary DNA. All these studies were made assuming that sequence-activity relationships are invariant, thus, ignoring the conditions of the measurement system.
At the same time, Sarai and co-workers (18,19) observed that point mutations in the OR1-operator act differently while binding with two antagonist proteins, Cro- and λ-repressors. Analogously, point mutations in the c-Myb binding site cause different free energy changes (ΔΔGs) in natural (20) and mutant target proteins (21). Analogous data were obtained for the natural, EREBP-2, and homologous proteins, EREBP-4 and AtEBP-1, binding the GCC box in plants (22). Similar evidence was obtained by comparing in vivo and in vitro systems of the Inr-element (16) and TATA box (23) activities. Analogously, Hyde-DeRuyscher at al. (24) found a discrepancy in activity measured in different cell systems and plasmid constructs. Also, Javahery et al. (25) found no correlation between the Inr/YY1-induction magnitudes and YY1/Inr-affinities.
The above data are better explained by a ‘jigsaw puzzle’ hypothesis (26) rather than by the ‘key/lock’ principle of intermolecular recognition (12,13). The ‘jigsaw puzzle’ hypothesis takes into account not only DNA–protein interactions, but also protein–protein ones. With this in mind, regulatory machine function is strongly dependent upon regulatory genome regions, measurement methods, genetic constructs, etc. Currently, the sensitivity of activity magnitudes to the measurement system is widely used in order to detect the cell-specificity of regulatory sites referring to SNP (27).
In this respect, the ACTIVITY database of DNA/RNA site sequences with known activities was supplemented by three sub-databases: (i) SYSTEM, measurement systems; (ii) KNOWLEDGE, sequence-activity relationship at fixed experimental conditions; and (iii) CROSS_TEST, procedures for the application of one measurement system to another. These databases are useful for molecular biology, pharmacogenetics, metabolic engineering, drug-design and biotechnology. They can be queried using SRS (28) and are available at http://wwwmgs.bionet.nsc.ru/systems/Activity/.
DATA REPRESENTATION
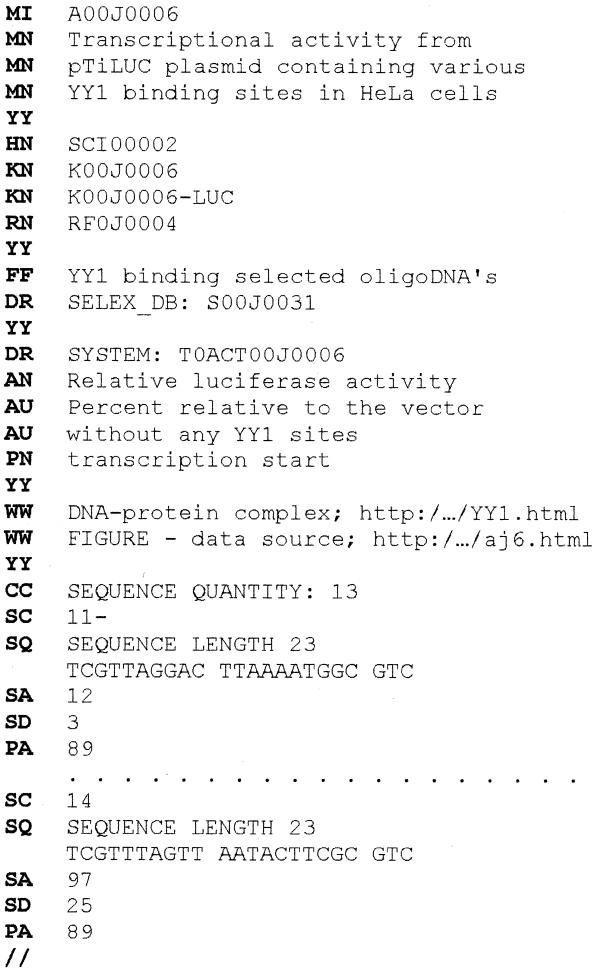
Each entry in the ACTIVITY database describes a set of ‘sequence-activity’ data measured in a fixed experimental system. For example, the entry on luciferase activity from reporter plasmids with selected YY1 binding sites at the –88 position relative to the transcription start site (24) is shown in Figure 1. Each line begins with a two-letter descriptor: MI, identifier; MN, entry name; HN, annotator’s name (linked to the SCIENTIST database); KN, KNOWLEDGE database entry; RN, reference (linked to REFERENCE database); FF, site’s name; OG, OS, OC, gene, species and taxon specificity (if the sequences are not synthetic); AN, type of activity’s measurement; AU, measurement units; PN, sequence phasing point; SC, site’s variant; SQ, site sequence; SA, activity magnitude; SD, standard deviation; PA, position of the phasing point relative to the sequence start; DR, links to the other databases, if any (SELEX_DB, TRRD, SYSTEM); WW, links to other web resources, if any. The entry presents the ‘sequence-activity’ data in a computable format.
Figure 1.
Example of an ACTIVITY entry.
The SYSTEM sub-database describes the measurement systems and experimental conditions (Fig. 2). Its entry is supplied by nine fields: MI, identifier; MN, name; EP, aim of the experiment given by the author; EC, system type; EM, conditions and materials; AM, measurement method; AC, control observation; EE, conclusion made by the author; DR, links to the other databases if any (SCIENTIST, REFERENCE, ACTIVITY, SELEX_DB). Also, SYSTEM contains information about limitations made by the author on sequence-activity data interpretations (EP and EE). These limits are set by experimental details causing the system-dependence of the data (EC, EM, AM and AC).
Figure 2.
Example of a SYSTEM entry.
The KNOWLEDGE sub-database documents the sequence-activity relationships revealed by experimental ‘sequence-activity’ data and treated by our knowledge discovery system (17). A KNOWLEDGE sub-database entry contains 12 fields (Fig. 3): MI, identifier; MN, name; HN, researcher (linked to the SCIENTIST database); DA, ACTIVITY entry; WW, web resource; CF, mathematical model; CT, computational method; PV, DNA property; AB, sequence region; LC, linear correlation coefficient; AL, significance α; C-, C-code procedure calculating the value of this relationship in an arbitrary DNA. The entry gives information, which could be applied by using well tested and documented computational procedures (C-, LC and AL).
Figure 3.
Example of a KNOWLEDGE entry.
The CROSS_TEST sub-database integrates both ACTIVITY and relevant database entries by cross-testing the KNOWLEDGE-documented computational procedure on independent data (Fig. 4). Each CROSS_TEST sub-database entry has 12 fields: MI, identifier; MN, name; WW, web resource; DR, database; MD, adaptation procedure; AB, sequence region; LC, linear correlation coefficient; XI, χ2-coefficient of the site/random DNA discrimination; ST, means, standard deviation, false negatives; NT, means, standard deviation, false positives; AL, significance α; C-, computational procedure adapting the sequence-activity relationship from one measurement system to another (Fig. 4). As can be seen, this entry gives the statistical reasoning why one system could be adaptive to another (LC, XI, ST, NT and AL). Within these statistical limits, one may adapt computational procedures by implementing a C-coded program (C-). To provide the query for the measurement system cross-test results, there are two keyword descriptor fields (AB and MD).
Figure 4.
Example of a CROSS-TEST entry.
DATABASE CONTENT
This version of the ACTIVITY database contains 554 entries citing 265 original publications. Since the influence of the measurement system on sequence-activity relationships is not well studied yet, only 70 entries are examples of the most well studied sites (Inr-element, TATA-box, YY1-binding site, OR1-operator, etc.) and were selected for inclusion into the current SYSTEM sub-database release. Twenty-three entries, exemplifying activity-measurement systems and referring only to selected sites, were treated by the knowledge discovery system (17). The results are stored in the KNOWLEDGE sub-database. The CROSS_TEST accumulates over 100 cross-tests clustered by sequence-activity relationships.
All these cross-tests were statistically significant. However, only half of them correspond to both key/lock intermolecular recognition and statistical-mechanical theory of DNA–protein interactions (12,13). The other adaptation methods take into account the various surrounding site-dependent statistics, i.e., means, minimal, maximal activity estimates and the differences between them. These surround-dependent adaptations are in accordance with the ‘jigsaw puzzle’ concept (26), which states that DNA–protein and protein–protein-interaction co-exist and co-adapt with each other in a multivariate regulatory machine. Since protein–protein interactions may influence DNA–protein interactions, the surround-dependent statistics describe the regulatory machine more flexibly by the ‘jigsaw puzzle’ concept than by the inflexible positional estimates. This reasoning is consistent with recent work (29) demonstrating the necessity of surround-dependent estimates in addition to a Weight Matrix Score for prediction of the CTF/NFI–DNA affinity, which could not be predicted just by a positional estimate (30). All the cross-test results given in our work indicate that the basis of a sequence-activity relationship is system-invariant, whereas relationships between the site and its surroundings could be system-dependent and lead to varying activity values. This approach may be useful for pharmacogenetics and for drug design.
AVAILABILITY
ACTIVITY is available through the Web, http://wwwmgs.bionet.nsc.ru/systems/Activity/. Please email all ACTIVITY applications to Mrs J. V. Ponomarenko (jpon@bionet.nsc.ru) or request collaborations through Prof. N. A. Kolchanov (kol@bionet.nsc.ru). No inclusion of ACTIVITY into other databases is permitted without explicit permission of the authors. Please send comments, corrections and requests by email or fax (+7 3832 331278). We kindly ask that this article be cited when reporting the results based on ACTIVITY usage.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The work is supported by a grant, RFBR-98-07-910126 (Russia), and STA Fellowship #499042 (Japan).
References
- 1.Haussler D. (1998) Computational genefinding. Trends Guide in Bioinformatics, 1, 12–15. [Google Scholar]
- 2.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Prüß,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. Updated article in this issue: Nucleic Acids Res. (2001), 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolchanov N.A., Podkolodnaya,O.A., Ananko,E.A., Ignatieva,E.V., Stepanenko,I.L., Kel-Margoulis,O.V., Kel,A.E., Merkulova,T.I., Goryachkovskaya,T.N., Busygina,T.V. et al., (2000) Transcription regulatory regions database (TRRD): its status in 2000. Nucleic Acids Res., 28, 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponomarenko J.V., Orlova,G.V., Ponomarenko,M.P., Lavryushev,S.V., Frolov,A.S., Zybova,S.V. and Kolchanov,N.A. (2000) SELEX_DB: an activated database on selected randomized DNA/RNA sequences addressed to genomic sequence annotation. Nucleic Acids Res., 28, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schug J. and Overton,G.C. (1997) TESS: Transcription Element Search Software on the WWW. Technical Report CBIL-TR-1997-1001-v0.0. Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania.
- 6.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector—New fast and sensitive tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Q., Hertz,G. and Stormo,G. (1995) Matrix search 1.0: a computer program that scans DNA sequences for transcriptional elements using a database of weight matrices. Comput. Appl. Biosci., 11, 563–566. [DOI] [PubMed] [Google Scholar]
- 8.Roses A.D. (2000) Pharmacogenetics and the practice of medicine. Nature, 405, 857–865. [DOI] [PubMed] [Google Scholar]
- 9.Vasiliev G.V., Merkulov,V.M., Kobzev,V.F., Merkulova,T.I., Ponomarenko,M.P., and Kolchanov,N.A. (1999) Point mutations within 663–666 bp of intron 6 of the human TDO2 gene, associated with a number of psychiatric disorders, damage the YY-1 transcription factor binding site. FEBS Lett. 462, 85. [DOI] [PubMed] [Google Scholar]
- 10.Mulligan M.E., Hawley,D.K., Entriken,R. McClure,W.R. (1984) Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res., 12, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonsson J., Norberg,T., Carlsson,L., Gustafsson,C. and Wold,S. (1993) Quantitative sequence-activity models (QSAM)—tools for sequence design. Nucleic Acids Res., 21, 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg O.G. and von Hippel,P.H. (1987) Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol., 193, 723–750. [DOI] [PubMed] [Google Scholar]
- 13.Berg O.G. and von Hippel,P.H. (1988) Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J. Mol. Biol., 200, 709–723. [DOI] [PubMed] [Google Scholar]
- 14.Stormo G.D., Schneider,T.D. and Gold,L. (1986) Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res., 14, 6661–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrick D., Villanueba,K., Childs,J., Kalil,R., Schneider,T.D., Lawrence,C.E., Gold,L. and Stormo,G.D. (1994) Quantitative analysis of ribosome binding sites in E.coli. Nucleic Acids Res., 22, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraus R.J., Murray,E.E., Wiley,S.R., Zink,N.M., Loritz,K., Gelembiuk,G.W. and Mertz,J.E. (1996) Experimentally determined weight matrix definitions of the initiator and TBP binding site elements of promoters. Nucleic Acids Res., 24, 1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ponomarenko M.P., Ponomarenko,J.V., Frolov,A.S., Podkolodny,N.L., Savinkova,L.K., Kolchanov,N.A. and Overton,G.C. (1999) Identification of sequence-dependent DNA features correlating to activity of DNA sites interacting with proteins. Bioinformatics, 15, 687–703. [DOI] [PubMed] [Google Scholar]
- 18.Takeda Y., Sarai,A. and Rivera,V.M. (1989) Analysis of the sequence-specific interactions between Cro repressor and operator DNA by systematic base substitution experiments. Proc. Natl Acad. Sci. USA, 86, 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarai A. and Takeda,Y. (1989) Lambda repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proc. Natl Acad. Sci. USA, 86, 6513–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanikawa J., Yasukawa,T., Enari,M., Ogata,K., Nishimura,Y., Ishii,S. and Sarai,A. (1993) Recognition of specific DNA sequences by the c-myb protooncogene product: role of three repeat units in the DNA-binding domain.Proc. Natl Acad. Sci. USA, 90, 9320–9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogata K., Kanei-Ishii,C., Sasaki,M., Hatanaka,H., Nagadoi,A., Enari,M., Nakamura,H., Nishimura,Y., Ishii,S. and Sarai,A. (1996) The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol., 3, 178–187. [DOI] [PubMed] [Google Scholar]
- 22.Hao D., Ohme-Takagi,M. and Sarai,A. (1998) Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem., 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- 23.McCormick A., Brady,H., Fukushima,J. and Karin,M., (1991) The pituitary-specific regulatory gene GHF1 contains a minimal cell type-specific promoter centered around its TATA box. Genes Dev., 5, 1490–1503. [DOI] [PubMed] [Google Scholar]
- 24.Hyde-DeRuyscher R., Jennings,E. and Shenk,T. (1995) DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res., 23, 4457–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Javahery R., Khachi,A., Lo,K., Zenzie-Gregory,B. and Smale,S.T. (1994) DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol., 14, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson P.F. and McKhight,S. (1989) Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem., 58, 799–829. [DOI] [PubMed] [Google Scholar]
- 27.Ludlow L.B., Schick,B.P., Budarf,M.L., Driscoll,D.A., Zackai,E.H., Cohen,A. and Konkle,B.A. (1996) Identification of a mutation in a GATA binding site of the platelet glycoprotein Ibbeta promoter resulting in the Bernard-Soulier syndrome. J. Biol. Chem., 271, 22076–22080. [DOI] [PubMed] [Google Scholar]
- 28.Etzold T. and Argos,P. (1993) SRS—an indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci., 9, 49–57. [DOI] [PubMed] [Google Scholar]
- 29.Roulet E., Bucher,P., Schneider,R., Wingender,E., Dusserre,Y., Werner,T. and Mermod,N. (2000) Experimental analysis and computer prediction of CTF/NFI transcription factor DNA binding sites. J. Mol. Biol., 297, 833–848. [DOI] [PubMed] [Google Scholar]
- 30.Roulet E., Fisch,I., Bucher,P. and Mermod,N. (1998) Evaluation of computer tools for prediction of transcription factor binding sites on genomic DNA. In Silico Biology, 1, 21–28. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.