Abstract
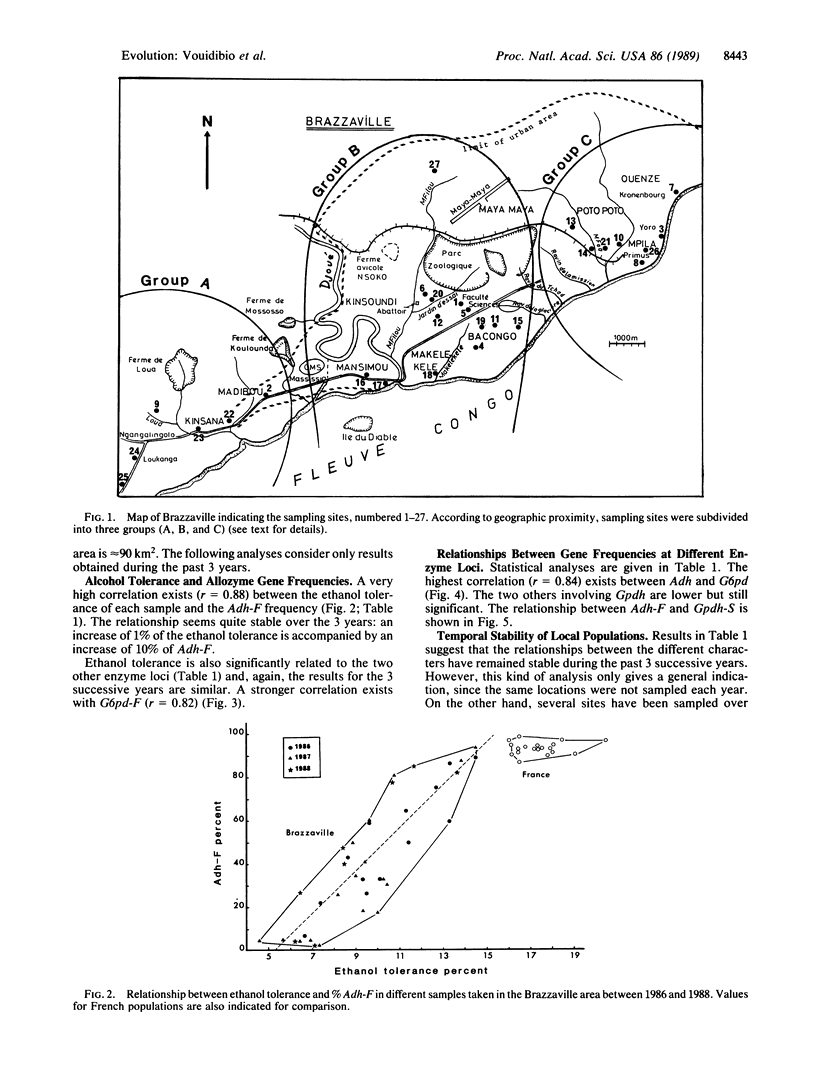
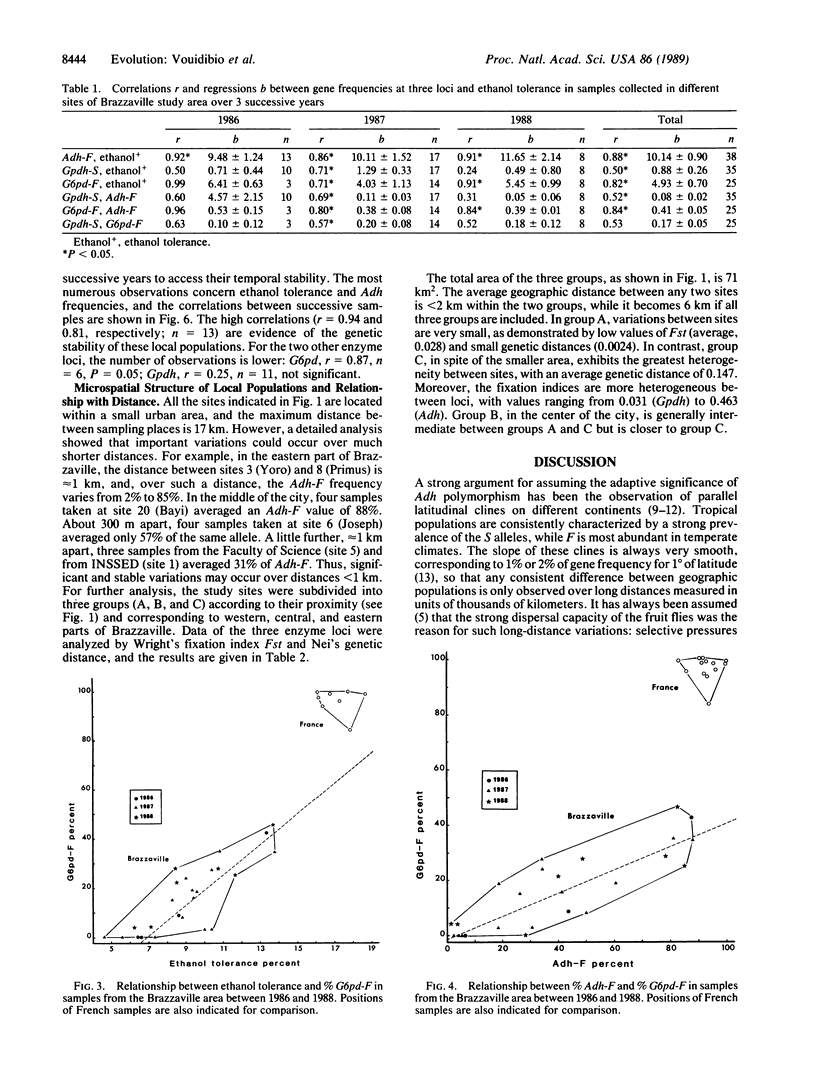
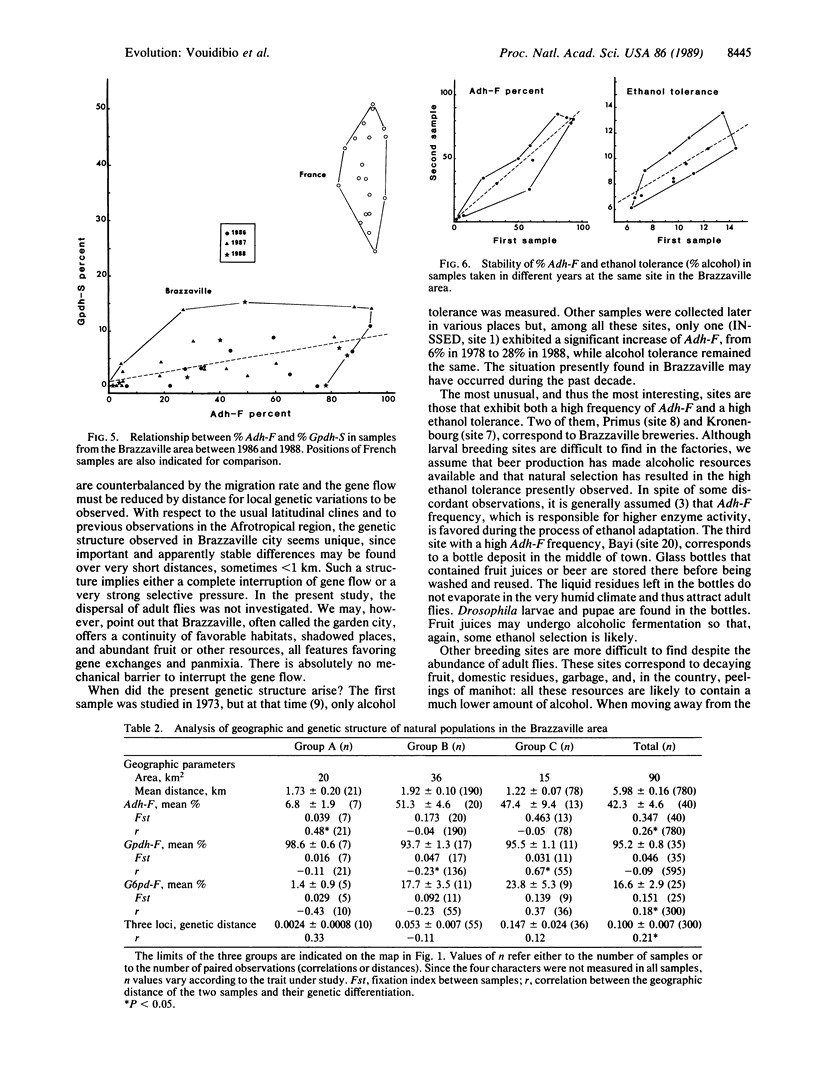
Alcohol dehydrogenase (Adh) (alcohol:NAD+ oxidoreductase, EC 1.1.1.1) gene frequencies and ethanol tolerance in Drosophila melanogaster are known to exhibit long-range latitudinal variations on different continents; this has led to the argument that the clines are adaptive. Accordingly, tropical populations are characterized both by a low frequency of Adh-F and by a low ethanol tolerance. In the urban area of Brazzaville (Congo) under an equatorial African climate, an original genetic structure of local populations has been found: Adh-F frequency varies from 3% to 90% when countryside and brewery populations are compared. This variation is accompanied by an increase of ethanol tolerance (from 6% to 13% alcohol). Such differences, which have remained stable for the past 3 years, were observed between collection sites less than 1 km apart. Two other enzyme loci exhibited a correlated variation with Adh-F--i.e., an increase of the S allele of glycerol-3-phosphate dehydrogenase (NAD+) (sn-glycerol-3-phosphate:NAD+ 2-oxidoreductase, EC 1.1.1.8) and of the F allele of glucose-6-phosphate dehydrogenase (D-glucose-6-phosphate:NADP+ 1-oxidoreductase, EC 1.1.1.49). Such observations suggest very strong selective pressures exerted by environmental ethanol that oppose the gene flow due to adult dispersal between contiguous habitats. A functional relationship between the polymorphisms of the three enzyme loci seems likely, and a metabolic interaction involving NAD and NADP cofactors is proposed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anxolabéhère D., Nouaud D., Périquet G., Tchen P. P-element distribution in Eurasian populations of Drosophila melanogaster: A genetic and molecular analysis. Proc Natl Acad Sci U S A. 1985 Aug;82(16):5418–5422. doi: 10.1073/pnas.82.16.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener D. R., Clegg M. T. Dynamics of correlated genetic systems. IV. Multilocus effects of ethanol stress environments. Genetics. 1978 Nov;90(3):629–644. doi: 10.1093/genetics/90.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener D. R., Clegg M. T. Evidence for biochemical and physiological differences between enzyme genotypes in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4444–4447. doi: 10.1073/pnas.78.7.4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J. R., Alonso-Moraga A., Borai F., Capy P., Merçot H., McEvey S. F., Munoz-Serrano A., Tsakas S. Latitudinal variation of Adh gene frequencies in Drosophila melanogaster: a Mediterranean instability. Heredity (Edinb) 1989 Feb;62(Pt 1):11–16. doi: 10.1038/hdy.1989.2. [DOI] [PubMed] [Google Scholar]
- David J. R., Bocquet C. Similarities and differences in latitudinal adaptation of two Drosophila sibling species. Nature. 1975 Oct 16;257(5527):588–590. doi: 10.1038/257588a0. [DOI] [PubMed] [Google Scholar]
- David J. R., Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 1988 Apr;4(4):106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- David J. R. Latitudinal variability of Drosophila melanogaster: allozyme frequencies divergence between European and Afrotropical populations. Biochem Genet. 1982 Aug;20(7-8):747–762. doi: 10.1007/BF00483971. [DOI] [PubMed] [Google Scholar]
- Eanes W. F., Bingham B., Hey J., Houle D. Targeted selection experiments and enzyme polymorphism: negative evidence for octanoate selection at the G6PD locus in Drosophila melanogaster. Genetics. 1985 Feb;109(2):379–391. doi: 10.1093/genetics/109.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer B. W., Langevin M. L., McKechnie S. W. Dietary ethanol and lipid synthesis in Drosophila melanogaster. Biochem Genet. 1985 Aug;23(7-8):607–622. doi: 10.1007/BF00504295. [DOI] [PubMed] [Google Scholar]
- Jallon J. M. A few chemical words exchanged by Drosophila during courtship and mating. Behav Genet. 1984 Sep;14(5):441–478. doi: 10.1007/BF01065444. [DOI] [PubMed] [Google Scholar]
- Laurie-Ahlberg C. C., Maroni G., Bewley G. C., Lucchesi J. C., Weir B. S. Quantitative genetic variation of enzyme activities in natural populations of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1073–1077. doi: 10.1073/pnas.77.2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElfresh K. C., McDonald J. F. The effect of alcohol stress on nicotinamide adenine dinucleotide (NAD+) levels in Drosophila. Biochem Genet. 1983 Apr;21(3-4):365–374. doi: 10.1007/BF00499145. [DOI] [PubMed] [Google Scholar]
- McKechnie S. W., Geer B. W. sn-Glycerol-3-phosphate oxidase and alcohol tolerance in Drosophila melanogaster larvae. Biochem Genet. 1986 Dec;24(11-12):859–872. doi: 10.1007/BF00554525. [DOI] [PubMed] [Google Scholar]
- Oakeshott J. G., Chambers G. K., Gibson J. B., Eanes W. F., Willcocks D. A. Geographic variation in G6pd and Pgd allele frequencies in Drosophila melanogaster. Heredity (Edinb) 1983 Feb;50(Pt 1):67–72. doi: 10.1038/hdy.1983.7. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987 May 15;236(4803):787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Stalker H. D. Chromosome studies in wild populations of D. melanogaster. Genetics. 1976 Feb;82(2):323–347. doi: 10.1093/genetics/82.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]


