Abstract
Granulin (GRN) is a potent mitogen and growth factor implicated in many human cancers, but its regulation is poorly understood. Recent findings indicate that GRN is regulated strongly by the microRNA miR-107, which functionally overlap with miR-15, miR-16, and miR-195 due to a common 5' sequence critical for target specificity. In this study, we queried whether miR-107 and paralogs regulated GRN in human cancers. In cultured cells, anti-Argonaute RIP-ChIP experiments indicate that GRN mRNA is directly targeted by numerous miR-15/107 miRNAs. Further tests of this association in human tumors. MiR-15 and miR-16 are known to be downregulated in chronic lymphocytic leukemia (CLL). Using pre-existing microarray datasets, we found that GRN expression is higher in CLL relative to non-neoplastic lymphocytes (P>0.00001). By contrast, other prospective miR-15/miR-16 targets in the dataset (BCL-2 and cyclin D1) were not up-regulated in CLL. Unlike in CLL, GRN was not up-regulated in chronic myelogenous leukemia (CML) where miR-107 paralogs are not known to be dysregulated. Prior studies have shown that GRN is also up-regulated, and miR-107 down-regulated, in prostate carcinoma. Our results indicate that multiple members of the miR-107 gene group indeed repress GRN protein levels when transfected into prostate cancer cells. At least a dozen distinct types of cancer have the pattern of increased GRN and decreased miR-107 expression. These findings indicate for the first time that the mitogen and growth factor GRN is dysregulated via the miR-15/107 gene group in multiple human cancers, which may provide a potential common therapeutic target.
Keywords: Blood, miRNAs, miR-16, miR-103, FTD, PGRN
Introduction
Granulin (GRN) contributes to multiple human cancers. This gene product potentiates neoplastic transformation, stimulates tumor growth, metastases, and tissue invasion, inhibits anti-apoptotic mechanisms, and adversely impacts therapeutic responses (1, 2). GRN is a pleiotropic but evolutionarily-conserved protein that has been given multiple names, including progranulin (PGRN), acrogranin, gp88, proepithelin, PC cell-derived growth factor (PCDGF), and granulin-epithelin precursor (1). To avoid confusion, we refer here to “GRN” for the protein and “GRN” for the gene or mRNA.
Recent work demonstrated that GRN expression is suppressed post-transcriptionally by miR-107 (3), a member of a microRNA (miRNA) gene group that also includes miR-15, miR-16, miR-103, miR-195, miR-424, miR-497, miR-503, and miR-646 (4). All of these miRNAs are moderate-to-highly expressed in many human tissues and share a common 5' seed sequence, AGCAGC. The 5' end sequence homology confers similar specificity in terms of targeting mRNAs for post-transcriptional decay and/or translational inhibition (4). Paralogous miRNAs, miR-15 and miR-16 genes, reside within the human chromosome 13q minimal deletion region that confers chronic lymphocytic leukemia (CLL) susceptibility (5). We hypothesized that since miR-15 and miR-16 are capable of strongly suppressing GRN, that CLL lymphocytes may have increased expression of oncogenic GRN. We also note that other human cancers including carcinoma of the prostate show increased GRN expression with decreased miR-107 expression (6, 7). We sought to evaluate whether prior studies using gene expression methods, and our own experiments with cultured cancer cell lines, would substantiate the hypothesis that miR-107 dysregulation may correlate with increased GRN expression in human cancers.
Materials and Methods
RIP-ChiP studies
RIP-ChIP methods have been described in detail previously (8). Briefly, H4 cells (American Type Culture Collection, Manassas, VA), cultured under the vendor's recommended conditions, were plated in 10-cm culture plates at a density of 2.5×106/plate day before transfections. Cells were transfected with 25 nM of “Pre-miRNA” (siRNA-like reagents from Ambion, Austin, TX) referent to hsa-miR-103, hsa-miR-107, hsa-miR-15b* (antisense strand), hsa-miR-16, hsa-miR-195, hsa-miR-320, Negative Control miRNA #1 using siRMAX (Invitrogen, Carlsbad, CA), and mutated miRNAs as shown below, according to manufacturer's instructions. Statistical tests were performed using the Student's t-test with a P<0.01 cutoff.
Analyses of National Institute of Health Gene Expression Omnibus (GEO) data
Data from the GEO database was assessed to allow us to infer whether GRN mRNA is decreased in cancers where miR-15/107 gene group members show increased expression. Information about the individual datasets used is provided below.
Tissue culture cell transfections with miR-15/107 gene group members and controls
Transfections were performed as noted above. Western blots were performed as previously described (8).
Results and Discussion
We previously performed the requisite experiments including reporter assays with recognition site mutation and miRNA inhibitor studies to show that miR-107 can target specifically GRN mRNA with resulting decreased GRN protein (3). Here we tested the hypothesis that this mechanism is relevant to human cancers. Specifically, we investigated whether down-regulation of miR-15/107 gene group miRNAs (4) contribute to human cancers through increasing expression of GRN.
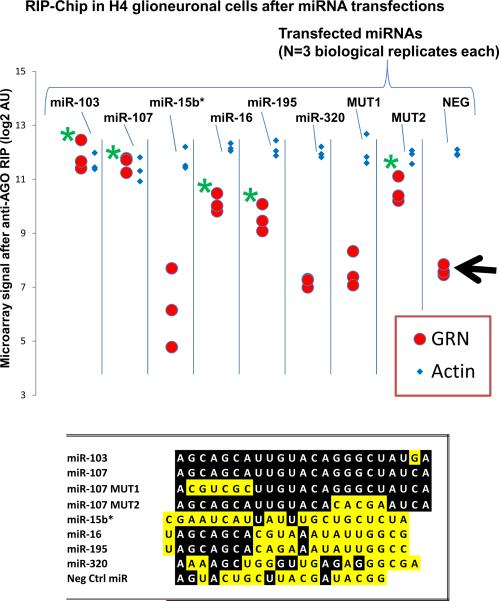
We first used a cell culture system to test multiple members of the miR-15/107 gene group directly, along with controls miRNAs, to ascertain whether the miR-15/107 group miRNAs cause specific incorporation of GRN mRNA into the microribonucleoparticle (miRNP) that contains the Argonaute (AGO) protein for mRNA targeting. We used our Anti-AGO antibody and the RNA co-immunoprecipitation with downstream microarray analyses (RIP-Chip) experimental design as described previously (8). In accordance with our previous findings for miR-107, we observed incorporation of GRN mRNA into the AGO-miRNP following transfections with miR-103, miR-16, and miR-195, but not miR-15b* (an anti-sense oriented control), miR-320, or a negative control miRNA. We also transfected the cells with non-physiological “mutant” miRNAs that are related to but distinct from miR-107, with changes in the 5' portion (MUT1) or 3' portion (MUT2) as shown in Figure 2. Note that incorporation of GRN into the miRNP is abolished by MUT1 but not MUT2, which underscores the importance of the 5' seed portion of the miRNA for target specificity. Because BCL2 and CCND1 have been shown to be targets of miR-16, we evaluated them in the RIP-ChiP assay and they showed a far lesser degree of enrichment in the miRNP following miR-16 transfection relative to GRN (data not shown).
Figure 2.
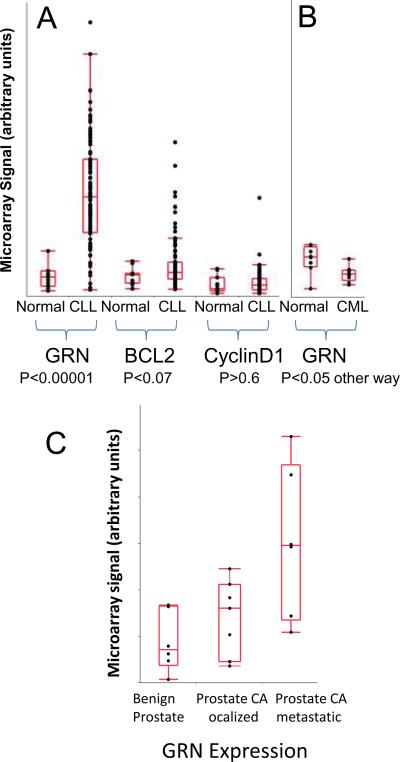
Data from multiple laboratories, accessed via the National Institute of Health Gene Expression Omnibus (GEO) database, show that GRN mRNA levels are elevated in cancers (chronic lymphocytic leukemia/CLL, and prostate carcinoma) where miR-15/107 gene expression also is decreased. A. Data from the Christian Stratowa laboratory used mRNA from mononuclear cells in blood samples followed by downstream Affymetrix human genome U95A or U95Av2 microarrays to test the differences in expression between normal (N=11) and CLL (N=100) cells (13). Note that GRN levels are dramatically increased in CLL cells (P<0.00001 using a Student's t-test) whereas other putative miR-15/16 targets are not significantly increased in this study. B. The findings in CLL appear to be specific. Data assessing myelogenous cells from bone marrow or peripheral blood, from Lucy Crossman and colleagues (14) assessed bone marrow and peripheral blood samples from normal (N=8) and chronic myelogenous leukemia (CML; N=9) patients using Affymetrix human genome U95Av2 microarrays. MiR-15/107 gene group member expressions are not decreased in CML (19, 20), while GRN levels are actually downregulated in CML cancer cells relative to controls. Results with peripheral blood and bone marrow are pooled. C. The increased GRN expression in prostate cancer result is in accordance with data from the Arul Chinnaiyan lab (16) using Affymetrix HGU133 plus 2 microarrays. Here, it is demonstrated that metastatic prostate cancer cells express increased GRN. These data helped motivate further work on prostate cancer cells to show directly that miR-15/107 gene group members downregulated GRN.
After showing that multiple miR-15/107 gene group paralogs can target GRN, we next analyzed publicly accessible data from the Gene Expression Omnibus (GEO) database (9) to see if potentially oncogenic target mRNAs are up-regulated in cancers with known miRNA down-regulation. CLL is the best established example of a tumor with decreased miR-15a and miR-16 expression since these tandem genes reside in the CLL minimal deletion region (10). There have been putative target genes described as oncogenic through the miRNA pathways, i.e. miR-15 and miR-16 targets oncogenically derepressed after the miRNAs are deleted. These putative miR-15/16 targets are BCL-2 and Cyclin D1 (11, 12). We used previously developed high-quality datasets from the GEO database showing mRNAs in normal mononuclear blood cells and CLL cells assessed by Christian Stratowa and colleagues (13) at the Department of Lead Discovery, Boehringer Ingelheim Austria (Figure 1A). Based on these data we pursued the assessment of GRN and other putative miR-15/16 targets. Consistent to our hypothesis, levels of GRN mRNA in CLL cells are much higher than the normal lymphocytes (P<0.0001). In comparison, the differences in gene expression between normal lymphocytes and CLL cells for other putative miR-15/16 target mRNAs are much more marginal (BCL-2), or nonexistent (Cyclin-D1).
Figure 1.
RNA co-immunoprecipitation using anti-Argonaute (AGO) with downstream microarray analyses with Affymetrix Human Gene ST 1.0 (RIP-Chip) shows that multiple members of the miR-15/107 miRNA group recruit GRN mRNA to the AGO-microribonucleoparticle (miRNP). The vast majority of mRNAs (β-Actin is representative) were not enriched in the miRNP following miR-107 transfection. The RIP-Chip results from different transfections can be compared (each performed in triplicate; miRNA sequences shown at bottom). The negative control miRNA is indicated on the right side with the black arrow. Only after transfection with miR-103, miR-107, miR-16, miR-195, and MUT2, which all have common 5' sequence AGCAGCA motif, GRN mRNA is recruited to the AGO-miRNP. Statistical tests were performed using the Student's t-test with a P<0.01 cutoff (green asterisk indicates significant difference versus the control miRNA transfection).
To determine if this were a nonspecific effect in human leukemias, we subsequently evaluated data about chronic myelogenous leukemia (CML) from Dr. Lucy Crossman's group at the Oregon Health and Science University Cancer Institute (Portland, OR) (14), also available from the GEO database. MiR-15 and miR-16 have not been shown to be downregulated in CML. We found that indeed, the level of GRN mRNA is actually higher in normal leukocytes than CML cells. These data are compatible with the hypothesis that miR-15 and miR-16 down-regulation in CLL contributes specifically to increased expression of the oncogene GRN.
There have been other human cancers where miR-15/107 gene group members have been shown to be decreased, and independently, GRN levels are found to be up-regulated. These cancers are listed in Table 1. Note that these are only cancers where both of these findings have been described; in the vast majority of human cancers one or both of these (GRN and miRNAs) levels have not been queried reliably.
Table 1.
Human neoplasms where miR-15/107 gene members are down-regulated and granulin expression is up-regulated.
Studies in multiple subtypes of human neoplasms have observed miR-15/107 gene member down-regulation and granulin up-regulation. Both miR-15/107 genes and GRN have been found to be dysregulated in other tumor types, but here are shown the tumor types where both were independently verified. The miR-15/107 gene group comprises miR-15a, miR-15b, miR-16, miR-103, miR-107, miR-195, miR-424, miR-497, miR-503, and miR-646. The methods used for the various studies are indicated in parentheses. ″Other″ methods to detect downregulated miRNAs include Northern blots, specialized high-throughput methods, and in situ hybridization.
| Carcinomas | miR-15/107 group member downregulated ? (METHODS) | miRNA downregulated in this cancer: | Granulin upregulated ? (METHODS) | REFS |
|---|---|---|---|---|
| Bladder carcinoma | YES (A) | miR-195 | YES (G) | 1 |
| Endometrial carcinoma | YES (C) | miR-424 | YES (G) | 2 |
| Gastric carcinoma | YES (A,D) | miR-195 | YES (F) | 3 |
| Head and neck carcinoma | YES (A,B) | miR-16, -195 | YES (G) | 4 |
| Hepatocellular carcinoma | YES (B, D) | miR-15b, -16 | YES (G) | 5 |
| Breast carcinoma | YES (B) | miR-16, -497 | YES (G) | 6 |
| Non-small cell lung carcinoma | YES (B) | Multiple | YES (F) | 7 |
| Ovarian carcinoma | YES (B) | miR-195 | YES (G) | 8 |
| Prostatic carcinoma | YES (A) | miR-15,-16,-107 | YES (G) | 9 |
| Hematological malignancies | ||||
| Chronic lymphocytic leukemia | YES (C,D) | miR-15a, 16 | YES (E) | 10 |
| Multiple myeloma | YES (A) | miR-15a | YES (G) | 11 |
To directly test whether multiple members of the miR-15/107 gene group can regulate GRN in cancer, we used human prostate cancer cells. We chose prostate cancer because independent laboratories have found that miR-15/107 gene group member expression is decreased in prostate cancer, and GRN plays a potent role in prostate cancer tumorogenesis and malignancy (6, 7, 15). To confirm the up-regulation of GRN in prostate cancer we used more data from the GEO database (Figure 2C), testing mRNA from different prostate samples based on the data reported by Dr. Arul Chinnaiyan's laboratory (16) at the University of Michigan. These data indeed show that GRN levels are increased in metastatic prostate cancer.
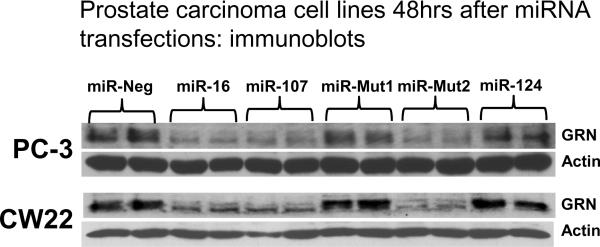
Finally, to directly test the hypothesis that GRN protein levels are regulated through members of the miR-15/107 gene group, we transfected prostate cancer cell lines PC-3 and CW22 (17) with specific miRNAs. These data are shown in Figure 3. Note that miR-107MUT2 (with the 5' seed portion intact) inhibits GRN expression whereas the miR-107MUT2 miRNA (with the 3' part of the miRNA intact) does not.
Figure 3.
In cell culture transfection experiments, miR-15/107 gene group members specifically knock down endogenous GRN in prostate cancer cell lines. Note that all the miRNAs with the miR-107 gene group seed, including the artificial MUT2 miRNA (see Figure 2), knock down GRN protein levels but not the control protein β-Actin. By contrast, miRNAs without the seed sequence do not knock down GRN.
These data show collectively that multiple members of the miR-15/107 gene group can regulate GRN expression and that the GRN expression is exerted through the 5' seed sequence of the miRNAs. However, this does not exclude the possibility that in other systems other portions of miRNAs can regulate GRN (or other mRNA targets) differentially. It is also unknown how the different miR-15/107 group members interact in vivo in terms of combinatorial effects. There is a fast-expanding research that has focused on the miR-15/107 group of genes and their effects on metabolism, cell cycle functions, and cell stress (4).
In conclusion, the present study support the hypothesis that regulation of GRN through members of the miR-15/107 gene group may have an important oncogenic impact on multiple human cancers. This is a biological phenomenon with potential therapeutic implications. Theoretically, a therapeutic strategy that “replaces” miR-15/107 gene group expression would attenuate GRN expression and possible decrease malignant potential of tumors. We note that methods have been developed for delivering miR-16 systemically, which was helpful in reducing prostate cancer burden in a mouse model (18), and we hypothesize that this could have been accomplished through GRN. We hope that in the future this and other methods will be tried for human therapies that work through this novel mechanism.
Acknowledgement
We thank Ms Willa Huang for technical and collegial assistance in the project. This research was supported by grants R01 NS061933, K08 NS050110, and P30-AG028383, from the NIH, Bethesda, MD.
References
- 1.Bateman A, Bennett HP. The granulin gene family: from cancer to dementia. Bioessays. 2009;31:1245–54. doi: 10.1002/bies.200900086. [DOI] [PubMed] [Google Scholar]
- 2.Ong CH, Bateman A. Progranulin (granulin-epithelin precursor, PC-cell derived growth factor, acrogranin) in proliferation and tumorigenesis. Histol Histopathol. 2003;18:1275–88. doi: 10.14670/HH-18.1275. [DOI] [PubMed] [Google Scholar]
- 3.Wang W-X, Wilfred BR, Madathil SK, et al. MiR-107 regulates Granulin/Progranulin with implicatins for traumatic brain injury and neurodegenerative disease. Am J Pathol. 2010 doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 Group of MicroRNA Genes: Evolutionary Biology, Cellular Functions, and Roles in Human Diseases. J Mol Biol. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonci D, Coppola V, Musumeci M, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 7.Pan CX, Kinch MS, Kiener PA, et al. PC cell-derived growth factor expression in prostatic intraepithelial neoplasia and prostatic adenocarcinoma. Clin Cancer Res. 2004;10:1333–7. doi: 10.1158/1078-0432.ccr-1123-03. [DOI] [PubMed] [Google Scholar]
- 8.Wang WX, Wilfred BR, Hu Y, Stromberg AJ, Nelson PT. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA. 2010;16:394–404. doi: 10.1261/rna.1905910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:11755–60. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Q, Fu H, Sun F, et al. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia L, Zhang D, Du R, et al. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–9. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 13.Haslinger C, Schweifer N, Stilgenbauer S, et al. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22:3937–49. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 14.Crossman LC, Mori M, Hsieh YC, et al. In chronic myeloid leukemia white cells from cytogenetic responders and non-responders to imatinib have very similar gene expression signatures. Haematologica. 2005;90:459–64. [PubMed] [Google Scholar]
- 15.Monami G, Emiliozzi V, Bitto A, et al. Proepithelin regulates prostate cancer cell biology by promoting cell growth, migration, and anchorage-independent growth. Am J Pathol. 2009;174:1037–47. doi: 10.2353/ajpath.2009.080735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varambally S, Yu J, Laxman B, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Zhu ML, Kyprianou N. Role of androgens and the androgen receptor in epithelial-mesenchymal transition and invasion of prostate cancer cells. FASEB J. 24:769–77. doi: 10.1096/fj.09-136994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeshita F, Patrawala L, Osaki M, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 18:181–7. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agirre X, Jimenez-Velasco A, San Jose-Eneriz E, et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res. 2008;6:1830–40. doi: 10.1158/1541-7786.MCR-08-0167. [DOI] [PubMed] [Google Scholar]
- 20.San Jose-Eneriz E, Roman-Gomez J, Jimenez-Velasco A, et al. MicroRNA expression profiling in Imatinib-resistant Chronic Myeloid Leukemia patients without clinically significant ABL1-mutations. Mol Cancer. 2009;8:69. doi: 10.1186/1476-4598-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichimi T, Enokida H, Okuno Y, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–52. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 22.Lovat F, Bitto A, Xu SQ, et al. Proepithelin is an autocrine growth factor for bladder cancer. Carcinogenesis. 2009;30:861–8. doi: 10.1093/carcin/bgp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones MB, Houwink AP, Freeman BK, et al. The granulin-epithelin precursor is a steroid-regulated growth factor in endometrial cancer. J Soc Gynecol Investig. 2006;13:304–11. doi: 10.1016/j.jsgi.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Cohn DE, Fabbri M, Valeri N, et al. Comprehensive miRNA profiling of surgically staged endometrial cancer. Am J Obstet Gynecol. doi: 10.1016/j.ajog.2010.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Miao Y, Xiao B, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–7. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 26.Line A, Stengrevics A, Slucka Z, Li G, Jankevics E, Rees RC. Serological identification and expression analysis of gastric cancer-associated genes. Br J Cancer. 2002;86:1824–30. doi: 10.1038/sj.bjc.6600321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hui AB, Lenarduzzi M, Krushel T, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 16:1129–39. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 28.Kong WJ, Zhang SL, Chen X, et al. PC cell-derived growth factor overexpression promotes proliferation and survival of laryngeal carcinoma. Anticancer Drugs. 2007;18:29–40. doi: 10.1097/01.cad.0000236315.96574.58. [DOI] [PubMed] [Google Scholar]
- 29.Tran N, O'Brien CJ, Clark J, Rose B. Potential role of micro-RNAs in head and neck tumorigenesis. Head Neck. 32:1099–111. doi: 10.1002/hed.21356. [DOI] [PubMed] [Google Scholar]
- 30.Cheung ST, Wong SY, Lee YT, Fan ST. GEP associates with wild-type p53 in hepatocellular carcinoma. Oncol Rep. 2006;15:1507–11. [PubMed] [Google Scholar]
- 31.Chung GE, Yoon JH, Myung SJ, et al. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 23:113–9. [PubMed] [Google Scholar]
- 32.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–21. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 33.Fassan M, Baffa R, Palazzo JP, et al. MicroRNA expression profiling of male breast cancer. Breast Cancer Res. 2009;11:R58. doi: 10.1186/bcr2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monami G, Gonzalez EM, Hellman M, et al. Proepithelin promotes migration and invasion of 5637 bladder cancer cells through the activation of ERK1/2 and the formation of a paxillin/FAK/ERK complex. Cancer Res. 2006;66:7103–10. doi: 10.1158/0008-5472.CAN-06-0633. [DOI] [PubMed] [Google Scholar]
- 35.Tangkeangsirisin W, Serrero G. PC cell-derived growth factor (PCDGF/GP88, progranulin) stimulates migration, invasiveness and VEGF expression in breast cancer cells. Carcinogenesis. 2004;25:1587–92. doi: 10.1093/carcin/bgh171. [DOI] [PubMed] [Google Scholar]
- 36.Xu F, Zhang X, Lei Y, et al. Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma. J Cell Biochem. doi: 10.1002/jcb.22762. [DOI] [PubMed] [Google Scholar]
- 37.Stewart DJ. Tumor and host factors that may limit efficacy of chemotherapy in non-small cell and small cell lung cancer. Crit Rev Oncol Hematol. doi: 10.1016/j.critrevonc.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Feng FY, Chen SJ, Gao YN, Xiao T, Liu YN. Correlation between the expression of PCDGF in serum and the chemotherapeutic sensitivity in NSCLC. Zhonghua Zhong Liu Za Zhi. 2006;28:603–5. [PubMed] [Google Scholar]
- 39.Du L, Schageman JJ, Irnov, et al. MicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCs. J Exp Clin Cancer Res. 29:75. doi: 10.1186/1756-9966-29-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones MB, Spooner M, Kohn EC. The granulin-epithelin precursor: a putative new growth factor for ovarian cancer. Gynecol Oncol. 2003;88:S136–9. doi: 10.1006/gyno.2002.6704. [DOI] [PubMed] [Google Scholar]
- 41.Dahiya N, Sherman-Baust CA, Wang TL, et al. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cuevas-Antonio R, Cancino C, Arechavaleta-Velasco F, et al. Expression of Progranulin (Acrogranin/PCDGF/Granulin-Epithelin Precursor) in Benign and Malignant Ovarian Tumors and Activation of MAPK Signaling in Ovarian Cancer Cell Line. Cancer Invest. 2009 doi: 10.3109/07357900903346455. [DOI] [PubMed] [Google Scholar]
- 43.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Hayashi J, Kim WE, Serrero G. PC cell-derived growth factor (granulin precursor) expression and action in human multiple myeloma. Clin Cancer Res. 2003;9:2221–8. [PubMed] [Google Scholar]
- 45.Gutierrez NC, Sarasquete ME, Misiewicz-Krzeminska I, et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia. 24:629–37. doi: 10.1038/leu.2009.274. [DOI] [PubMed] [Google Scholar]