SUMOylation of TopoIIα inhibits its decatenation activity, preventing resolution of tangled DNA at centromeres before anaphase onset.
Abstract
DNA topoisomerase IIα (TopoIIα) is an essential chromosome-associated enzyme with activity implicated in the resolution of tangled DNA at centromeres before anaphase onset. However, the regulatory mechanism of TopoIIα activity is not understood. Here, we show that PIASy-mediated small ubiquitin-like modifier 2/3 (SUMO2/3) modification of TopoIIα strongly inhibits TopoIIα decatenation activity. Using mass spectrometry and biochemical analysis, we demonstrate that TopoIIα is SUMOylated at lysine 660 (Lys660), a residue located in the DNA gate domain, where both DNA cleavage and religation take place. Remarkably, loss of SUMOylation on Lys660 eliminates SUMOylation-dependent inhibition of TopoIIα, which indicates that Lys660 SUMOylation is critical for PIASy-mediated inhibition of TopoIIα activity. Together, our findings provide evidence for the regulation of TopoIIα activity on mitotic chromosomes by SUMOylation. Therefore, we propose a novel mechanism for regulation of centromeric DNA catenation during mitosis by PIASy-mediated SUMOylation of TopoIIα.
Introduction
Resolution of sister chromatid cohesion is a fundamental process required for faithful chromosome segregation. Together with cohesin-mediated cohesion, DNA catenation maintains chromosome cohesion in the early stages of mitosis (Díaz-Martínez et al., 2008; Yanagida, 2009). By the onset of anaphase, however, the cohesin complex is removed by separase through anaphase-promoting complex/cyclosome (APC/C)-dependent ubiquitination and degradation of securin, and the catenation of centromeric DNA is resolved by the action of a specialized enzyme called DNA topoisomerase IIα (TopoIIα; Porter and Farr, 2004; Lee and Bachant, 2009). Several early studies showed that TopoIIα relocalizes from chromosome arms to the centromere during mitosis (Gorbsky, 1994; Ishida et al., 1994; Tavormina et al., 2002), and further studies using self-primed in situ labeling revealed that catalytically active TopoIIα accumulates primarily at the centromere (Andersen et al., 2002). In addition, recent studies have shown that ultrafine bridges originating from tangled DNA in metaphase chromosomes were resolved by TopoIIα activity after removal of the cohesin complex (Wang et al., 2010), which indicates a role for TopoIIα activity in mitosis. This evidence strongly suggests tight regulation of TopoIIα activity in space and time. Although extensive biochemical studies have elucidated the molecular mechanism of TopoII family proteins’ enzymatic reactions (Schoeffler and Berger, 2008), how the catalytic activity of TopoIIα is regulated at the centromere in such a specific manner is unknown.
Studies examining the relationship between TopoIIα activity and posttranslational modification (PTM) have not clearly demonstrated that TopoIIα activity is regulated by PTM (Isaacs et al., 1998; Ishida et al., 2001). Yet, one PTM of TopoIIα, SUMOylation, has been suggested as a potential regulator of TopoIIα activity given that TopoIIα SUMOylation is mitosis specific and occurs near centromeres (Bachant et al., 2002). SUMO (small ubiquitin-like modifier) is a conserved ubiquitin family protein in eukaryotes (Johnson, 2004; Geiss-Friedlander and Melchior, 2007). Vertebrates typically express three SUMO paralogues designated as SUMO1, -2, and -3. SUMO2 and -3 are ∼95% identical, whereas SUMO1 has ∼45% identity with both SUMO2 and -3. (In this paper, we refer to SUMO2 and -3 as SUMO2/3 when they are indistinguishable.) SUMO proteins contain a C-terminal di-glycine motif that is exposed by a hydrolase before a SUMOylation reaction of target proteins. The biochemical process of SUMOylation requires unique components but is somewhat similar to the ubiquitination pathway. First, SUMO proteins are activated by the E1 enzyme (Aos1/Uba2 heterodimer); then, they are transferred to the E2 enzyme (Ubc9) and finally conjugated to cellular substrates via an E3 ligase enzyme. Defects in the SUMOylation pathway have been found to cause faulty mitosis (Watts, 2007; Dasso, 2008), typically represented in most organisms by failure of proper chromosome segregation (Biggins et al., 2001; Hari et al., 2001; Nacerddine et al., 2005).
Siz1p and Siz2p, which are conserved eukaryotic SUMO E3 ligases, are responsible for the SUMOylation of TopoII in budding yeast, and the loss of Siz-mediated TopoII SUMOylation decreases chromosome transmission fidelity (Takahashi et al., 2006). Using a Xenopus egg extract (XEE) cell-free assay, we previously showed that PIASy, a member of the PIAS/Siz family of SUMO ligases, is an essential chromosomal component for promoting TopoIIα SUMO2/3 modification in vertebrates, and suggested a role for PIASy in chromosome segregation (Azuma et al., 2005). Moreover, studies using HeLa cells revealed that PIASy is required for faithful chromosomal separation, which is not dependent on centromeric cohesin but is related to TopoIIα localization at the centromere (Díaz-Martínez et al., 2006). Together, this evidence indicates that the PIAS/Siz family of E3 ligases has a conserved role in chromosome segregation in eukaryotes through regulation of TopoII SUMOylation. In contrast, studies using lysates from mouse embryonic fibroblasts (MEFs) that were deficient in RanBP2/Nup358, a SUMO E3 ligase that is also a component of the nuclear pore complex in vertebrates (Pichler et al., 2002), provided evidence that RanBP2 facilitates SUMOylation of TopoIIα through SUMO1 conjugation (Dawlaty et al., 2008). The discrepancy between these models remains to be examined.
For this paper, we established an in vitro SUMOylation assay using recombinant TopoIIα as substrate in order to elucidate the biochemical consequence of PIASy-mediated SUMO2/3 modification on TopoIIα activity. Consistent with our previous results using XEE assays (Azuma et al., 2005), we demonstrate that PIASy robustly facilitates SUMOylation of TopoIIα, and this modification is SUMO2/3 specific in vitro. We also observed that SUMOylated TopoIIα exhibits much less DNA decatenation activity, which indicates a potential mechanism for inhibition of TopoIIα activity by SUMOylation. Using mass spectrometric analysis of TopoIIα isolated from mitotic chromosomes, we identified lysine 660 (Lys660) as a novel SUMOylation site of TopoIIα. Further biochemical studies demonstrated that elimination of SUMOylation at Lys660 suppressed SUMOylation-dependent inhibition of TopoIIα activity, independent of other SUMOylation sites. Finally, we show that SUMOylation of Lys660 is stimulated by DNA binding of TopoIIα, which suggests that the enzymatically active TopoIIα might be a primary target of Lys660 SUMOylation.
Our findings strongly suggest that TopoIIα SUMOylation regulates decatenation of centromeric DNA. The colocalization of TopoIIα and PIASy at centromeres and the stimulation of inhibitory SUMOylation at Lys660 by DNA binding of TopoIIα further suggest that this novel regulation of TopoIIα activity is controlled spatiotemporally during mitosis. Therefore, we propose that the SUMO ligase PIASy catalyzes SUMO2/3 modification of TopoIIα that regulates TopoIIα activity during mitosis.
Results
PIASy promotes SUMO2/3 conjugation of TopoIIα in XEEs
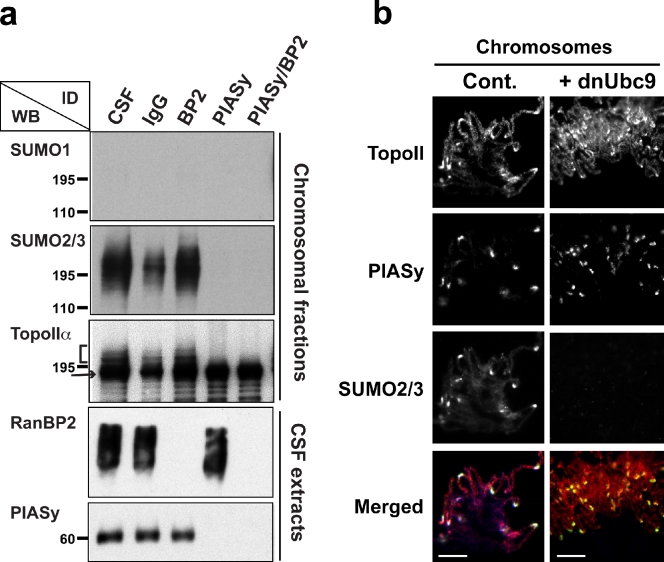
We previously demonstrated, using an XEE cell-free assay, that TopoIIα is modified by SUMO2/3 in a PIASy-dependent manner (Azuma et al., 2005). More recently, studies in MEFs suggested that RanBP2 promotes SUMOylation of TopoIIα through SUMO1 conjugation (Dawlaty et al., 2008). To investigate whether RanBP2 has a role in SUMOylation of TopoIIα in XEE assays, we immunodepleted XEE of specific E3 enzymes (RanBP2, PIASy, or RanBP2/PIASy) and examined TopoIIα for alterations in SUMOylation. After immunodepletion of RanBP2 from XEE, SUMO2/3 modification was still intact in the chromosomal fractions (indicated by the bracket in Fig. 1 a). In contrast, no detectable TopoIIα SUMOylation was observed in the absence of PIASy (Fig. 1 a). The lack of SUMO1-modified proteins associated with mitotic chromosomes indicates that TopoIIα on mitotic chromosomes is exclusively modified by SUMO2/3 (Fig. 1 a).
Figure 1.
PIASy but not RanBP2 is required for SUMO2/3 conjugation of TopoIIα in XEE. (a) XEE were immunodepleted using antibodies against RanBP2, PIASy, or RanBP2/PIASy. The depleted extracts were incubated with 10,000 sperm nuclei/µl for 1 h at 25°C. Non- or mock (IgG)-depleted extracts were also subjected to the same procedure. Isolated chromosomes from each reaction were analyzed by Western blotting (WB) for the indicated protein. Immunodepletion (ID) of RanBP2 had no effect on the SUMOylation of TopoIIα, whereas PIASy ID eliminated TopoIIα SUMOylation. The arrow and bracket indicate unmodified and SUMO2/3-modified TopoIIα, respectively. Positions of molecular mass standards (kD) are indicated on the left. (b) The mitotic chromosomes were prepared as in Materials and methods and were analyzed by immunostaining with the indicated antibodies: TopoIIα is shown in red, PIASy is shown in green, and SUMO2/3 is shown in blue in merged panel. TopoIIα colocalized with PIASy and SUMO2/3 at the centromeres. The addition of dnUbc9 eliminated SUMO2/3 modification but did not alter the localization of TopoIIα at the centromeres of mitotic chromosomes. Bars, 10 µm.
We further analyzed PIASy-dependent SUMO2/3 modification of TopoIIα on mitotic chromosomes by immunofluorescence microscopy. Using EGFP-fused SUMO2 in XEE assays, we previously showed that SUMO2/3-modified proteins are localized on inner centromeric regions (Azuma et al., 2005). For the current study, we prepared an antibody to enable visualization of endogenous SUMO2/3 on chromosomes. Mitotic chromosomes prepared from replicated chromatin in XEE were fixed and immunostained using antibodies specific for TopoIIα, PIASy, and SUMO2/3 (Fig. 1 b). We observed that PIASy localizes to distinct foci on mitotic chromosomes, and these foci overlap with SUMO2/3 localization (Fig. 1 b). Because endogenous SUMO2/3 also colocalizes with an inner centromeric marker, Aurora B (Fig. S1), we concluded that PIASy displays centromeric localization. Finally, we verified the localization of TopoIIα using two different antibodies. Both antibodies revealed that TopoIIα is located throughout the chromosome axis, with clear accumulation at the centromeres of mitotic chromosomes as previously shown in other species (Fig. 1 b and Fig. S1; Gorbsky, 1994; Chang et al., 2003). Overall, the colocalization of TopoIIα and SUMO2/3 was clearly apparent at the centromeric regions but not in other parts of the chromosome, which suggests that SUMOylation of TopoIIα mainly occurs at the centromere and in proximity to PIASy (Fig. 1 b). The staining pattern of TopoIIα did not change after addition of dominant-negative Ubc9 (dnUbc9), which prevents SUMOylation, indicating that localization of TopoIIα was not altered by the perturbation of SUMOylation (Figs. 1 b and S1). Because we found no evidence for RanBP2 involvement in SUMO2/3 modification of TopoIIα in our assay system, we focused our subsequent studies on PIASy-mediated TopoIIα SUMOylation.
PIASy promotes SUMO2/3 conjugation of TopoIIα in reconstituted in vitro SUMOylation assays
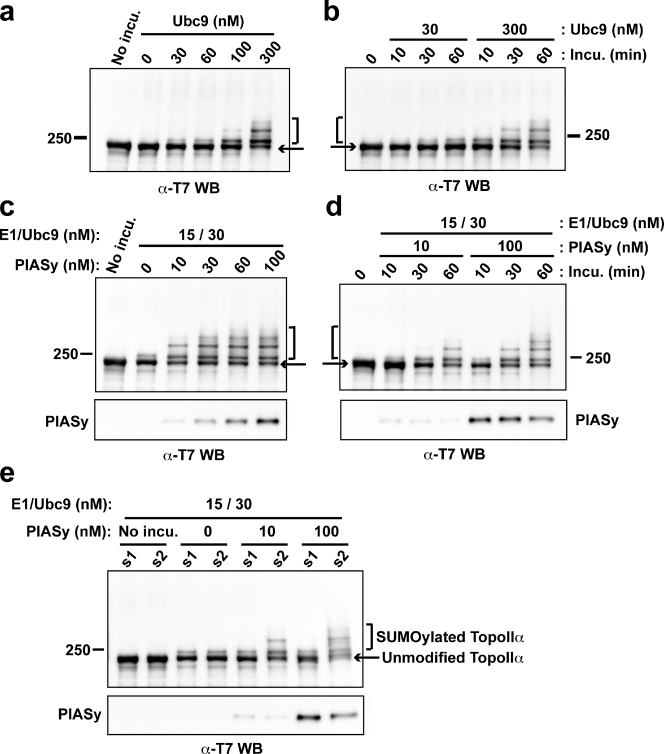
We previously demonstrated the requirement of PIASy activity for SUMOylation of chromosomal TopoIIα in XEE assays (Azuma et al., 2003). However, because of the extreme instability of the SUMOylation reaction, experimentally managing the state of SUMOylation in vivo was difficult, limiting the use of XEE assays for further investigations of TopoIIα SUMOylation. Therefore, we next sought to examine the PIASy-dependent SUMOylation of TopoIIα in an in vitro assay. For these studies, all of the protein components required for SUMOylation were prepared as described in Materials and methods. For most in vitro SUMOylation reactions, we used 500 nM of T7-tagged TopoIIα, 5 µM SUMO, and 15 nM E1 (Aos1/Uba2), and the SUMOylation was detected by Western blotting using the T7 tag.
Ubc9, a SUMO E2 conjugating enzyme, directly binds to its consensus sequence on substrates without E3 ligase (Bernier-Villamor et al., 2002; Geiss-Friedlander and Melchior, 2007), and, in fact, major SUMOylation sites of budding yeast TopoII were identified based on this consensus sequence (Bachant et al., 2002). Hence, we sought to identify whether SUMOylation of Xenopus laevis TopoIIα is dependent on Ubc9. Purified X. laevis TopoIIα was incubated with various concentrations of Ubc9 and analyzed by Western blotting. Our results indicated that Ubc9 could promote SUMOylation of TopoIIα in a dose-dependent manner (Fig. 2 a), and 300 nM Ubc9 was sufficient to promote SUMOylation of >50% of the TopoIIα. However, when the concentration of Ubc9 was close to the endogenous levels found in XEE, 30 nM, the efficiency of TopoIIα SUMOylation dropped to barely detectable levels (Fig. 2, a and b).
Figure 2.
PIASy is required for the efficient SUMOylation of TopoIIα and for the selection of SUMO paralogues. (a) Ubc9 dosage-dependent SUMOylation. T7 tagged-TopoIIα was incubated in a reaction containing various concentrations of Ubc9 (0–300 nM) in the presence of SUMO2. The amount of SUMO2-conjugated TopoIIα was similar to that seen in XEE only when 300 nM Ubc9 was added. (b) Time course experiment with physiological (30 nM) and higher (300 nM) concentration of Ubc9. (c) PIASy dosage-dependent SUMOylation. T7-TopoIIα was incubated as in panel a, except with various concentrations of PIASy (0–100 nM) and with the physiological concentration of Ubc9 (30 nM). PIASy efficiently facilitated SUMOylation of TopoIIα under conditions using 30 nM Ubc9, where SUMOylation had barely appeared in the absence of PIASy. SUMOylation was saturated using >60 nM PIASy. (d) Time course experiment of PIASy-dependent SUMOylation. The reactions were performed with physiological (10 nM) or higher (100 nM) concentrations of PIASy in the presence of 30 nM Ubc9. (e) T7-TopoIIα was incubated with either SUMO1 (s1) or SUMO2 (s2) in the presence of PIASy as indicated. PIASy showed a preference for SUMO2 over SUMO1. Positions of molecular mass standards (kD) are indicated on the sides of the gel blots.
Next, to assess the contribution of PIASy to SUMOylation, TopoIIα was incubated with various concentrations of PIASy using a physiologically relevant concentration of Ubc9 (30 nM). Under such conditions, TopoIIα was efficiently SUMOylated in the presence of PIASy, and could be SUMOylated even when PIASy concentration was 10 nM, which is near the endogenous level in XEE (Fig. 2, c and d).
The observation that TopoIIα is specifically modified by SUMO2/3 in XEE led us to hypothesize that PIASy may select the SUMO paralogue for the conjugation. To test this hypothesis, TopoIIα was incubated with either SUMO1 or SUMO2 in the presence of PIASy and physiological concentrations of Ubc9. This assay revealed that PIASy mediates modification of TopoIIα by SUMO2 but not by SUMO1 (Fig. 2 e), even though SUMO1 was highly active with RanGAP1 as the substrate (not depicted), as previously shown (Azuma et al., 2001). This indicates that PIASy plays a role in the selection of SUMO paralogues. We also confirmed that PIASy mediates modification of TopoIIα with SUMO3 in similar level as with SUMO2 (unpublished data). Together, we conclude that PIASy is an essential element for SUMO2/3 modification of TopoIIα under physiological conditions.
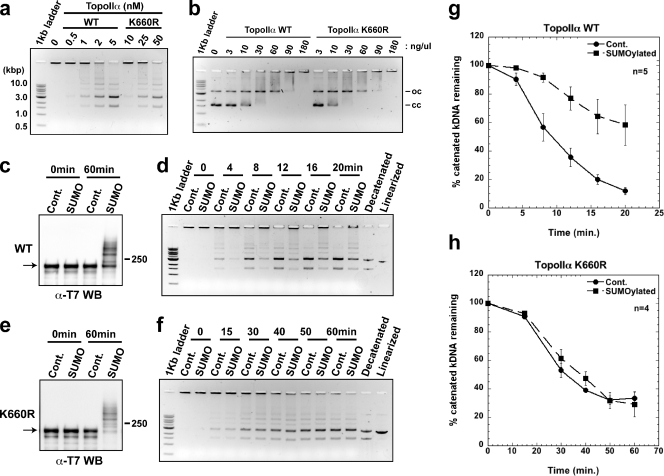
PIASy-mediated SUMOylation inhibits the decatenation activity of TopoIIα
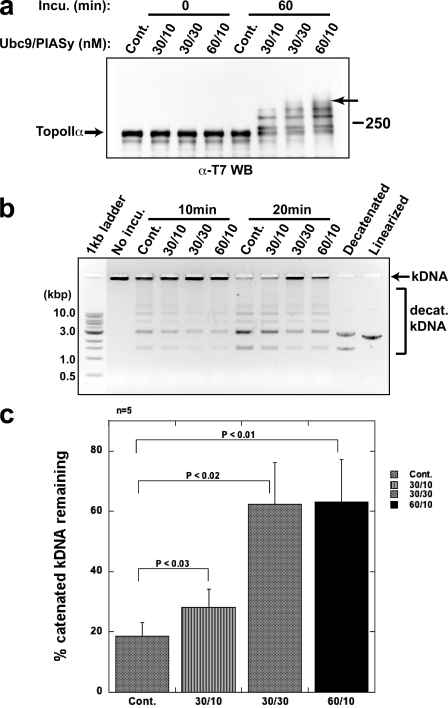
Previous studies in HeLa cells have suggested that PIASy is required for regulation of cohesin-independent cohesion of centromeres (Díaz-Martínez et al., 2006). We speculated that PIASy might play a role in localizing TopoIIα to the centromere by regulating its SUMOylation; however, we found that inhibition of SUMOylation did not alter TopoIIα localization in XEE assays (Fig. 1 b). Therefore, we examined whether PIASy-dependent SUMOylation alters TopoIIα activity, which is required for proper chromosome segregation. To this end, TopoIIα was incubated in an in vitro SUMOylation reaction with either a processed form of SUMO2 (SUMO2-GG) or a truncated form of SUMO2 (SUMO2-G) that cannot be conjugated to substrates because of a lack of one C-terminal glycine. SUMOylated and non-SUMOylated TopoIIα were then analyzed for decatenation activity using kinetoplast DNA (kDNA) as a substrate. Once decatenated by TopoIIα, the interlocking chain of circular kDNA releases to form minicircles. As shown in Fig. 3 a, TopoIIα was differentially SUMOylated in 60 min in vitro using various concentrations of Ubc9 and PIASy in the presence of SUMO2-GG. A control reaction that contained Ubc9 and PIASy in the presence of SUMO2-G showed no SUMOylation. Western blot analysis revealed that the intensity of TopoIIα SUMOylation could be controlled by the in vitro reaction conditions and that >50% of the TopoIIα was modified under each condition. Subsequent assays indicated that the decatenation activity of TopoIIα is markedly inhibited by SUMO conjugation (Fig. 3, b and c). One interesting aspect of this analysis is that, despite subtle differences in TopoIIα SUMOylation among the series of reactions, the presence of the highest molecular weight band (indicated by the arrow in Fig. 3 a) appeared to correlate with strong inhibition of TopoIIα decatenation activity. Together, our results indicate that PIASy-mediated SUMO modification of TopoIIα inhibits TopoIIα decatenation activity.
Figure 3.
SUMO modification affects the decatenation activity of TopoIIα. (a) T7-TopoIIα was incubated with various combinations of Ubc9/PIASy as indicated to obtain a series of SUMOylation profiles. All control reactions (Cont.) were performed with 60 nM Ubc9/10 nM PIASy and SUMO2-G, which could not be conjugated. The samples were analyzed by Western blotting for the T7 tag. The arrow indicates maximal SUMO modification of TopoIIα (seen in 30/30 and 60/10). Positions of molecular mass standards (kD) are indicated on the right. (b) Representative data of decatenation assay. Samples in a were further incubated with decatenation buffer that contained kDNA for 10 or 20 min, and the products were resolved in an agarose gel. Decatenated and linearized markers are designated. (c) Band intensity data from five independent experiments performed as in b are presented as the percentage of catenated kDNA remaining after a 20-min incubation, with standard error (error bars) and probability value from a Student’s t test. SUMO2 modification of TopoIIα decreased its decatenation activity.
Lysine at 660 is one SUMOylation site of TopoIIα in XEE
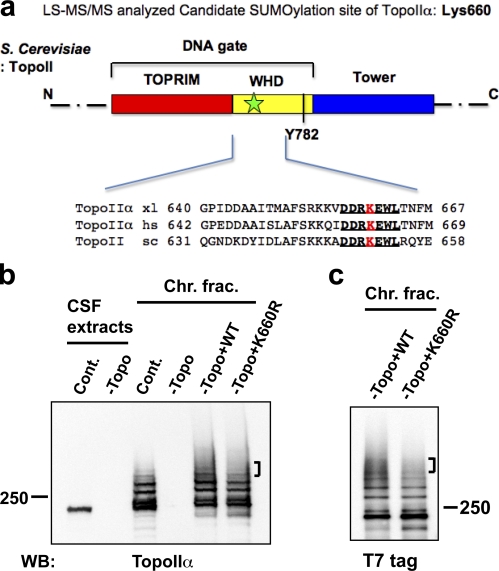
To better understand the molecular basis of TopoIIα inhibition by SUMOylation, we sought to identify SUMOylation sites of TopoIIα. To this end, a SUMOylated form of endogenous TopoIIα was purified from mitotic chromosomes prepared in XEE (Fig. S2). Isolated bands were double-digested with trypsin and chymotrypsin followed by mass spectrometric analysis using the same method as used to identify the SUMOylation site of poly (ADP-ribose) polymerase I (PARP1), another mitotic chromosomal substrate (Ryu et al., 2010). Double digestion with trypsin and chymotrypsin generates a remnant of the SUMO sequence (QQQTGG, with a mass of 599.2663 D) on the lysine of a SUMO modified peptide. With ∼50% sequence coverage, Lys660 (K*EWLTNFMQDR, where the asterisk refers to QQQTGG) was shown to have the SUMO signature in the digested pool of SUMO2/3-TopoIIα (Fig. S3). This result was unexpected given that (a) the sequences surrounding Lys660 do not match the canonical or noncanonical consensus sequences predicted using a SUMO prediction program (Xue et al., 2006), and (b) the genetically determined SUMOylation sites in budding yeast were all located in the C-terminal domain (Bachant et al., 2002; Takahashi et al., 2006), whereas Lys660 is located in the core active domain (DNA gate) of TopoIIα (Fig. 4 a; Berger et al., 1996; Schoeffler and Berger, 2005). Also, it was striking that the sequences including and surrounding Lys660 are highly conserved, from yeast to human (Fig. 4 a).
Figure 4.
TopoIIα K660R, a candidate SUMOylation mutant, shows incomplete SUMOylation in XEE. (a) Schematic diagram of S. cerevisiae TopoIIα primary structure. Domains are denoted by color. This panel was modified from Schoeffler and Berger (2008), with permission from Cambridge University Press. TOPRIM, Topoisomerase-primase fold domain; WHD, Winged-helix domain; Tower, adjacent domain to WHD. The black bar indicates the catalytic tyrosine (Y782) for DNA cleavage in the WHD domain. Lys660 in X. laevis TopoIIα was designated as a potential SUMOylation site by mass spectrometric analysis. The approximate position of the candidate lysine is shown by a green star in the DNA gate domain of TopoIIα. The sequences near TopoIIα Lys660 from X. laevis (xl), Homo sapiens (hs), and S. cerevisiae (sc) are conserved (indicated with bold and underlined text). (b) XEE were immunodepleted using nonspecific IgG (Cont.) or an anti-TopoIIα antibody (−Topo). Efficiency of TopoIIα depletion was confirmed by comparison of mock-depleted (Cont.) to TopoIIα-depleted (−Topo) CSF extracts (left two lanes, labeled CSF extracts). WT nontagged TopoIIα (WT) or mutant TopoIIα, with substitution of arginine for Lys660 (K660R), was added to the TopoIIα-depleted extracts (−Topo). After 1 h incubation at 25°C, mitotic chromosomes were isolated and analyzed by anti-TopoIIα Western blotting. Analyzed chromosome samples were from mock-depleted (Cont.), TopoII-depleted (−Topo), and recombinant TopoIIα added-back extracts (−Topo+WT or –Topo+K660R). (c) Same examination as in b except that the recombinant TopoIIα proteins had a T7 tag at the N terminus. Both nontagged and T7-tagged K660R mutant showed subtle but reproducible reduction in higher shifted bands (indicated by brackets) of SUMOylation compared with WT. Positions of molecular mass standards (kD) are indicated on the sides of the gel blots.
To confirm the mapping result, recombinant wild-type TopoIIα (WT) or TopoIIα with an arginine substitution for Lys660 (K660R) were prepared as described in Materials and methods, and the purity of these recombinant proteins was confirmed, as shown in Fig. S4. Add-back experiments of the recombinant proteins to TopoIIα-immunodepleted XEE indicated that the K660R mutant exhibited a slight reduction in higher shifted species of SUMOylated TopoIIα compared with that of WT. Although major SUMOylations still occurred (Fig. 4, b and c) using both an untagged (Fig. 4 b) and T7-tagged (Fig. 4 c) recombinant TopoIIα, K660R showed a reproducible deficiency in generating SUMOylation represented by higher shifted bands. In summary, Lys660 is a SUMOylation site of TopoIIα associated with mitotic chromosomes in XEE, and other SUMOylation sites remain to be identified.
Lack of TopoIIα SUMOylation at Lys660 abolishes SUMOylation-dependent inhibition of TopoIIα activity
Available x-ray crystal structure information for TopoIIα indicates that Lys660 faces DNA (Dong and Berger, 2007). Therefore, we predicted that alteration of Lys660 would affect TopoIIα activity. Indeed, when we compared the decatenation activity of recombinant, unSUMOylated WT and K660R TopoIIα, K660R was ∼20 times less active than WT (Fig. 5 a). Yet, gel mobility shift assays showed that both WT and K660R bind to DNA with similar affinity (Fig. 5 b), which suggests that lower decatenation activity of K660R is not simply caused by the deficiency of DNA binding. Given that the relatively minor alteration, substituting Lys660 with arginine, reduces TopoIIα decatenation activity, we speculated that SUMO conjugation of Lys660 might have a significant impact on the activity of TopoIIα.
Figure 5.
The elimination of TopoIIα SUMOylation at Lys660 blocks SUMOylation-dependent inhibition of TopoIIα activity. (a) Unmodified TopoIIα WT and K660R proteins were incubated with kDNA to determine relative activity. K660R had ∼20 times less activity than WT. (b) Electrophoretic mobility shift assay. Unmodified TopoIIα WT and K660R were incubated with plasmid DNA to determine relative DNA binding affinity. Both WT and K660R displayed similar binding affinity to DNA. oc and cc stand for open and closed circle, respectively. (c and e) The TopoIIα WT and K660R were SUMO2-modified in vitro with 60 nM of Ubc9 and 30 nM of PIASy. Control reactions (Cont.) using the same condition except for SUMO2-G were also performed. Non-SUMOylated or SUMOylated TopoIIα samples were assayed for decatenation activity. (d and f) Representative results of decatenation activity assays with TopoIIα WT (d) and K660R (f) are shown. The mean decatenation activity from five independent experiments with TopoIIα WT (g) and four independent experiments with TopoIIα K660R (h) are displayed as the percentage of catenated kDNA remaining, with standard error (error bars). The strong inhibition of TopoIIα decatenation activity by SUMOylation was abolished in reactions using TopoIIα K660R. Positions of molecular mass standards (kD) are indicated on the sides of the gel blots.
To test this, TopoIIα WT and K660R were applied to the in vitro SUMOylation–decatenation coupled assay. For SUMOylation reactions (Fig. 5, c and e), we used 60/30 nM of Ubc9/PIASy to better observe the inhibition of TopoIIα WT decatenation activity by SUMOylation and the potential alteration in the SUMOylation-dependent regulation of TopoIIα activity for the K660R mutant. Consistent with our earlier results, the decatenation activity of TopoIIα WT was efficiently inhibited by SUMO modification (Fig. 5, d and g). However, it was striking that the decatenation activity of TopoIIα K660R was no longer inhibited by SUMO modification (Fig. 5, f and h), despite the fact that both TopoIIα WT (Fig. 5 c) and K660R mutant (Fig. 5 e) were robustly modified by SUMO2. Similar results were obtained using different tagged TopoIIα WT and K660R for the analysis (Fig. S5). These results suggest that PIASy-mediated SUMOylation on Lys660 has a crucial role in the regulation of TopoIIα activity.
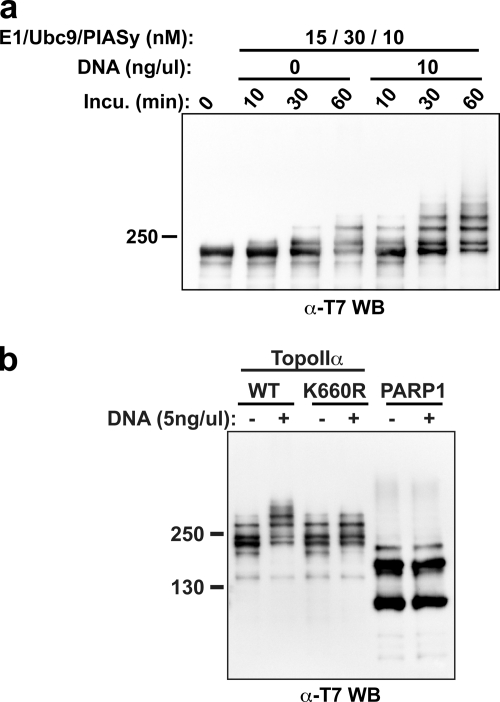
SUMOylation of TopoIIα Lys660 is regulated by DNA
Based on x-ray crystallographic structure analysis, TopoIIα Lys660 lies near the DNA backbone and appears not easily accessible for SUMOylation (Dong and Berger, 2007). We speculated that this limited accessibility might correlate with our inability to see a dramatic difference in the in vitro SUMOylation profile of WT and K660R TopoIIα, even though we observed subtle but reproducible reduction of K660R TopoIIα SUMOylation in XEE assays (Fig. 4, b and c). We suspected that when TopoIIα binds to DNA, it changes conformation, making it susceptible to SUMOylation of Lys660 in the XEE assay. To examine this hypothesis, we performed in vitro SUMOylation assays with WT TopoIIα in the presence or absence of DNA. As shown in Fig. 6 a, WT TopoIIα was extremely susceptible to SUMOylation in the presence of DNA. Addition of DNA to the in vitro reaction increased the amount of TopoIIα SUMOylation as well as the rate of modification, such that the amount of SUMOylated WT TopoIIα after 1 h of incubation without DNA was comparable to that formed after 10 min with DNA (Fig. 6 a). Because both TopoIIα and PIASy can bind DNA, we also considered the possibility that DNA acts as an adaptor to increase the binding affinity of TopoIIα and PIASy, leading to acceleration of SUMOylation. However, because the SUMOylation of PARP1, another chromosomal substrate of SUMO2/3 found in XEE (Ryu et al., 2010), is barely affected by the addition of an equivalent amount of DNA (Fig. 6 b), this possibility seems unlikely, which supports the idea that DNA-dependent enhancement of TopoIIα SUMOylation results from exposure of SUMOylation sites by DNA binding. Remarkably, we found that the K660R mutant displayed a reduction of SUMOylation in the presence of DNA, although there was no significant difference in the SUMOylation profile between WT and K660R in the absence of DNA (Fig. 6 b). Considering the comparable DNA binding affinity of both proteins (Fig. 5 b), it is therefore reasonable to conclude that the alteration of TopoIIα conformation by DNA binding (Berger et al., 1996; Roca et al., 1996; Dong and Berger, 2007) increases the efficiency of TopoIIα SUMOylation. Together, our results suggest that there are SUMOylation sites of TopoIIα whose availability for SUMOylation depends on the conformational change of TopoIIα resulting from DNA binding, and that the SUMOylation of Lys660, which plays a key role in SUMOylation-dependent TopoIIα inhibition, is one of those sites.
Figure 6.
DNA binding of TopoIIα increases susceptibility of SUMOylation at Lys660. (a) TopoIIα WT in vitro SUMOylation reactions were performed with or without DNA. The samples were analyzed with anti-T7 tag antibody Western blots. The presence of DNA in the SUMOylation reactions significantly stimulates TopoIIα WT SUMOylation. (b) TopoIIα WT and TopoIIα K660R were subjected to in vitro reactions under the same condition as in a except for using 5 ng/µl of DNA. PARP1, a mitotic chromosomal SUMO2/3 substrate, was used as a control. A deficiency of TopoIIα K660R SUMOylation was observed in the presence of DNA compared with TopoIIα WT. Positions of molecular mass standards (kD) are indicated on the left.
Discussion
Our central achievement here is the finding of SUMOylation-dependent regulation of TopoIIα activity. We previously found that SUMOylation of TopoIIα did not significantly alter its enzymatic activity when using TopoIIα-containing fractions from XEE (Azuma et al., 2003). However, we anticipated a potential role for SUMOylation in the regulation of TopoIIα activity based on the difficulty separating SUMOylated TopoIIα associated with centromeres from unSUMOylated TopoIIα associated with other chromosome regions. The in vitro SUMOylation assay established for the current study allowed us to overcome the time and space obstacles of SUMOylation of TopoIIα, and thus to reexamine the possible role of SUMOylation in regulating TopoIIα activity during mitosis. Our assay clearly reveals that SUMOylation significantly inhibits the decatenation activity of TopoIIα and that this inhibition is correlated with the TopoIIα SUMOylation profile. In other words, the existence of the highest shifted species of SUMOylated TopoIIα, which appear when relatively high concentration of enzymes (Fig. 3 a) or DNA are added to the reactions (Fig. 6 a), correlates with strong inhibition of TopoIIα activity.
The identification of Lys660 as a SUMOylation site further supports the premise that TopoIIα activity is SUMOylation dependent. Lys660 is located within the DNA gate, which plays a key role in manipulation of double helical DNA strands (Dong and Berger, 2007; Schoeffler and Berger, 2008). Considering the importance of this region for TopoIIα function, it was predictable that even slight alteration of Lys660 would impact the activity of TopoIIα. Supporting this idea, replacement of Lys660 with arginine resulted in a substantial reduction of the TopoIIα decatenation activity (Fig. 5 a), although no loss of DNA binding is observed (Fig. 5 b). Notably, the abolition of SUMO modification at Lys660 eliminated the SUMOylation-dependent inhibition of TopoIIα decatenation activity. The interpretation of this result is obviously limited by the lower catalytic activity of the Lys660 mutant, which might render partial inhibition caused by SUMOylation of other sites of TopoIIα imperceptible. Therefore, detailed kinetic analyses of TopoIIα reactions, combined with analysis of other currently unidentified SUMOylation sites, will be necessary to clarify the function of each TopoIIα SUMOylation site.
We further observed that Lys660 SUMOylation is enhanced in the presence of DNA, and this is likely caused by exposure of the Lys660 site during the catalytic action of TopoIIα (Fig. 6 b). The hypothesis that an active TopoIIα conformation makes Lys660 more susceptible to SUMOylation is supported by an earlier finding that etoposide (or VP16) treatment of human cells induces hyper SUMOylation of TopoIIα at the centromere (Agostinho et al., 2008). Etoposide immobilizes TopoIIα in an intermediate structure with cleaved DNA (Baldwin and Osheroff, 2005), thus potentially exposing a SUMOylation site, as shown in Fig. 6. Collectively, our results strongly suggest that SUMOylation of Lys660 is responsible for the control of decatenation activity. However, it is possible that other TopoIIα SUMOylation sites contribute to changes in TopoIIα activity.
The internal repeat (IR) domain of RanBP2 possesses SUMO E3 ligase activity in vitro, and studies in RanBP2-deficient MEFs have implicated RanBP2 in SUMO1 conjugation of TopoIIα (Dawlaty et al., 2008). We found that the IR domain of RanBP2 could enhance TopoIIα SUMOylation in vitro (unpublished data), but paralogue specificity and SUMOylation site selection under these conditions were different from TopoIIα SUMOylation in XEEs. There are several other findings that argue against a role of RanBP2 as the primary E3 enzyme for TopoIIα in mitosis: First, the addition of Nocodazole, which disrupts the localization of RanBP2 from the centromere (Joseph et al., 2002), does not eliminate SUMOylation of TopoIIα (Azuma, 2009). Second, although Dawlaty et al. (2008) found that RanBP2 promotes SUMO1 conjugation of TopoIIα, TopoIIα is exclusively conjugated to SUMO2/3 in XEEs unless ectopic SUMO1 is supplied (Azuma et al., 2003). Finally, we observed that SUMO2/3 modification of TopoIIα on mitotic chromosomes was intact in RanBP2-immunodepleted XEEs unless PIASy was co-depleted (Fig. 1 a), which strongly suggests that RanBP2 is dispensable. Additionally, endogenous TopoIIα, PIASy, and SUMO2/3 colocalized at the centromeres of mitotic chromosomes (Fig. 1 b), which is consistent with the notion that PIASy is the E3 enzyme for TopoIIα SUMOylation at this site. In contrast, RanBP2 localizes to the outer kinetochore (Joseph et al., 2002). It is possible that the discrepancy between findings in XEEs and MEFs simply reflects the difference in experimental systems, and that different SUMO ligases mediate TopoIIα conjugation in mice and frogs. Alternatively, we have recently shown that centromeric SUMOylation results from precise localization of PIASy and its substrates (Ryu and Azuma, 2010). RanBP2 might indirectly affect the SUMOylation of TopoIIα by regulating the localization of TopoIIα to centromeres in MEFs through a mechanism that is not used in XEEs. The failure to localize TopoIIα could thus impair its subsequent PIASy-dependent SUMO2/3 modification.
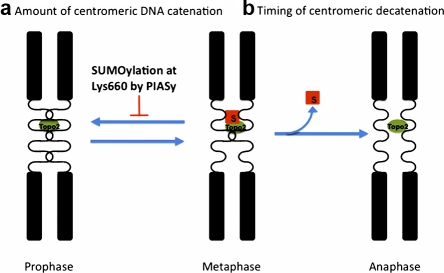
Our findings that (a) PIASy colocalizes with TopoIIα at the centromere, where enzymatically active TopoIIα is thought to accumulate (Andersen et al., 2002); (b) the DNA-bound form of TopoIIα is more susceptible to SUMOylation on Lys660; and (c) the SUMOylation of Lys660 inhibits TopoIIα activity lead us to propose that PIASy-dependent SUMOylation of TopoIIα regulates centromeric catenation. In this model, when SUMOylation is depressed, overly active TopoIIα leads to DNA recatenation at the centromeres, where sister chromatids are highly compact and close to each other. With proper SUMOylation, active TopoIIα is rendered temporarily inert to prevent recatenation (Fig. 7 a), and so only the proper amount of catenated DNA remains, avoiding early disjunction of sister chromatids before anaphase. This model explains why the perturbation of SUMOylation by either the elimination of PIASy or addition of dnUbc9 causes abnormal chromosomal segregation as represented by anaphase bridges, which could be the result of hypercatenation of centromeric DNA (Azuma et al., 2003, 2005). The model also explain why depletion of PIASy in HeLa cells produces cohesin-independent sister chromatid cohesion (Díaz-Martínez et al., 2006).
Figure 7.
Implications of SUMOylation in regulating the resolution of centromeric DNA. (a) Regulating the amount of catenated centromeric DNA. Active TopoIIα resolves catenated DNA at the centromere and SUMOylation reduces the activity of TopoIIα that has completed the decatenation of DNA. Without SUMOylation, overly active TopoIIα could recatenate DNA at the centromere. (b) Regulation of the timing of decatenation. TopoIIα SUMOylation keeps centromeric TopoIIα temporally inert until anaphase, when decatenation of centromeric DNA must take place. Without proper deSUMOylation of TopoIIα, decatenation of centromeric DNA will be compromised.
There are several pieces of evidence that support a requirement of TopoIIα activity for proper anaphase execution. According to a recent study of PICH (Plk1-interacting checkpoint helicase)-positive DNA threads, centromeric DNA catenation was resolved at the onset of anaphase (Wang et al., 2008). Wang et al., (2010) also demonstrated that TopoIIα decatenates centromeric DNA after removal of the cohesin complex. Lastly, TopoIIα SUMOylation is highly dynamic, with TopoIIα heavily modified by SUMO during metaphase and the rapid disappearance of modified TopoIIα at the onset of anaphase (Azuma et al., 2003). In this context, it is possible that deSUMOylation of TopoIIα regulates the timing of resolution of the catenated DNA at the centromere at the onset of anaphase. At anaphase, when the centromeres of sister chromatids are distal enough, deSUMOyation of TopoIIα allows the preferential decatenation of the last tangled sister chromatids. As such, we further propose that deSUMOylation of TopoIIα is critical to control the timing of the final decatenation at anaphase (Fig. 7 b). Extensive analysis using specific deSUMOylation enzymes of TopoIIα must be performed to directly test this hypothesis.
In summary, our finding that TopoIIα activity is inhibited by PIASy-mediated SUMOylation allows us to answer a long-standing question of how the catalytic activity of TopoIIα is tightly regulated in a space- and time-dependent manner. Future studies using somatic cells to observe consequential phenotypes caused by SUMOylation-deficient TopoIIα and an examination of specific deSUMOylation mechanisms of TopoIIα will strengthen the functional significance of SUMO-modified TopoIIα.
Materials and methods
DNA subcloning, site-directed mutagenesis, recombinant protein expression and purification, and antibodies
The cDNA of TopoIIα was cloned from X. laevis tadpole cDNA (provided by T. Amano and Y.B. Shi, National Institute of Child Health and Human Development, Bethesda, MD) using PCR amplification. The X. laevis TopoIIα coding sequence was subcloned into a pPIC 3.5 Kb vector in which either calmodulin-binding protein (CBP)-ZZ, CBP-T7, or T7-ZZ tag sequences were inserted (the CBP and ZZ TAP tag plasmids were provided by H. Yoon and K. Gould, Vanderbilt University, Nashville, TN). The lysine-to-arginine substitution of TopoIIα was generated by site-directed mutagenesis using a QuikChange kit (Agilent) according to manufacturer’s instructions. All constructs were verified by DNA sequencing.
For preparation of recombinant TopoIIα proteins, the plasmids were transformed into the GS115 strain of Pichia pastoris yeast and expressed according to the manufacturer’s instructions (Invitrogen). Protein purification with the CBP and ZZ tags was performed using a modified TAP protocol (EMBL Heidelberg). In brief, yeast cells expressing TopoIIα were frozen and ground with dry ice in a coffee mill, then mixed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM CaCl2, 1 mM MgCl2, 0.1% Triton X-100, 5% glycerol, 1 mM DTT, and 10 mM PMSF). Samples were then centrifuged at 25,000 g for 40 min. To capture the CBP-tagged TopoIIα, the supernatant was mixed with calmodulin-sepharose resin (GE Healthcare) for 90 min at 4°C. The resin was then washed with lysis buffer, and TopoIIα was eluted with buffer containing 10 mM EGTA. For ZZ-tagged TopoIIα, proteins were captured on IgG-Sepharose (GE Healthcare), and elution was performed by cleaving with PreScission Protease (GE Healthcare) according to the manufacturer’s instructions. The elution was further purified by Mono Q anion-exchange chromatography (GE Healthcare). The E1 complex (Aos1/Uba2 heterodimer; expression plasmids were provided by F. Melchior, Zentrum für Molekulare Biologie der Universität Heidelberg/Heidelberg University), PIASy, Ubc9, dnUbc9, and SUMO paralogues were expressed in BL21(DE3) or Rosetta(DE3) bacteria and purified as described previously.
Anti-TopoIIα/β monoclonal antibody was obtained from the Marine Biological Laboratory. Anti-SUMO2/3 polyclonal antibody was generated in guinea pigs (Azuma et al., 2003), and the polyclonal antibody against TopoIIα, C terminus (aa 1358–1579), was prepared in rabbits by injection with a recombinant His-T7 fused fragment, and was then affinity purified (Azuma et al., 2005). The anti-PIASy antibody used in this study has been described previously (Azuma et al., 2005).
XEEs, immunodepletion, and add-back assays
Sperm nuclei and low-speed extracts of X. laevis eggs arrested in metaphase by cytostatic factor (CSF) were prepared according to standard protocols (Kornbluth et al., 2001; Azuma, 2009). Immunodepletion was performed as described previously with protein A–conjugated magnetic beads (Dynal; Arnaoutov and Dasso, 2003). For add-back experiments, the purified recombinant TopoIIα proteins were added to the immunodepleted extracts at a final concentration of ∼300 nM, which is comparable to the concentration of endogenous TopoIIα measured by quantitative Western blotting. Chromosome isolation and analysis of SUMOylation were performed as described previously (Azuma, 2009). In brief, mitotic chromosomes were prepared by incubating sperm nuclei with CSF-XEE at the final concentration of ∼5,000 sperm nuclei/µl at room temperature. After observation of assembly of mitotic chromosome, which takes an ∼45–60-min incubation, chromosomal fractions were isolated from XEE by centrifugation through a 40% glycerol cushion at 10,000 g for 5 min at 4°C. The precipitated chromosomes were boiled in SDS-PAGE sample buffer, and the extracted proteins were resolved in 8–16% gradient SDS-PAGE gel followed by Western blotting analysis with the indicated antibodies.
Immunofluorescence
The mitotic chromosomes for immunofluorescence analysis were prepared as described previously (Arnaoutov and Dasso, 2003). In brief, CSF extracts were driven into interphase by 0.6 mM CaCl2. 500 sperm/µl was incubated with the interphase extracts for ∼60 min at 23°C to allow for complete DNA replication. Then, a volume of fresh CSF extract equal to half of the original volume was added to induce mitosis. For inhibition of mitotic SUMOylation, dnUbc9 was added just before induction of mitosis, at a final concentration of 150 ng/µl. After a 30-min incubation, the reactions were diluted threefold and fixed with PFA at a final concentration of 2%. The samples were spun onto coverslips through a 35% glycerol cushion, postfixed in 1.6% PFA, and analyzed by immunostaining using antibodies against TopoIIα (either mouse monoclonal or rabbit polyclonal), SUMO2/3 (guinea pig polyclonal), PIASy (rabbit polyclonal), and Aurora B (rabbit polyclonal; A. Arnaoutov and M. Dasso, National Institute of Child Health and Human Development/National Institutes of Health). Anti–rabbit Alexa Fluor 568, anti–guinea pig Alexa Fluor 684, and anti–mouse Alexa Fluor 488 were used as secondary antibodies. DNA was counterstained by Hoechst 33342 (EMD) and samples were mounted in Vectashield medium (Vector Laboratories). Specimens were observed using Volocity Imaging Software (PerkinElmer) on a microscope (TE2000-U; Nikon) with a Plan-Apochromat 100×/1.40 NA objective lens, and images were taken with a Retiga SRV charge-coupled device camera (QImaging) at room temperature. Photoshop and Illustrator software (Adobe) were used for processing the obtained images to intensities and sizes according to JCB policy.
Purification of SUMOylated TopoIIα from mitotic chromosomes
After mitotic chromosomes were assembled and isolated from CSF extracts, chromosomal proteins were subsequently extracted by boiling the chromosome fractions with SDS-PAGE sample buffer. Immunoprecipitation was performed on extracted denatured proteins as described previously (Kane et al., 2002) using affinity-purified anti-TopoIIα antibody that had been covalently cross-linked to protein A Dynabeads (Invitrogen) by dimethyl pimelimidate 2 HCl (Thermo Fisher Scientific). Isolated proteins were separated on 8–16% Tris-HCl gradient gels (Invitrogen) by SDS-PAGE, and visualized by silver staining (Owl kit; Daiichi). For liquid chromatography tandem mass spectrometry (LS-MS/MS) analysis, the isolated proteins were stained with CBB-R250.
In vitro SUMOylation assays
SUMOylation reactions were incubated at 25°C for 1 h unless otherwise indicated. SUMO2 was used in most of reactions except where noted. The reactions contained 15 nM E1, 5 µM SUMO2-GG, 500 nM T7-tagged TopoIIα, 2.5 mM ATP, and various concentrations of Ubc9 and PIASy as indicated in Fig. 5. Reaction buffers were composed of 20 mM Hepes, pH 7.8, 100 mM NaCl, 5 mM MgCl2, 0.05% Tween 20, 5% glycerol, 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), and 1 mM DTT. The reactions were stopped with half volumes of 3× SDS-PAGE sample buffer, and the samples were resolved on 8–16% Tris-HCl gradient gels by SDS-PAGE, then analyzed by Western blotting with HRP-conjugated anti-T7 monoclonal antibody (EMD).
In vitro SUMOylation–decatenation coupled assays
These assays were performed with 60 nM Ubc9 and 30 nM PIASy together with other protein components as described in “In vitro SUMOylation assays.” A control was performed under the same conditions except that SUMO2-G was used instead of SUMO2-GG. After a 1-h incubation at 25°C, the non-SUMOylated and SUMOylated samples were further incubated with 6.2 ng/µl of kDNA (TopoGEN, Inc.) in decatenation buffer at 25°C for the indicated time periods. Decatenation buffer consisted of 50 mM Tris-HCl, pH 8.0, 120 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 30 µg BSA/ml, and 2 mM ATP. The reactions were stopped by adding one third volume of 6× DNA dye (30% glycerol, 0.1% SDS, 10 mM EDTA, and 0.2 µg/µl bromophenol blue), and samples were loaded on a 1% agarose gel and electrophoresed at 100 V in TAE buffer (Tris-acetate-EDTA) until the samples reached the middle of the gel. The amount of kDNA remaining in the wells was measured using an Image Station 4000R (Kodak), and standard error was calculated using KaleidaGraph (Synergy Software).
Electrophoretic mobility shift assay
The assay was performed as described previously (Walker et al., 2004), with slight modifications. In brief, various amounts of TopoIIα WT and K660R (as indicated in Fig. 5 b) were incubated with 90 ng of pBS KS(+) DNA for 5 min at 25°C. In vitro SUMOylation reaction buffer was used as the reaction buffer in these assays to keep protein–DNA binding conditions similar to those used in in vitro SUMOylation reactions. The reactions were stopped by adding one half volume of loading buffer (50% glycerol, 10 mM EDTA, and 0.2% Bromophenol blue), and the samples were subjected to electrophoresis at 40 V for 2 h at 4°C in a 1% agarose gel containing ethidium bromide. The DNA was documented using an Image Station 4000R.
Online supplemental material
Fig. S1 shows colocalization of TopoIIα with Aurora B regardless of SUMOylation. Fig. S2 shows the isolated endogenous TopoIIα from mitotic chromosomes that were subjected to analysis of SUMOylation sites. Fig. S3 shows the result of LC-MS/MS analysis of SUMOylated TopoIIα, which indicates Lys 660 as the candidate SUMOylation site. Fig. S4 shows the purified recombinant TopoIIα proteins that were used in this study. Fig. S5 shows the decatenation assays with the recombinant TopoIIα proteins that were purified using a CBP tag and ZZ tag affinity column. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201004033/DC1.
Acknowledgments
We appreciate the gifts of anti-Aurora B antibody from A. Arnaoutov and M. Dasso, the X. laevis cDNA from T. Amano and Y. Shi, the CBP-ZZ TAP-tag plasmid from H. Yoon and K. Gould, and plasmids for E1 expression from F. Melchior. We thank K. L. Neufeld (University of Kansas), A. Arnaoutov, and M. Dasso for critical reading of this manuscript and for providing editorial assistance. We thank to Ms. L. J. Embree (National Cancer Institute) for editorial assistance.
M. Furuta was supported as a Japan Society for the Promotion of Science Research Fellow in Biomedical and Behavioral Research at the National Institutes of Health. This project was supported in part by a start-up grant from the Department of Molecular Biosciences at the University of Kansas and National Institutes of Health/National Center for Research Resources, Center For Cancer Experimental Therapeutics at the Centers of Biomedical Research Excellence (CCET-COBRE; P20 RR015563), and is currently supported by National Institutes of Health/National Institute of General Medical Sciences grants GM80278 and GM67945 (to S.P. Gygi).
Footnotes
Abbreviations used in this paper:
- CBP
- calmodulin-binding protein
- CSF
- cytostatic factor
- dnUbc9
- dominant-negative Ubc9
- kDNA
- kinetoplast DNA
- MEF
- mouse embryonic fibroblast
- PTM
- posttranslational modification
- SUMO
- small ubiquitin-like modifier
- TopoIIα
- DNA Topoisomerase IIα
- WT
- wild type
- XEE
- Xenopus egg extract
References
- Agostinho M., Santos V., Ferreira F., Costa R., Cardoso J., Pinheiro I., Rino J., Jaffray E., Hay R.T., Ferreira J. 2008. Conjugation of human topoisomerase 2 alpha with small ubiquitin-like modifiers 2/3 in response to topoisomerase inhibitors: cell cycle stage and chromosome domain specificity. Cancer Res. 68:2409–2418 10.1158/0008-5472.CAN-07-2092 [DOI] [PubMed] [Google Scholar]
- Andersen C.L., Wandall A., Kjeldsen E., Mielke C., Koch J. 2002. Active, but not inactive, human centromeres display topoisomerase II activity in vivo. Chromosome Res. 10:305–312 10.1023/A:1016571825025 [DOI] [PubMed] [Google Scholar]
- Arnaoutov A., Dasso M. 2003. The Ran GTPase regulates kinetochore function. Dev. Cell. 5:99–111 10.1016/S1534-5807(03)00194-1 [DOI] [PubMed] [Google Scholar]
- Azuma Y. 2009. Analysis of SUMOylation of topoisomerase IIalpha with Xenopus egg extracts. Methods Mol. Biol. 582:221–231 10.1007/978-1-60761-340-4_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Tan S.H., Cavenagh M.M., Ainsztein A.M., Saitoh H., Dasso M. 2001. Expression and regulation of the mammalian SUMO-1 E1 enzyme. FASEB J. 15:1825–1827 [DOI] [PubMed] [Google Scholar]
- Azuma Y., Arnaoutov A., Dasso M. 2003. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 163:477–487 10.1083/jcb.200304088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Arnaoutov A., Anan T., Dasso M. 2005. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 24:2172–2182 10.1038/sj.emboj.7600700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J., Alcasabas A., Blat Y., Kleckner N., Elledge S.J. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 9:1169–1182 10.1016/S1097-2765(02)00543-9 [DOI] [PubMed] [Google Scholar]
- Baldwin E.L., Osheroff N. 2005. Etoposide, topoisomerase II and cancer. Curr Med Chem Anticancer Agents. 5:363–372 10.2174/1568011054222364 [DOI] [PubMed] [Google Scholar]
- Berger J.M., Gamblin S.J., Harrison S.C., Wang J.C. 1996. Structure and mechanism of DNA topoisomerase II. Nature. 379:225–232 10.1038/379225a0 [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor V., Sampson D.A., Matunis M.J., Lima C.D. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 108:345–356 10.1016/S0092-8674(02)00630-X [DOI] [PubMed] [Google Scholar]
- Biggins S., Bhalla N., Chang A., Smith D.L., Murray A.W. 2001. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 159:453–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.J., Goulding S., Earnshaw W.C., Carmena M. 2003. RNAi analysis reveals an unexpected role for topoisomerase II in chromosome arm congression to a metaphase plate. J. Cell Sci. 116:4715–4726 10.1242/jcs.00797 [DOI] [PubMed] [Google Scholar]
- Dasso M. 2008. Emerging roles of the SUMO pathway in mitosis. Cell Div. 3:5 10.1186/1747-1028-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawlaty M.M., Malureanu L., Jeganathan K.B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J.M. 2008. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 133:103–115 10.1016/j.cell.2008.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Martínez L.A., Giménez-Abián J.F., Azuma Y., Guacci V., Giménez-Martín G., Lanier L.M., Clarke D.J. 2006. PIASgamma is required for faithful chromosome segregation in human cells. PLoS One. 1:e53 10.1371/journal.pone.0000053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Martínez L.A., Giménez-Abián J.F., Clarke D.J. 2008. Chromosome cohesion - rings, knots, orcs and fellowship. J. Cell Sci. 121:2107–2114 10.1242/jcs.029132 [DOI] [PubMed] [Google Scholar]
- Dong K.C., Berger J.M. 2007. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 450:1201–1205 10.1038/nature06396 [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8:947–956 10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- Gorbsky G.J. 1994. Cell cycle progression and chromosome segregation in mammalian cells cultured in the presence of the topoisomerase II inhibitors ICRF-187 [(+)-1,2-bis(3,5-dioxopiperazinyl-1-yl)propane; ADR-529] and ICRF-159 (Razoxane). Cancer Res. 54:1042–1048 [PubMed] [Google Scholar]
- Hari K.L., Cook K.R., Karpen G.H. 2001. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 15:1334–1348 10.1101/gad.877901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs R.J., Davies S.L., Sandri M.I., Redwood C., Wells N.J., Hickson I.D. 1998. Physiological regulation of eukaryotic topoisomerase II. Biochim. Biophys. Acta. 1400:121–137 [DOI] [PubMed] [Google Scholar]
- Ishida R., Sato M., Narita T., Utsumi K.R., Nishimoto T., Morita T., Nagata H., Andoh T. 1994. Inhibition of DNA topoisomerase II by ICRF-193 induces polyploidization by uncoupling chromosome dynamics from other cell cycle events. J. Cell Biol. 126:1341–1351 10.1083/jcb.126.6.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida R., Takashima R., Koujin T., Shibata M., Nozaki N., Seto M., Mori H., Haraguchi T., Hiraoka Y. 2001. Mitotic specific phosphorylation of serine-1212 in human DNA topoisomerase IIalpha. Cell Struct. Funct. 26:215–226 10.1247/csf.26.215 [DOI] [PubMed] [Google Scholar]
- Johnson E.S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355–382 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- Joseph J., Tan S.H., Karpova T.S., McNally J.G., Dasso M. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595–602 10.1083/jcb.200110109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S., Sano H., Liu S.C., Asara J.M., Lane W.S., Garner C.C., Lienhard G.E. 2002. A method to identify serine kinase substrates. Akt phosphorylates a novel adipocyte protein with a Rab GTPase-activating protein (GAP) domain. J. Biol. Chem. 277:22115–22118 10.1074/jbc.C200198200 [DOI] [PubMed] [Google Scholar]
- Kornbluth S., Yang J., Powers M. 2001. Analysis of the cell cycle using Xenopus egg extracts. In Current Protocols in Cell Biology Yamada K. M., editor John Wiley & Sons, Inc, New York, NY: 11.11.1–11.11.13 [DOI] [PubMed] [Google Scholar]
- Lee M.T., Bachant J. 2009. SUMO modification of DNA topoisomerase II: trying to get a CENse of it all. DNA Repair (Amst.). 8:557–568 10.1016/j.dnarep.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacerddine K., Lehembre F., Bhaumik M., Artus J., Cohen-Tannoudji M., Babinet C., Pandolfi P.P., Dejean A. 2005. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell. 9:769–779 10.1016/j.devcel.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Pichler A., Gast A., Seeler J.S., Dejean A., Melchior F. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 108:109–120 10.1016/S0092-8674(01)00633-X [DOI] [PubMed] [Google Scholar]
- Porter A.C., Farr C.J. 2004. Topoisomerase II: untangling its contribution at the centromere. Chromosome Res. 12:569–583 10.1023/B:CHRO.0000036608.91085.d1 [DOI] [PubMed] [Google Scholar]
- Roca J., Berger J.M., Harrison S.C., Wang J.C. 1996. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl. Acad. Sci. USA. 93:4057–4062 10.1073/pnas.93.9.4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Azuma Y. 2010. Rod/Zw10 complex is required for PIASy-dependent centromeric SUMOylation. J. Biol. Chem. 285:32576–32585 10.1074/jbc.M110.153817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Al-Ani G., Deckert K., Kirkpatrick D., Gygi S.P., Dasso M., Azuma Y. 2010. PIASy mediates SUMO-2/3 conjugation of poly(ADP-ribose) polymerase 1 (PARP1) on mitotic chromosomes. J. Biol. Chem. 285:14415–14423 10.1074/jbc.M109.074583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeffler A.J., Berger J.M. 2005. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 33:1465–1470 10.1042/BST20051465 [DOI] [PubMed] [Google Scholar]
- Schoeffler A.J., Berger J.M. 2008. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 41:41–101 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Yong-Gonzalez V., Kikuchi Y., Strunnikov A. 2006. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 172:783–794 10.1534/genetics.105.047167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina P.A., Côme M.G., Hudson J.R., Mo Y.Y., Beck W.T., Gorbsky G.J. 2002. Rapid exchange of mammalian topoisomerase II α at kinetochores and chromosome arms in mitosis. J. Cell Biol. 158:23–29 10.1083/jcb.200202053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.V., Nitiss K.C., Jensen L.H., Mayne C., Hu T., Jensen P.B., Sehested M., Hsieh T., Nitiss J.L. 2004. A mutation in human topoisomerase II alpha whose expression is lethal in DNA repair-deficient yeast cells. J. Biol. Chem. 279:25947–25954 10.1074/jbc.M312314200 [DOI] [PubMed] [Google Scholar]
- Wang L.H., Schwarzbraun T., Speicher M.R., Nigg E.A. 2008. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 117:123–135 10.1007/s00412-007-0131-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.H., Mayer B., Stemmann O., Nigg E.A. 2010. Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J. Cell Sci. 123:806–813 10.1242/jcs.058255 [DOI] [PubMed] [Google Scholar]
- Watts F.Z. 2007. The role of SUMO in chromosome segregation. Chromosoma. 116:15–20 10.1007/s00412-006-0079-z [DOI] [PubMed] [Google Scholar]
- Xue Y., Zhou F., Fu C., Xu Y., Yao X. 2006. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 34:W254–W257 10.1093/nar/gkl207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M. 2009. Clearing the way for mitosis: is cohesin a target? Nat. Rev. Mol. Cell Biol. 10:489–496 10.1038/nrm2712 [DOI] [PubMed] [Google Scholar]