Abstract
Unlike normal cells, which metabolize glucose by oxidative phosphorylation for efficient energy production, tumor cells preferentially metabolize glucose by aerobic glycolysis, which produces less energy but facilitates the incorporation of more glycolytic metabolites into the biomass needed for rapid proliferation. The metabolic shift from oxidative phosphorylation to aerobic glycolysis is partly achieved by a switch in the splice isoforms of the glycolytic enzyme pyruvate kinase. While normal cells express the pyruvate kinase M1 isoform (PKM1), tumor cells predominantly express the M2 isoform (PKM2). Switching from PKM1 to PKM2 promotes aerobic glycolysis and provides a selective advantage for tumor formation. The PKM1/M2 isoforms are generated through alternative splicing of two mutually exclusive exons. A recent study demonstrates that the alternative splicing event is controlled by heterogeneous nuclear ribonucleoprotein (hnRNP) family members hnRNPA1, hnRNPA2, and polypyrimidine tract binding protein (PTB; also known as hnRNPI). These findings not only provide additional evidence that alternative splicing plays an important role in tumorigenesis, but also shed light on the molecular mechanism by which hnRNP proteins regulate cell proliferation in cancer.
One characteristic that distinguishes cancer cells from normal cells is their metabolic regulation. Most adult tissues, in the presence of oxygen, use a large fraction of nutrients for maximal energy production, through the citric acid cycle and oxidative phosphorylation. Fast growing cells, such as embryonic cells and cancer cells, utilize a different metabolic regulation from most adult tissues in that they convert a large amount of glucose into lactate even when oxygen is abundant. This phenomenon is termed as the Warburg effect (1), or aerobic glycolysis, which is an inefficient means of producing energy but is thought to enable growing cells to incorporate metabolites from glycolysis into synthesis of macromolecules for cell growth.
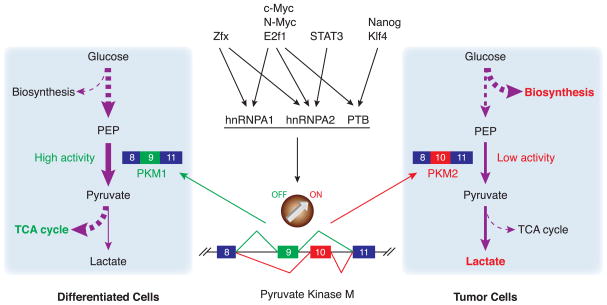
One of the mechanisms that controls the glycolytic phenotype is the tight regulation of the enzyme pyruvate kinase (PK). PK catalyzes the dephosphorylation of phosphoenolpyruvate (PEP) to convert it into pyruvate, and has been implicated as a critical determinant of metabolic phenotype (2). Pyruvate kinase has four isoforms, produced from two distinct genes, which are specifically expressed in tissues with different metabolic functions. Pyruvate kinase L is expressed in tissues with gluconeogenesis, such as liver, and pyruvate kinase R is found in erythrocytes (3). These two isoforms are expressed from the same gene under the control of two different promoters. The other two isoforms are pyruvate kinase M1 (PKM1) and pyruvate kinase M2 (PKM2), which are produced by alternative splicing of transcripts of the PKM gene. PKM1 is expressed in adult tissues in which a large amount of energy is produced, such as muscle and brain, whereas PKM2 is expressed in some differentiated tissues, such as fat tissues and lung, and tissues or cells with a high rate of nucleic acid synthesis, such as embryonic cells, stem cells and tumor cells (3). During tissue differentiation in development, embryonic PKM2 is replaced by tissue-specific isoforms. However, PKM1 and other isozymes disappear during tumorigenesis and PKM2 reappears, a reversion that is nearly universal (4). Recently, Cantley and colleagues showed that replacing PKM2 with PKM1 greatly reduced both lactate production in tumor cells and tumor size, suggesting that the choice of PKM1 or PKM2 is directly connected to tumor metabolic phenotype (2). PKM1 and PKM2 mRNAs differ only by inclusion of one or another of two mutually exclusive exons (see Figure 1), so the regulation of PKM alternative splicing is of great importance for understanding tumor metabolic regulation.
Figure 1. hnRNP proteins control the metabolic switch between oxidative phosphorylation and aerobic glycolysis by regulating the PKM alternative splicing.
In cancer cells, transcription of hnRNPA1, A2 and PTB genes is upregulated, by c-Myc and likely one or more of the other factors indicated. Binding of the hnRNPs to the splice sites flanking exon 9 in PKM transcripts results in exon 9 exclusion and exon 10 inclusion, generating PKM2. PKM2 converts PEP to pyruvate less efficiently than PKM1, leading to the accumulation of glycolytic metabolites for anabolic metabolism (Efficiencies indicated by thickness of arrows).
The molecular and kinetic characteristics of PKM1 and PKM2 determine their specific functions in differentiated or growing cells (3). PKM1 forms a tetramer that has high affinity for PEP and converts PEP efficiently into pyruvate, and it is not allosterically regulated. In addition, the pyruvate produced by PKM1 is preferentially used in oxidative phosphorylation. On the other hand, PKM2 can function as both a tetramer with high affinity for PEP and also as a dimer with low affinity to PEP (3), and the tetramer/dimer ratio is regulated by metabolic intermediates, such as fructose 1,6-biphosphate (5). In tumor cells, PKM2 is primarily found as the dimeric form and this has the advantage that the glycolic intermediates above pyruvate accumulate for synthetic processes. Therefore, a high level of PKM2 dimer increases the levels of glycolytic intermediates, such as fructose 1,6-biphosphate. When PKM2 dimers are bound by fructose 1,6-biphosphate, an allosterical regulator, the tetramer forms and converts PEP into pyruvate. Interestingly, the pyruvate produced by PKM2 is directly converted into lactate instead of going into the citric acid cycle, possibly because PKM2 tetrameric form may be associated with other glycolytic enzymes (3). In addition, the activity of PKM2 is also regulated by tyrosine-phosphorylated peptides, the binding of which leads to dissociation of fructose 1,6-biphosphate and therefore dissociation of the tetramer (5). Indeed, it was shown recently that tyrosine 105 of PKM2 can be phosphorylated by fibroblast growth factor receptor type 1, and this leads to inactivation of PKM2 (6).
In light of the above, understanding the regulation of switching between PKM1 and PKM2 pre-mRNA splicing is of great importance. Whether an alternative exon is included in an mRNA is often determined by cis-regulatory elements, which are in turn recognized by transacting regulatory proteins (7). Therefore, David et al. investigated the mechanism of PKM alternative splicing, first by searching for proteins that bind to the two alternative exons and/or their flanking regions (8). Using UV crosslinking and RNA affinity purification with HeLa cell extracts, heterogeneous nuclear ribonucleoprotein (hnRNP) protein family members hnRNP A1, hnRNP A2 and polypyrimidine tract binding protein (PTB, also known as hnRNP I) were found to bind to intronic sequences flanking exon 9 (contained in PKM1) but not exon 10 (contained in PKM2) (See Figure 1). HnRNP proteins often bind to RNA elements known as exonic or intronic splicing silencers (ESS or ISSs) to function as splicing repressors (7), and the sequences identified thus function exon 9-specific ISSs.
To determine if the identified hnRNPs indeed function in PKM splicing control, David et al. next showed that hnRNP A1, hnRNP A2 and PTB indeed repress inclusion of PKM exon 9 in a variety of cancer cell lines by depleting these proteins using siRNAs (8). In siRNA treated cells, exon 9 exclusion was significantly relieved and PKM1 mRNA levels were increased, indicating that the expression levels of hnRNP A1, hnRNP A2 and PTB are critical for PKM2 expression in cancer cells.
David et al. next examined the expression of these hnRNP proteins and PKM1/2 in different cell types. Changes in concentration of splicing factors in different tissues can be one means of regulating tissue-specific alternative splicing (7). The mouse myoblast cell line C2C12 serves as an excellent model for studying PKM1/2 conversion in growing cells versus differentiated cells, because when they differentiate into mature myotubes, a switch from PKM2 to PKM1 occurs (9). David et al. showed that hnRNP A1 and PTB levels decrease in differentiated C2C12 cells, consistent with a model that higher expression levels of these proteins are critical for exon 9 exclusion and PKM2 expression in growing cells. To examine further the correlation between PKM2 expression and hnRNP A1, hnRNP A2 and PTB levels, different classes of human glioma tumor samples were examined. Consistently, low expression of hnRNP A1/A2 and PTB was observed in normal brains, higher expression of these proteins was detected in pilocytic astrocytoma samples, and the highest levels of hnRNP A1/A2 and PTB, which correlated with the highest level of PKM2, was found in aggressive glioblastoma multiforme samples. In addition, the fact that no correlation between the levels of other splicing factors, such as the SR protein SRSF1 (10), with PKM2 expression was observed indicates that the correlation between PKM2 and hnRNP A1/A2/PTB expression is specific (8, 11). A previous study suggested that SRSF1 has the potential to function as an oncogene (12); however, it does not seem to play a role in regulation of PKM alternative splicing.
A subsequent study by Clower et al. reached similar conclusions regarding the role of hnRNP A1/A2 and PTB in PKM splicing (11). These authors also observed a correlation between the levels of the three proteins in C2C12 and several other tissues and cell lines and PKM splicing, and that depleting them with shRNAs, in a glioblastoma cell line, resulted in a switch in PKM splicing to favor PKM1 production. They also showed that depletion led to a decrease in lactate production, as would be expected from a switch to PKM1.
An important question then became the mechanism responsible for overexpression of the three hnRNP proteins in cancer cells. One possibility was that transcription of the genes expressing these splicing regulatory proteins is under the control of a proliferation-associated transcription factor, due to the tight coupling of PKM2 expression and cell growth. c-Myc is a potent regulator of multiple metabolic pathways essential for cancer growth (13). Furthermore, it has been shown by genome wide-chromatin ChIP experiments to bind to the PTB, hnRNP A1 and hnRNP A2 promoter regions and, by siRNA experiments, to upregulate their expression levels (14–16). David et al. examined the expression of c-Myc in C2C12 cells and the glioma samples and showed that the expression of c-Myc correlates with hnRNP A1/A2 and PTB overexpression almost perfectly. However, N-Myc, which also binds to the three promoters, was only highly overexpressed in pilocytic astrocytomas (8). More importantly, c-Myc siRNA-depleted NIH3T3 cells showed decreases in hnRNP A1/A2 and PTB levels and a sharp increase in the PKM1/PKM2 mRNA ratio. However, no decrease of hnRNP A1/A2 and PTB RNA levels was observed by depleting other proliferation associated transcription factors, such as E2F1 in HeLa cells and Rb in MCF-7 cells. Taken together, these experiments indicate that c-Myc regulates PKM alternative splicing by directly controlling expression of hnRNP A1, A2 and PTB (8).
The tight link between hnRNP expression, PKM alternative splicing and cell proliferation implies a critical role of hnRNP proteins in tumorigenesis. Because of their capability of binding a wide range of RNA, and DNA, sequences, hnRNPA1 and hnRNPA2 have been suggested to be involved in multiple cellular processes, including DNA replication and repair, telomere biogenesis, transcription and mRNA export, as well as pre-mRNA splicing regulation (17). hnRNPA1 and hnRNPA2 have been found to be overexpressed consistently in a wide variety of cancers. Evidence of their direct involvement in cancer cell proliferation was first suggested by an RNAi experiment in which the simultaneous knockdown of hnRNPA1 and hnRNPA2 lowered the growth rate of Colo16 skin cancer cells (18). Possible molecular mechanisms underlying their stimulatory effect on cancer cell proliferation were unknown until the demonstration that hnRNPA1 and hnRNPA2 (together with PTB) regulate PKM alternative splicing (8). PTB is also involved in multiple processes, including polyadenylation, mRNA stability and translation initiation in addition to splicing (19). Knockdown of PTB alone suppresses ovarian tumor cell growth (20), which may be due in part to switching PKM2 to M1.
Regulation of splicing of a single target mRNA by multiple hnRNP proteins is uncommon. For example, Venables et al. showed by down-regulating the expression of each of 14 individual hnRNP proteins that there is little overlap in their targets (21). The fact that PTB, hnRNPA1 and hnRNPA2 are all required to regulate PKM exon 9 splicing reinforces the importance of producing PKM2 in proliferating cells, both during embryogenesis and in tumors. Given their role in tumorigenesis, hnRNPA1, hnRNPA2 and PTB have the potential to be therapeutic targets. Reducing the expression of these proteins in cancer cells is promising, because RNAi-mediated knockdown of both hnRNPA1 and A2 induced apoptosis specifically in cancer cells but not in normal mortal cell lines (22). This may be an intriguing approach to regulating PKM2 expression, given that high-specificity isoenzyme-selective inhibitors of PKM2 are lacking.
It is possible, perhaps likely, that the roles of hnRNPA1, A2, and PTB in cancer extend beyond the switch of the PKM1/M2 isoforms. For example, He et al. showed recently that knockdown of hnRNPA2 affects transcript abundance of 123 genes (out of 22,283 human genes examined in microarray analyses), many of which are cell proliferation associated (23). A genome-wide analysis of PTB-RNA interactions revealed that PTB binds to 10,372 out of the 24,378 annotated human genes, with 58% of the putative PTB-binding sites localized in introns (24). Although the functional significance of all these interactions is largely unknown, as expected, the intron preceding exon 9 of PKM was identified as one of these targets. The global range of hnRNPA1 and A2 regulation on alternative splicing is less studied. Venables et al. showed that hnRNPA1 and A2 control 5% of the alternative splicing events specific for apoptotic genes in HeLa cells and PC-3 cells (21). Given the importance of alternative splicing in cancer and other human disorders, identification of all the targets of these hnRNP proteins is an important goal. By comparing splicing variants in normal and cancer cells it will be possible to identify additional cancer-associated splicing targets.
Alternative splicing is a major mechanism generating the proteome diversity. More than 90% of human genes are now believed to produce alternatively spliced transcripts (25, 26). Large scale effects of hnRNP proteins on alternative splicing could lead to significant changes in proteomic diversity in a cell population, perhaps increasing the chances for variant cells to overcome normal homeostatic controls, thereby promoting cell transformation. It is not known whether hnRNPA1, A2 or PTB has tumor-inducing potential. hnRNPA1 and hnRNPA2 are highly expressed in the basal layer of skin tissue and are moderately expressed in a few normal cell types (22), indicating that in these cellular environments their expression is not sufficient to induce tumor. However, this does not rule out the possibility that they participate in early transforming events of tumorigenesis. For example, Zerbe et al. observed aberrant expression of hnRNPA1 at early stages of mouse lung tumorigenesis, nine weeks before adenocarcinomas with increased disorganization were observed (27). This suggests that hnRNPA1 may help to regulate initial stages of neoplastic transformation. It will be important to determine whether overexpression of these proteins (alone or in combination) in normal differentiated cells can initiate transformation and, if so, under what growth conditions. However, overexpression of hnRNPA1, A2 and PTB in a variety of cancers must result from upstream transforming events, for example c-Myc overexpression. ChIP-seq analyses revealed that the promoter regions of hnRNPA1, A2 and PTB contain binding sites for oncogenic Myc genes and for several transcription factors that connect with external signaling pathways (14; see Figure 1). Just as PKM splicing is controlled by multiple hnRNP proteins, expression of the hnRNP proteins themselves is likely regulated by multiple transcription factors, thereby ensuring PKM2 expression in a variety of different cancers and cell types. Further analysis of the hnRNP protein gene promoters will provide insights into how production of these proteins responds to a variety of cell growth signals, and importantly how they are deregulated in cancer.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 3.Mazurek S. Pyruvate kinase type M2: A key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2010 doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–8. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–6. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 6.Hitosugi T, Kang S, Vander Heiden MG, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–54. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harada Y, Nakamura M, Asano A. Temporally distinctive changes of alternative splicing patterns during myogenic differentiation of C2C12 cells. J Biochem. 1995;118:780–90. doi: 10.1093/oxfordjournals.jbchem.a124980. [DOI] [PubMed] [Google Scholar]
- 10.Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–4. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–9. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer. 2008;8:51–6. doi: 10.1038/nrc2274. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 15.Shiio Y, Donohoe S, Yi EC, Goodlett DR, Aebersold R, Eisenman RN. Quantitative proteomic analysis of Myc oncoprotein function. EMBO J. 2002;21:5088–96. doi: 10.1093/emboj/cdf525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlosser I, Holzel M, Hoffmann R, et al. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2005;24:520–4. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- 17.He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66:1239–56. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Brown MA, Rothnagel JA, Saunders NA, Smith R. Roles of heterogeneous nuclear ribonucleoproteins A and B in cell proliferation. J Cell Sci. 2005;118:3173–83. doi: 10.1242/jcs.02448. [DOI] [PubMed] [Google Scholar]
- 19.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36:641–7. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 20.He X, Pool M, Darcy KM, et al. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–8. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venables JP, Koh CS, Froehlich U, et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol. 2008;28:6033–43. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patry C, Bouchard L, Labrecque P, et al. Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 2003;63:7679–88. [PubMed] [Google Scholar]
- 23.He Y, Rothnagel JA, Epis MR, Leedman PJ, Smith R. Downstream targets of heterogeneous nuclear ribonucleoprotein A2 mediate cell proliferation. Mol Carcinog. 2009;48:167–79. doi: 10.1002/mc.20467. [DOI] [PubMed] [Google Scholar]
- 24.Xue Y, Zhou Y, Wu T, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 27.Zerbe LK, Pino I, Pio R, et al. Relative amounts of antagonistic splicing factors, hnRNP A1 and ASF/SF2, change during neoplastic lung growth: implications for pre-mRNA processing. Mol Carcinog. 2004;41:187–96. doi: 10.1002/mc.20053. [DOI] [PubMed] [Google Scholar]