Summary
The health of organisms and cells depends on appropriate responses to diverse internal and external cues, stimuli, or challenges, such as changes in hormone or cytokine levels, or exposure to a pathogen. Cellular responses must be tailored to the identity and intensity of the stimulus and therefore intracellular signals must carry information about both. However, signaling mediators often form intricate networks that react to multiple stimuli yet manage to produce stimulus-specific responses. The multi-functionality (“functional pleiotropism”) of signaling nodes suggests that biological networks have evolved ways of passing physiologically relevant stimulus information through shared channels. Increasing evidence supports the notion that this is achieved in part through temporal regulation of signaling mediators’ activities. The present challenge is to identify the features of temporal activity profile that represent information about a given stimulus and understand how cells read the temporal codes to control their responses.
What's in a signal?
The striking temporal control of signaling mediators’ activity revealed by recent studies suggest that dynamics (understood as the spatiotemporal patterns of activity) are an intrinsic part of a signal [1,2] that, together with the chemical identity of the mediator, carries information about the stimulus. Pronounced temporal control of signaling mediators is particularly pervasive in stress [3,4] and immune responses [5,6], triggered by stimuli that have a well-defined starting point, at least in cell culture studies.
In general, signal dynamics often depend on one or more properties of the stimulus, such as its identity, its amplitude, rate of increase, duration, or rate of decrease. Particular stimulus features do not necessarily translate into equivalent signal features; for example, stimulus amplitude may determine signal duration or vice versa. Signals are often classified as amplitude or frequency modulated (AM or FM). Signals are thought to be amplitude modulated when their amplitude, duration, or a combination of both is modulated by the stimulus. Frequency modulation is ascribed to particular cases when the frequency of a periodic signal is a function of the stimulus. In this review, we advance the view that to understand temporal signaling codes, we must focus on not only on the encoding mechanism that relates stimulus to signal, but also on the decoding mechanism that relates signal to cellular response.
The signal-as-information paradigm
In several model systems, signal dynamics have been shown to control the specificity of a response. For example, it has been shown that different inflammatory stimuli generate distinct temporal profiles of the activity of the central node kinase IKK or transcription factor NFκB and that temporal regulation plays a key role determining which subset of target genes are activated [5,7-9].
In the “signal-as-information” paradigm, information about the stimulus is “encoded” into a set of “coding features”; these are relayed and potentially post-processed until they are “decoded” to generate the cellular response (Figure 1A and B). Alternative approaches de-emphasize the interpretation of the spatiotemporal dynamics of signaling mediators in favor of correlating input-output relationships linking stimuli, signals, and responses [10]. A cursory glance at a typical interaction network map [11] may indeed suggest that attempting to track information as it propagates through a cascade full of feedback and feed-forward loops is a hopeless task. However, the complexity of these maps is deceiving, as different parts operate in different cell types, and within the same cell different parts operate on different time scales. Thus, it is often possible to isolate relevant functional modules [2,12-15] and study the propagation of signals within and in between them. In those cases for which a modular biology approach is possible, the “signal-as-information” paradigm provides a sensible framework as the encoding mechanism for one signaling module may represent the decoding mechanism of the previous one.
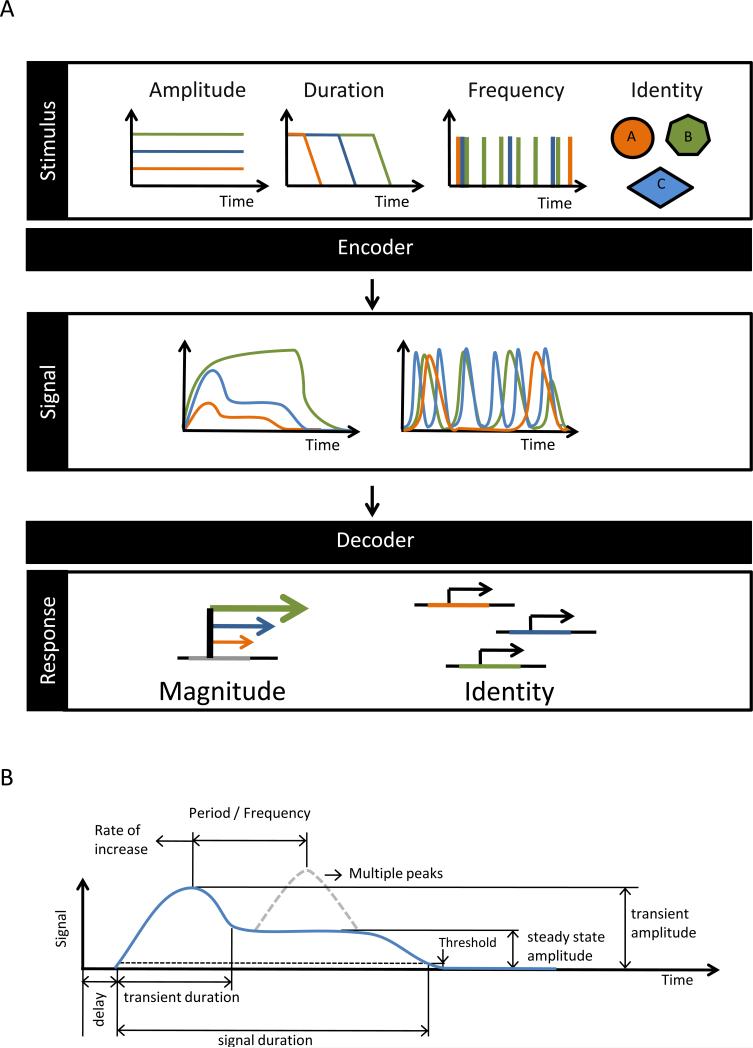
Figure 1. Features of the Stimulus are encoded in features of the signal, determine the expression of genes.
(A) Features of the stimulus, such as its type (identity), amplitude, duration, or frequency, are encoded by receptor-associated signaling networks into different features of the intra-cellular signal's temporal profile. Gene regulatory networks decode the information contained in the features of the signal's temporal profile and thus determine whether a specific gene is activated and by how much. (B) An example of a timecourse of an amplitude-modulated intra-cellular signal and potential coding features: Transient and steady state amplitudes, transient duration, duration over a threshold, number and time between peaks, etc.
How can we extract the information present in a signal in order to understand the operable temporal signaling codes? Phenomenological investigations of signal dynamics allow us merely to speculate on the information content of specific features or aspects of dynamical signal profiles. Instead, we suggest that understanding a temporal “code” and its associated “coding features” must involve studies of the mechanisms and properties of the decoding network. In the following sections we discuss selected literature to examine this view point.
Amplitude modulated (AM) codes
In AM signals, stimulus information may be represented in the signal's amplitude, duration, or in a complex combination of both. In pure amplitude encoding, the relevant aspects of the stimulus are reflected into the “amplitude” of the signal. Complex signals may carry amplitude-encoded information in multiple ways. For example in a biphasic signal, information about the stimulus could be reflected in the amplitude of the early transient peak or in the level of activity the signaling mediator settles in after the initial overshoot (steady state), or both (Figure 1B). Depending on the characteristic response time of the decoding mechanism only one of those amplitudes may act as a coding feature with physiological meaning; this emphasizes the importance of characterizing decoding mechanisms in order to identify the signaling code within AM signals.
Stimulus amplitude being encoded into the enzymatic activity of successive kinases or other signaling mediators has been documented in numerous systems, some dating back to the earliest experiments on receptors [16-18]. Amplitude-into-amplitude encoding occurs naturally when the stimulus is within the subsaturating regime of the receptor dose-response curve. When the signaling mediator is allowed to reach a steady state, the amplitude code is unambiguous regarding stimulus dose and duration. Signal amplitude can also encode information about the stimulus duration. This may occur, for example, when a component of the pathway has a longer characteristic activation timescale than the duration of the stimulus, such that the maximum signal activity will reflect the duration of an activating stimulus of known amplitude [19]. Signals may also be amplified or attenuated as they travel through the network [19,20], and in some cases, the activity of a signaling mediator can significantly outlast the stimulus, thus providing for temporal amplification or a short-term memory of a transient stimulus.
Whether signal amplitude is the relevant coding feature is determined by the decoding mechanism that controls the response. In fact, pure amplitude decoding provides for a simple information code, as substrate-product relationships or interaction affinities readily function as decoding mechanisms. In gene expression, the activity of many promoters depends on the nuclear concentration of the corresponding transcription factors because DNA-protein interactions are subsaturated and fast [21]. A recent study of the TNF-NFκB axis identified A20 as a rheostat for amplitude encoding [22], another revealed that clustered NFκB binding sites are important for amplitude decoding [23]. These studies address the TNF regime above 0.1ng/ml when the majority of cells are responsive, whereas below that concentration the cellular response appears to be thresholded [9].
The duration of an AM signal may sometimes be the relevant “coding feature”. Signal duration is a relative concept because the activity level of a mediator rarely remains constant. Duration usually refers to the time duration for which a particular signaling mediator activity remains above a biologically relevant threshold. However, when duration is the sole coding feature, the amplitude of the signal is irrelevant, either because it is constant, or because any variation in amplitude is not detected by the decoding mechanism (possibly because it is easily saturable).
In the simplest of cases signal duration may reflect the stimulus duration. However, encoding amplitude into duration can also be achieved in several ways. Transient stimuli may drive the production or modification of a pathway component in a dose-dependent manner. The time it takes the active species to decay to below the biologically relevant threshold once the stimulus abates depends on the amplitude reached by the signal, which, in turn, is a function of the stimulus dose and duration. There is potential ambiguity in this encoding strategy, but it is avoided if the duration of the stimulus is a fixed physiological parameter or it is longer than the characteristic time for the signaling mediator or decoding mechanism. Persistent stimuli can undergo dose-to-duration encoding through the action of some types of adaptive systems [24,25]. For example, it has been shown that slow negative feedback can perform this conversion because the time it takes to shut down the signal depends on the intensity of the stimulus [25-27]. Information about the identity of a stimulus can be encoded into signals of different durations by receptor associated signaling networks or positive feedback mechanisms [8].
Duration information can be decoded by virtue of the kinetics of the pathway's components. For example, decoding can be performed at the gene expression level when the gene product abundance is determined by the duration of active transcription. Differences in the time it takes promoters to become accessible or for complexes necessary for transcription initiation to form or simple mRNA stability could function as basis for duration decoding [28-30]. Within a network, signal duration can determine specificity by selectively activating targets (decoding circuits) according to their kinetics [31].
There are biological examples for which duration appears to be the main coding feature [32-35]. One of the most studied is the developmental switch in PC12 cells [34,35]. Exposure of these cells to NGF triggers sustained ERK activity and leads to differentiation, whereas exposure to EGF results in transient ERK activity and promotes proliferation. A similar ERK-dependent effect involving entry into S-phase has been observed in 3T3 fibroblasts in response to PDGF and EGF [36,37]. In these examples, the stimulus identity is encoded in the duration of ERK activity by the action of the receptors specific for each growth factor [38]. Decoding is achieved in the 3T3 system through the ERK-dependent stabilization of immediate early genes products [36,37,39,40] occurring only in the presence of a sustained signal. In yeast, it has been proposed that the switch between the mating and filamentous growth phenotype depends on the duration of MAP kinase Kss1 activity [41], which also has been linked to a related morphogenic decision-making process [42]. Interestingly, the concentration of mating pheromone in this system appears to be encoded into the duration of the signal at the MAPKK level (and possibly upstream) but decoded and converted into maximum amplitude by a MAPK with slow activation kinetics [26]. Aderem and collaborators recently uncovered a feedforward circuit underlying the decoding of persistent and transient TLR4-induced signals in macrophages [43] that could have important implications for the control of immune responses.
Experimentally, strict duration and to a lesser degree strict amplitude encoding is hard to demonstrate because amplitude and duration of a signal can rarely be encoded or decoded independently. While signals appear amplitude-modulated in the above-cited examples, the unequivocal characterization of the operable code requires a deeper understanding of the decoding mechanisms.
Frequency modulated codes
The presence of calcium oscillations in a variety of cell types coupled with the fact that many stimuli seem to affect the frequency rather than the amplitude [44-46] led Berridge and Galioni [47] to propose the notion of “frequency encoded” signals. In frequency-modulated signals, the frequency, or the number of pulses per unit time, carries the information about the stimulus and is read by a decoding mechanism to determine the cellular response. From a theoretical standpoint, biochemical oscillations can be generated whenever there is a delayed negative feedback, although non-linear kinetics are necessary for sustained oscillations [48,49]. However, to operate as a frequency encoder a delayed negative feedback circuits must generate signals with stimulus-dependent frequency modulation, and unless the delay is stimulus-dependent, this usually requires additional positive and negative feedback loops [see ref. 50 for an example] .
Mechanisms based on desensitization and recovery of a mediator, in which high and low frequency signals can result in low and high activity respectively, have been proposed as decoders of frequency-encoded information [51]. Initially studied in the context of inter-cellular communication, such mechanisms may also apply to intracellular processes when differences in the kinetics or stability of the decoding components are considered. Alternatively, the stability (i.e. half-life) of an active mediator can restrict the frequency response of a network. For example the rapid decay of a short-lived mediator may prevent a system from responding to input pulses that occur too far apart but maintain a sustained enough level in response to more frequent input pulses. Such decoding mechanisms have been proposed to explain the frequency-dependent response to pulses of Ca signals of the NFAT and NFκB transcription factors in T cells [52,53] and NFAT and gonadotropin components in gonadotrope signaling [54,55].
Because these mechanisms operate as integrators of repeated peaks of activity, they cannot distinguish between an increase in the number of peaks per unit time and an equivalent increase in pulse duration, so they are not considered true frequency decoders [56]. True frequency decoding can be achieved by networks containing incoherent feed-forward motifs can generate frequency-dependent responses largely insensitive to peak duration [56]. True frequency decoding may be performed by regulatory networks with natural resonant frequencies that therefore respond maximally to signals with frequencies close to the frequencies of their natural oscillations and may thus be classified as band pass filters [57].
To date, despite intense research, the details of the encoding and decoding mechanisms associated with the various types of periodic calcium signals are still not fully understood [58-65]. Periodic signals have been extensively studied also in the context of gonadotropin-releasing hormone, a stimulus that is itself periodic [54,56,66,67]. In this case, changes in the periodicity of the stimulus occurring normally during the female reproductive cycle lead to the production of alternative varieties of gonadotropin hormones [68,69]. Interestingly, the observation of a non-monotonic frequency response curve for the expression of the gonadotropin component genes led some to propose that yet unknown regulatory mechanisms must be operating in the this network as true frequency decoders [55].
In the absence of a well-documented frequency decoding mechanism, other biological functions have been proposed for periodic signals: first, spike-like signals may have evolved as way to prevent deleterious effects of tonic exposure to an active messenger or to prolong the effect of a species stored in small amounts. Second, and related, theoretical studies found that the utility of duration encoding in peaks may lie in a reduction of the effective activation threshold for the signal's targets [61]. Third, supported by a series of elegant experiments, Elowitz and collaborators showed that frequency-encoded signals ensure the coordinated expression of genes with different promoter dose-response curves regardless of the stimulus dose [70], thereby insulating gene expression control from variations in amplitude or affinity. However, in some cases oscillations may simply be an artifact of synthetic reporters inserted into cells, un-physiological stimulation regimes, such as the sudden exposure to an agonist, or they may be byproducts of the multiple layers of regulation present in most pathways (“ringing”) rather than a bona fide coding feature [7,71,72]. In fact, in many signaling networks, stochastic variation in the time delay associated with the de-novo production of a regulator (via transcriptional bursting for example) may induce pseudo-oscillations [73-75]. Because of their origin, these oscillations are not a function of the stimulus and are unlikely to carry information about it. Using periodic stimuli to excite a network [76] may be useful to probe dynamic properties, but they may reveal little about the information carried (if any) by intrinsically produced oscillations.
For Complex Signals, the Decoding Mechanism defines the Code
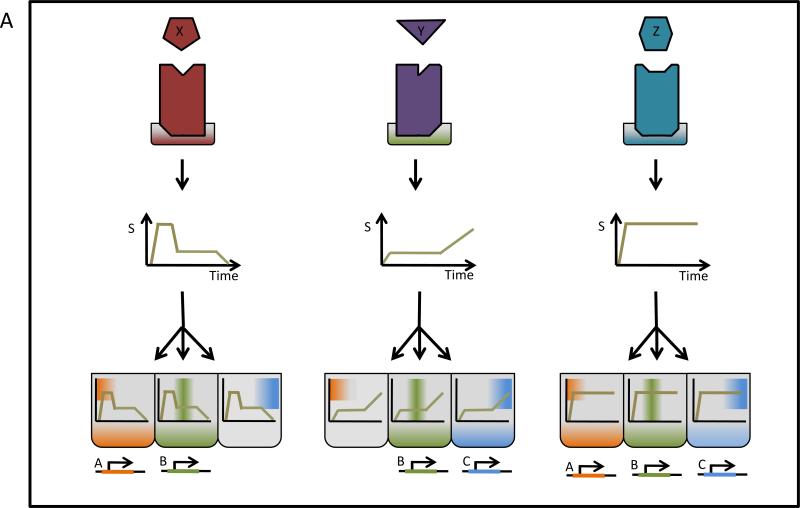
Unlike communications devices that broadcast signals using clearly defined protocols, most biological signaling networks produce complex dynamics and it is not always obvious which features of the temporal profile of the signal encode information. In fact, different encoding schemes might be used at different phases (early and late) or at different stages as the signal progresses through the network. The observation that a specific signal feature correlates with a particular aspect of a stimulus does not by itself demonstrate that is represents a physiologically relevant coding feature, just as the discovery of a specific post-translational modification by increasingly sensitive tools does not provide evidence for their physiological function. We argue that characterization of the operational signaling code requires a focus on the decoding mechanisms aiming to produce predictive mathematical descriptions. Indeed, a complex temporal profile of a pleiotropic regulator is likely to be read differently by different targets employing different decoding mechanisms to control distinct cellular responses (Figure 3).
Figure 3. Decoding mechanisms define the signaling code in complex signals.
Stimulus X, Y, and Z are encoded into complex signals. The presence or absence of specific dynamical features determines the what feature of the temporal signaling profile the decoders respond to. In this example, response B is common to stimuli A, Y., Z, but response A and C are more stimulus-sepcific.
In gene regulation, the promoter DNA provides a scaffold for diverse decoding mechanisms involving multiple constitutive and stimulus-responsive regulators. The highly specific regulation of gene expression in response to many stimuli is usually attributed to the combined action of multiple transcription factors on promoters and enhancers [77-80]. Combinatorial control is often thought as a static process in which the mere presence of the correct combination of factors is enough to elicit the correct specific response [78]. The picture, however, is not as simple not only because the adequate combination of transcription factors and co-activators must be present at the right times [28,30,81,82], but also because there may be additional layers of activity control via post-translational modifications and co-factors. The need for modifications of the chromatin environment emphasizes that gene regulation is a multi-step process, and each step may also contribute to combinatorial control in which dynamics play a role [36,37,83]. A particularly interesting example is the demonstration by Chung and coworkers of two distinct phases of signaling with different specificity driving the differentiation of PC12 cells [39]. The existence in a network of branches with distinct dynamics [3,26,42] could be necessary to ensure that the proper sequence of signals is present, signals that could be considered as bona fide components of a complex combinatorial signaling code.
Conclusions
A signaling code may consist of any of many potential coding features: The timing of a signal peak, its rate (i.e. derivative) of activation or inactivation, the phase, amplitude, and/or duration relationship between successive activity peaks could all encode information about the stimulus. However, the regulatory network that decodes the signal determines which feature is a functionally relevant coding feature. Hence, the decoder determines the signaling code. Kinetics, cooperativity, feedback and feedforward circuits may underlie decoders capable of responding only to specific family of temporal activity profiles among the complex universe of possible temporal profiles. Rich dynamics endow signaling networks with sufficient versatility to insulate information traveling through shared channels [31,84,85] and to control distinct responses (Figures 2 and 3). As the examples discussed here illustrate, the signals-as-information paradigm provides a useful framework to investigate these processes providing at the same time a rich descriptive framework to organize our understanding of signaling networks. We surmise that understanding the nature of the decoding mechanism is paramount as it defines the operational code for a specific target and downstream cellular response.
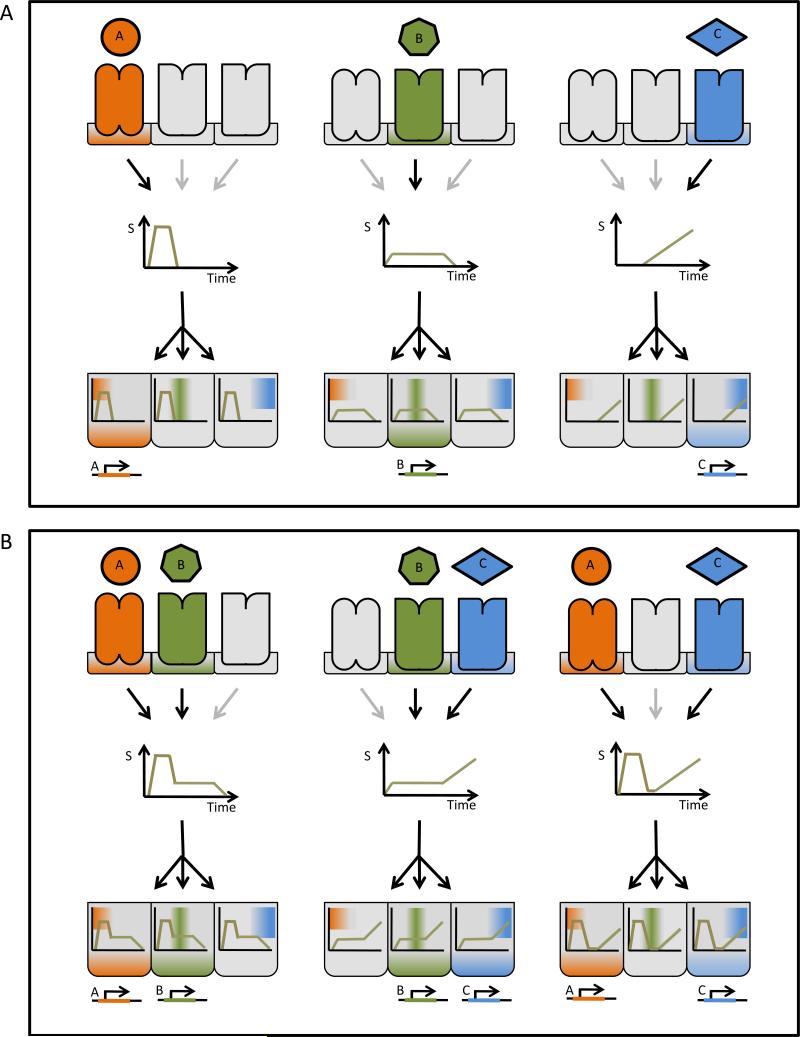
Figure 2. Temporal profiles can carry specific signal information for multiple pathways.
(A) Decoders A, B, and C, represented here by different gene regulatory networks, are sensitive to particular dynamical features of the signaling mediator's activity. Decoder A responds to early strong signals, decoder B is requires longer lasting signals regardless of the amplitude, whereas decoder C detects late strong signals. In this example, encoders associated with different receptors produce stimulus-specific temporal patterns that control the nature of the response. (B) The temporal code allows shared signaling mediators to respond properly to simultaneous stimuli.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brent R. Cell signaling: what is the signal and what information does it carry? FEBS Lett. 2009;583:4019–4024. doi: 10.1016/j.febslet.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 2 *.Alexander RP, Kim PM, Emonet T, Gerstein MB. Understanding modularity in molecular networks requires dynamics. Sci Signal. 2009;2:pe44. doi: 10.1126/scisignal.281pe44. [Thoughtfull perspective article stressing the role dynamics may play defining functional modules in biological systems] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao N, Behar M, Parnell SC, Torres MP, Borchers CH, Elston TC, Dohlman HG. A systems-biology analysis of feedback inhibition in the sho1 osmotic-stress-response pathway. Curr Biol. 2007;17:659–667. doi: 10.1016/j.cub.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 4.Klipp E, Nordlander B, Kruger R, Gennemark P, Hohmann S. Integrative model of the response of yeast to osmotic shock. Nat Biotechnol. 2005;23:975–982. doi: 10.1038/nbt1114. [DOI] [PubMed] [Google Scholar]
- 5 **.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [Experimenatl and computational modeling study of the roles of IkB proteins in tempral control of NFkB activity. Makes the case for duration encoding in NFkB pathway and that apparent oscillatory control of NFkB is a byproduct of potent negative feedback whose primary function is to ensure prompt signal termination upon stimulus removal] [DOI] [PubMed] [Google Scholar]
- 6 *.Bode KA, Schmitz F, Vargas L, Heeg K, Dalpke AH. Kinetic of RelA activation controls magnitude of TLR-mediated IL-12p40 induction. J Immunol. 2009;182:2176–2184. doi: 10.4049/jimmunol.0802560. [The authors show receptors of the TLR family activating the NFkB pathway with distinct dynamics upon stimulation with specific ligands affecting whether cytokine IL12 is produced or not as part of the response] [DOI] [PubMed] [Google Scholar]
- 7.Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8 *.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [Experimental demonstration that the stimulus identity elicits specific temporal patterns of IKK activity and that these are important in the specificity of the gene expression response] [DOI] [PubMed] [Google Scholar]
- 9.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saez-Rodriguez J, Alexopoulos LG, Epperlein J, Samaga R, Lauffenburger DA, Klamt S, Sorger PK. Discrete logic modelling as a means to link protein signalling networks with functional analysis of mammalian signal transduction. Mol Syst Biol. 2009;5:331. doi: 10.1038/msb.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol. 2006;2:2006 0015. doi: 10.1038/msb4100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmeyr JH, Westerhoff HV. Building the cellular puzzle: control in multi-level reaction networks. J Theor Biol. 2001;208:261–285. doi: 10.1006/jtbi.2000.2216. [DOI] [PubMed] [Google Scholar]
- 14.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 15.Hofmann KP, Spahn CM, Heinrich R, Heinemann U. Building functional modules from molecular interactions. Trends Biochem Sci. 2006;31:497–508. doi: 10.1016/j.tibs.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Black JW, Leff P. Operational models of pharmacological agonism. Proc R Soc Lond B Biol Sci. 1983;220:141–162. doi: 10.1098/rspb.1983.0093. [DOI] [PubMed] [Google Scholar]
- 17.Mackeigan JP, Murphy LO, Dimitri CA, Blenis J. Graded mitogen-activated protein kinase activity precedes switch-like c-Fos induction in mammalian cells. Mol Cell Biol. 2005;25:4676–4682. doi: 10.1128/MCB.25.11.4676-4682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18 **.Yu RC, Pesce CG, Colman-Lerner A, Lok L, Pincus D, Serra E, Holl M, Benjamin K, Gordon A, Brent R. Negative feedback that improves information transmission in yeast signalling. Nature. 2008;456:755–761. doi: 10.1038/nature07513. [Single cell measurement of response functions of succesive stages in the yeast mating pathway] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinrich R, Neel BG, Rapoport TA. Mathematical models of protein kinase signal transduction. Mol Cell. 2002;9:957–970. doi: 10.1016/s1097-2765(02)00528-2. [DOI] [PubMed] [Google Scholar]
- 20 *.Gomez-Uribe C, Verghese GC, Mirny LA. Operating regimes of signaling cycles: statics, dynamics, and noise filtering. PLoS Comput Biol. 2007;3:e246. doi: 10.1371/journal.pcbi.0030246. [Exhaustive analysis of the dynamic and static properties of the ubiquitous two-state switch] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kappaB-dependent gene activity. Embo J. 2006;25:798–810. doi: 10.1038/sj.emboj.7600977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner SL, Kearns JD, Zadorozhnaya V, Lynch C, O'Dea E, Boldin MP, Ma A, Baltimore D, Hoffmann A. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22:2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23 **.Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T, Pasparakis M, Milani P, Bulyk ML, Natoli G. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell. 37:418–428. doi: 10.1016/j.molcel.2010.01.016. [A fully documented example of amplitude encoding. The authors demonstrate separatelly that NF-kB activity follows the concentration of the stimulus and that the expression of some target genes correlates with the activity of NF-kB] [DOI] [PubMed] [Google Scholar]
- 24 *.Ma W, Trusina A, El-Samad H, Lim WA, Tang C. Defining network topologies that can achieve biochemical adaptation. Cell. 2009;138:760–773. doi: 10.1016/j.cell.2009.06.013. [Exhaustive analysis of adaptive regulatory motifs that could operate as encoders for AM signals] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behar M, Hao N, Dohlman HG, Elston TC. Mathematical and Computational Analysis of Adaptation via Feedback Inhibition in Signal Transduction Pathways. Biophys J. 2007;93:806–821. doi: 10.1529/biophysj.107.107516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26 *.Behar M, Hao N, Dohlman HG, Elston TC. Dose-to-duration encoding and signaling beyond saturation in intracellular signaling networks. PLoS Comput Biol. 2008;4:e1000197. doi: 10.1371/journal.pcbi.1000197. [The authors propose a mechanism for amplitude-to-duration encoding and make the case for duration encoding being used to represent the agonist concentration in the phermone pathway in yeast] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27 *.Locasale JW. Signal duration and the time scale dependence of signal integration in biochemical pathways. BMC Syst Biol. 2008;2:108. doi: 10.1186/1752-0509-2-108. [Mathematical analysis of the dynamic properties of adaptive systems containing negative feedback loops that can operate as filters, encoders, and decoders] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natoli G. Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol. 2009;1:a000224. doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29 *.Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [A review article on the role played by enhancers elements controlling gene expression and potentially decoding information contained in the temporal profiles of a signal] [DOI] [PubMed] [Google Scholar]
- 30.Michel D. How transcription factors can adjust the gene expression floodgates. Prog Biophys Mol Biol. 2010;102:16–37. doi: 10.1016/j.pbiomolbio.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 31 **.Behar M, Dohlman HG, Elston TC. Kinetic insulation as an effective mechanism for achieving pathway specificity in intracellular signaling networks. Proc Natl Acad Sci U S A. 2007;104:16146–16151. doi: 10.1073/pnas.0703894104. [A theoretical mechanism that uses dynamicall encoding and decoding to relay specificity information through shared pathway components] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tombes RM, Auer KL, Mikkelsen R, Valerie K, Wymann MP, Marshall CJ, McMahon M, Dent P. The mitogen-activated protein (MAP) kinase cascade can either stimulate or inhibit DNA synthesis in primary cultures of rat hepatocytes depending upon whether its activation is acute/phasic or chronic. Biochem J. 1998;330(Pt 3):1451–1460. doi: 10.1042/bj3301451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ventura JJ, Hubner A, Zhang C, Flavell RA, Shokat KM, Davis RJ. Chemical genetic analysis of the time course of signal transduction by JNK. Mol Cell. 2006;21:701–710. doi: 10.1016/j.molcel.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 34 *.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [Experimentally supported example of duration-encoded specificity] [DOI] [PubMed] [Google Scholar]
- 35.Qui MS, Green SH. PC12 cell neuronal differentiation is associated with prolonged p21ras activity and consequent prolonged ERK activity. Neuron. 1992;9:705–717. doi: 10.1016/0896-6273(92)90033-a. [DOI] [PubMed] [Google Scholar]
- 36 **.Murphy LO, MacKeigan JP, Blenis J. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol Cell Biol. 2004;24:144–153. doi: 10.1128/MCB.24.1.144-153.2004. [Experimentally supported instance of duration decoding during 3T3 cells differentiation. The authors show that sustained signaling is required to prevent c-Fos degradation and allow the progression of the response] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556–564. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 38 **.Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [Combining experimental and computational approaches, the authors that changes in the network topology can account for the dynamic encoding of specificity in PC12 exposed to NGF or EGF] [DOI] [PubMed] [Google Scholar]
- 39 **.Chung J, Kubota H, Ozaki Y, Uda S, Kuroda S. Timing-dependent actions of NGF required for cell differentiation. PLoS One. 2010;5:e9011. doi: 10.1371/journal.pone.0009011. [Experimental study of the action of specific ligands during the different phases of differentiation signals in the PC12 system] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chalmers CJ, Gilley R, March HN, Balmanno K, Cook SJ. The duration of ERK1/2 activity determines the activation of c-Fos and Fra-1 and the composition and quantitative transcriptional output of AP-1. Cell Signal. 2007;19:695–704. doi: 10.1016/j.cellsig.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 41 *.Sabbagh W, Jr., Flatauer LJ, Bardwell AJ, Bardwell L. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol Cell. 2001;8:683–691. doi: 10.1016/s1097-2765(01)00322-7. [The authors suggest the temporal activity profile of the pleiotropic MAPK Kss1 may play a role determining the response as yeast is exposed to specific conditions (also see ref. 26 and 84)] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG. Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell. 2008;30:649–656. doi: 10.1016/j.molcel.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy KA, Lampano AE, Nykter M, Shmulevich I, Aderem A. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schofl C, Brabant G, Hesch RD, von zur Muhlen A, Cobbold PH, Cuthbertson KS. Temporal patterns of alpha 1-receptor stimulation regulate amplitude and frequency of calcium transients. Am J Physiol. 1993;265:C1030–1036. doi: 10.1152/ajpcell.1993.265.4.C1030. [DOI] [PubMed] [Google Scholar]
- 45.D'Andrea P, Codazzi F, Zacchetti D, Meldolesi J, Grohovaz F. Oscillations of cytosolic calcium in rat chromaffin cells: dual modulation in frequency and amplitude. Biochem Biophys Res Commun. 1994;205:1264–1269. doi: 10.1006/bbrc.1994.2801. [DOI] [PubMed] [Google Scholar]
- 46.Berridge MJ. The AM and FM of calcium signalling. Nature. 1997;386:759–760. doi: 10.1038/386759a0. [DOI] [PubMed] [Google Scholar]
- 47.Berridge MJ, Galione A. Cytosolic calcium oscillators. Faseb J. 1988;2:3074–3082. doi: 10.1096/fasebj.2.15.2847949. [DOI] [PubMed] [Google Scholar]
- 48.Ferrell JE, Jr., Pomerening JR, Kim SY, Trunnell NB, Xiong W, Huang CY, Machleder EM. Simple, realistic models of complex biological processes: positive feedback and bistability in a cell fate switch and a cell cycle oscillator. FEBS Lett. 2009;583:3999–4005. doi: 10.1016/j.febslet.2009.10.068. [DOI] [PubMed] [Google Scholar]
- 49 *.Novak B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol. 2008;9:981–991. doi: 10.1038/nrm2530. [Survey of network motifs capable of producing oscillations and could operate as frequency encoders] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Goldbeter A. Frequency specificity in intercellular communication. Influence of patterns of periodic signaling on target cell responsiveness. Biophys J. 1989;55:125–145. doi: 10.1016/S0006-3495(89)82785-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52 **.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. [Experimental demonstration of calcium oscillations affecting gene expression in a frequency-dependent manner.] [DOI] [PubMed] [Google Scholar]
- 53.Tomida T, Hirose K, Takizawa A, Shibasaki F, Iino M. NFAT functions as a working memory of Ca2+ signals in decoding Ca2+ oscillation. Embo J. 2003;22:3825–3832. doi: 10.1093/emboj/cdg381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bedecarrats GY, Kaiser UB. Differential regulation of gonadotropin subunit gene promoter activity by pulsatile gonadotropin-releasing hormone (GnRH) in perifused L beta T2 cells: role of GnRH receptor concentration. Endocrinology. 2003;144:1802–1811. doi: 10.1210/en.2002-221140. [DOI] [PubMed] [Google Scholar]
- 55 **.Armstrong SP, Caunt CJ, Fowkes RC, Tsaneva-Atanasova K, McArdle CA. Pulsatile and sustained gonadotropin-releasing hormone (GnRH) receptor signaling: does the Ca2+/NFAT signaling pathway decode GnRH pulse frequency? J Biol Chem. 2009;284:35746–35757. doi: 10.1074/jbc.M109.063917. [Using live cell imaging this work demostrates that NFAT translocation acts as an integrator of oscillatory signals in the GnRH pathway rather than a true frequency decoder, suggesting that additional regulatory mechanisms may be at play] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56 **.Krakauer DC, Page KM, Sealfon S. Module dynamics of the GnRH signal transduction network. J Theor Biol. 2002;218:457–470. [Analysis of mechanisms capable of true frequency decoding in the context of gonadotrope signaling] [PubMed] [Google Scholar]
- 57 *.Hasty J, Dolnik M, Rottschafer V, Collins JJ. Synthetic gene network for entraining and amplifying cellular oscillations. Phys Rev Lett. 2002;88:148101. doi: 10.1103/PhysRevLett.88.148101. [A genetic network capable of producing oscillations with stimulus-dependent periodicity. The same architectural principles could potentially apply at the signaling level] [DOI] [PubMed] [Google Scholar]
- 58.Utzny C, Faroudi M, Valitutti S. Frequency encoding of T-cell receptor engagement dynamics in calcium time series. Biophys J. 2005;88:1–14. doi: 10.1529/biophysj.103.038216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larsen AZ, Olsen LF, Kummer U. On the encoding and decoding of calcium signals in hepatocytes. Biophys Chem. 2004;107:83–99. doi: 10.1016/j.bpc.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 60 *.Lewis RS. Calcium oscillations in T-cells: mechanisms and consequences for gene expression. Biochem Soc Trans. 2003;31:925–929. doi: 10.1042/bst0310925. [Lucid review of the encoding and decoding mechanisms though to underlie oscillatory calcium signals] [DOI] [PubMed] [Google Scholar]
- 61.Salazar C, Politi AZ, Hofer T. Decoding of calcium oscillations by phosphorylation cycles: analytic results. Biophys J. 2008;94:1203–1215. doi: 10.1529/biophysj.107.113084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quintana A, Griesemer D, Schwarz EC, Hoth M. Calcium-dependent activation of T-lymphocytes. Pflugers Arch. 2005;450:1–12. doi: 10.1007/s00424-004-1364-4. [DOI] [PubMed] [Google Scholar]
- 63.Tanimura A, Morita T, Nezu A, Shitara A, Hashimoto N, Tojyo Y. Use of Fluorescence Resonance Energy Transfer-based Biosensors for the Quantitative Analysis of Inositol 1,4,5-Trisphosphate Dynamics in Calcium Oscillations. J Biol Chem. 2009;284:8910–8917. doi: 10.1074/jbc.M805865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berridge MJ. Remodelling Ca2+ signalling systems and cardiac hypertrophy. Biochem Soc Trans. 2006;34:228–231. doi: 10.1042/BST20060228. [DOI] [PubMed] [Google Scholar]
- 65.Li W, Llopis J, Whitney M, Zlokarnik G, Tsien RY. Cell-permeant caged InsP3 ester shows that Ca2+ spike frequency can optimize gene expression. Nature. 1998;392:936–941. doi: 10.1038/31965. [DOI] [PubMed] [Google Scholar]
- 66.Tsutsumi R, Mistry D, Webster NJ. Signaling responses to pulsatile gonadotropin-releasing hormone in LbetaT2 gonadotrope cells. J Biol Chem. 2010;285:20262–20272. doi: 10.1074/jbc.M110.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lim S, Pnueli L, Tan JH, Naor Z, Rajagopal G, Melamed P. Negative feedback governs gonadotrope frequency-decoding of gonadotropin releasing hormone pulse-frequency. PLoS One. 2009;4:e7244. doi: 10.1371/journal.pone.0007244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crowley WF, Jr., Filicori M, Spratt DI, Santoro NF. The physiology of gonadotropin-releasing hormone (GnRH) secretion in men and women. Recent Prog Horm Res. 1985;41:473–531. doi: 10.1016/b978-0-12-571141-8.50015-9. [DOI] [PubMed] [Google Scholar]
- 69 *.Haisenleder DJ, Dalkin AC, Ortolano GA, Marshall JC, Shupnik MA. A pulsatile gonadotropin-releasing hormone stimulus is required to increase transcription of the gonadotropin subunit genes: evidence for differential regulation of transcription by pulse frequency in vivo. Endocrinology. 1991;128:509–517. doi: 10.1210/endo-128-1-509. [Early report of frequency encoding in gonadotrope signaling] [DOI] [PubMed] [Google Scholar]
- 70.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 72.Barken D, Wang CJ, Kearns J, Cheong R, Hoffmann A, Levchenko A. Comment on “Oscillations in NF-kappaB signaling control the dynamics of gene expression”. Science. 2005;308:52. doi: 10.1126/science.1107904. author reply 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lipniacki T, Paszek P, Brasier AR, Luxon BA, Kimmel M. Stochastic regulation in early immune response. Biophys J. 2006;90:725–742. doi: 10.1529/biophysj.104.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bratsun D, Volfson D, Tsimring LS, Hasty J. Delay-induced stochastic oscillations in gene regulation. Proc Natl Acad Sci U S A. 2005;102:14593–14598. doi: 10.1073/pnas.0503858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guido NJ, Lee P, Wang X, Elston TC, Collins JJ. A pathway and genetic factors contributing to elevated gene expression noise in stationary phase. Biophys J. 2007;93:L55–57. doi: 10.1529/biophysj.107.118687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elston TC. Probing pathways periodically. Sci Signal. 2008;1:pe47. doi: 10.1126/scisignal.142pe47. [DOI] [PubMed] [Google Scholar]
- 77.Lin YS, Carey M, Ptashne M, Green MR. How different eukaryotic transcriptional activators can cooperate promiscuously. Nature. 1990;345:359–361. doi: 10.1038/345359a0. [DOI] [PubMed] [Google Scholar]
- 78.Buchler NE, Gerland U, Hwa T. On schemes of combinatorial transcription logic. Proc Natl Acad Sci U S A. 2003;100:5136–5141. doi: 10.1073/pnas.0930314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 80.Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol. 2009;339:250–257. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81 *.White RJ, Sharrocks AD. Coordinated control of the gene expression machinery. Trends Genet. 26:214–220. doi: 10.1016/j.tig.2010.02.004. [Review surveying recent discoveries supporting the connection between signal dynamics and combinatorial control of gene expression] [DOI] [PubMed] [Google Scholar]
- 82.Murphy LO, Blenis J. MAPK signal specificity: the right place at the right time. Trends Biochem Sci. 2006;31:268–275. doi: 10.1016/j.tibs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 83.Natoli G. Tuning up inflammation: how DNA sequence and chromatin organization control the induction of inflammatory genes by NF-kappaB. FEBS Lett. 2006;580:2843–2849. doi: 10.1016/j.febslet.2006.02.072. [DOI] [PubMed] [Google Scholar]
- 84 *.Hao N, Zeng Y, Elston TC, Dohlman HG. Control of MAPK specificity by feedback phosphorylation of shared adaptor protein Ste50. J Biol Chem. 2008;283:33798–33802. doi: 10.1074/jbc.C800179200. [A potential example of dynamically encoded specificity in yeast] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85 *.Buckingham SD, Jones AK, Brown LA, Sattelle DB. Nicotinic acetylcholine receptor signalling: roles in Alzheimer's disease and amyloid neuroprotection. Pharmacol Rev. 2009;61:39–61. doi: 10.1124/pr.108.000562. [In this review the authors propose that differentiated dynamics may be behind the opposite effects resulting from different ligands of the nicotinic acteylcholine receptor in the context of Alzheimer's disease] [DOI] [PMC free article] [PubMed] [Google Scholar]