A large-scale analysis of spindle disassembly in budding yeast identifies factors required for disengagement of spindle halves, arrest of spindle elongation, and depolymerization of interpolar microtubules.
Abstract
The mitotic spindle is a complex and dynamic structure. Although much has been learned about how spindles assemble and mediate chromosome segregation, how spindles rapidly and irreversibly disassemble during telophase is less clear. We used synthetic lethal screens in budding yeast to identify mutants defective in spindle disassembly. Real-time, live cell imaging analysis of spindle disassembly was performed on nine mutants defective in this process. Results of this analysis suggest that spindle disassembly is achieved by mechanistically distinct but functionally overlapping subprocesses: disengagement of the spindle halves, arrest of spindle elongation, and initiation of interpolar microtubule depolymerization. These subprocesses are largely governed by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. Combinatorial inhibition of these subprocesses yielded cells with hyperstable spindle remnants and dramatic defects in cell cycle progression, establishing that rapid spindle disassembly is crucial for cell proliferation.
Introduction
The mitotic spindle comprises three sets of microtubules (MTs) that emanate from MT-organizing centers: astral MTs, which radiate toward the cell cortex, kinetochore MTs, which attach to chromosomes, and interpolar MTs (ipMTs), which interdigitate to form the central spindle bridge (for review see Glotzer, 2009). The size and integrity of the mitotic spindle are maintained by a vast array of MT-binding proteins that regulate MT dynamics (e.g., XMAP215, CLASP, and EB1) or cross-link the antiparallel ipMTs (e.g., Prc1 and BimC motors). During each cell cycle, the mitotic spindle is efficiently assembled to achieve DNA segregation and then disassembled.
Live cell fluorescence microscopy of budding yeast expressing the spindle marker GFP-Tub1 showed that spindle disassembly is characterized by depolymerization of the ipMTs from their plus ends and separation of the spindle halves. Once the spindle halves have separated, the ipMTs are no longer detectable after ∼2 min (Maddox et al., 2000). Thus, disassembly of the mitotic spindle is irreversible and swift. Moreover, these events are precisely coordinated during the cell cycle and occur only after chromosomes have cleared the plane of division but before cytokinesis (VerPlank and Li, 2005). This precise timing is governed in part by the mitotic exit network (MEN), which ultimately stimulates activity of Cdc14, a phosphatase that opposes Cdk1. Dephosphorylation of Cdk1 targets is critical for mitotic exit, but the connection between this process and spindle disassembly is poorly understood (for review see Sullivan and Morgan, 2007). Therefore, identifying the downstream targets of the MEN and their contributions to spindle midzone dissolution and spindle MT depolymerization are crucial for understanding spindle disassembly.
Other than MEN components, few proteins have been implicated in mediating spindle disassembly. One factor is the anaphase-promoting complex (APC), an E3 ubiquitin ligase critical for mitotic progression (for review see Peters, 2002). During mitotic exit, the APC, in association with the cofactor Cdh1 (APCCdh1), degrades an array of spindle-stabilizing proteins, including the BimC homologue Cin8 (Hildebrandt and Hoyt, 2001), the Prc1 homologue Ase1 (Juang et al., 1997), and Fin1 (Woodbury and Morgan, 2007). Other factors implicated in spindle disassembly include Kip3, a plus end–targeted MT depolymerase of the kinesin-8 family, and the Ipl1 kinase, the Aurora B homologue in yeast (Buvelot et al., 2003). Because Kip3 actively depolymerizes MTs in vitro and also regulates astral MT length in vivo, it has been speculated that Kip3 may accelerate ipMT shrinkage to help disassemble the spindle (Gupta et al., 2006; Varga et al., 2006). Ipl1 phosphorylates the MT-stabilizing protein Bim1 (yeast EB1) and promotes its dissociation from MTs in vitro, suggesting a possible mechanism by which Ipl1 mediates spindle disassembly in vivo (Zimniak et al., 2009). Bim1 is most likely not the sole target of Ipl1 during mitotic exit, considering that Ipl1 phosphorylates many kinetochore, spindle, and chromatin-associated proteins (e.g., the Dam1 and Ndc80 complexes, Ase1, and histone H3; Hsu et al., 2000; Cheeseman et al., 2002; Kotwaliwale et al., 2007). Therefore, to more clearly define the role of Ipl1 during spindle disassembly, it is crucial to test whether Bim1 phosphorylation drives spindle disassembly in vivo and to investigate the involvement of additional Ipl1 targets. Another unexplored issue is the relationship between Ipl1, Kip3, and the APC during spindle disassembly. Whether these proteins operate together in a single pathway or independently in multiple pathways and how their activities are coordinated are unclear.
We conducted a large-scale analysis of spindle disassembly in budding yeast. In addition to establishing a genetic method for identifying novel proteins important for spindle disassembly, we performed real-time analysis of spindle disassembly and assessed the effects of mutations in nine proteins critical for this process. Using our methods, we described distinct phenotypes that distinguish these mutants and inferred the normal contributions of these proteins to spindle disassembly.
Results
The APC and Kip3 cooperate in mitotic spindle disassembly via distinct mechanisms
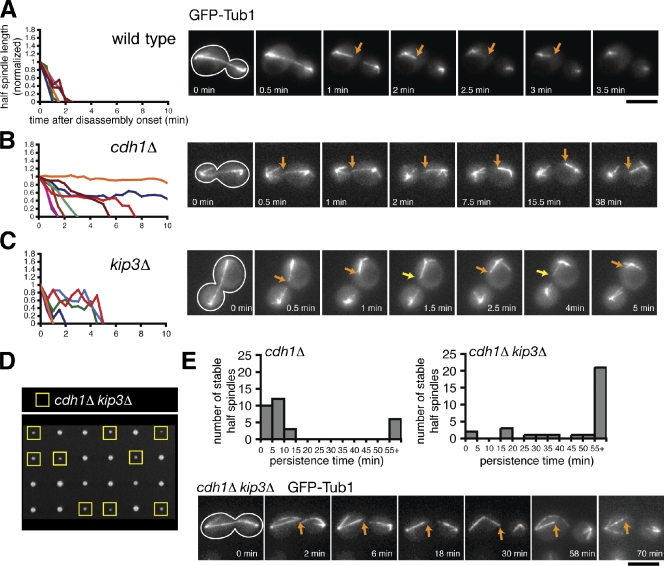
The enzymatic activities of the APCcdh1 and Kip3 are dramatically different, suggesting that they play distinct roles in spindle disassembly. To test this idea, we followed mitotic spindle elongation and disassembly using time-lapse fluorescence microscopy in cells expressing the MT marker GFP-Tub1. We found that depletion of Cdh1 or Kip3 had different effects on spindle morphology. Late anaphase spindles in cdh1Δ or kip3Δ cells were hyperextended relative to spindles in wild-type cells, which is indicative of a delay in spindle disassembly. However, spindles were consistently longer and more often fishhook shaped in cdh1Δ mutants relative to kip3Δ mutants (70 vs. 15% fishhook spindles; maximum spindle length = 7.59 ± 1.6 µm vs. 6.78 ± 1.0 µm [mean ± SD], respectively; n = 20; Fig. S1 A).
To analyze spindle disassembly in greater depth, we followed ipMT depolymerization after the spindle halves separated in late anaphase. In wild-type cells, ipMTs quickly depolymerized (shrinkage rate: R(hs) = 0.069 ± 0.006 µm × s−1; mean ± SEM) and rarely displayed recovery events (F(rec) = 0.0050 s−1; Fig. 1 A and Table I). In contrast, cdh1Δ cells displayed hyperstable spindle halves that changed very little in length after separation (R(hs) = 0.013 ± 0.004 µm × s−1; Fig. 1 B). Persistence time of these hyperstable spindle halves fits a bimodal distribution: ∼80% disassembled in <15 min, whereas ∼20% persisted >55 min (n = 30; Fig. 1 E). Intriguingly, in every case, only one spindle-half per cell remained hyperstable; the other spindle-half always depolymerized with only slightly slower kinetics relative to wild type (R(hs) = 0.051 ± 0.007 µm × s−1; F(rec) = 0.0054 s−1; Fig. 1 B, Table I, and Fig. S1 C). These data imply that APCCdh1 enhances ipMT depolymerization, likely through removal and degradation of midzone-stabilizing proteins. In support of this hypothesis, the midzone proteins Ase1-4GFP and Cin8-4GFP continued to decorate the mitotic spindle even after spindle breakage in cdh1Δ cells but not in wild-type cells (Fig. S1 B). In cdh1Δ cells, the spindle broke apart in a region outside of the midzone, leaving only one of the spindle halves decorated with midzone-localized Ase1-4GFP or Cin8-4GFP. Consistently, the decorated spindle halves were hyperstable (unpublished data). These results strongly suggest that the hyperstable half-spindle structures seen in cdh1Δ cells result from failure to remove stabilizing proteins like Ase1 and Cin8 from the spindle midzone and degrade them.
Figure 1.
The combined activities of APCCdh1 and Kip3 are required to depolymerize ipMTs completely. (A–C) Time-lapse fluorescence images of wild-type, cdh1Δ, and kip3Δ cells expressing GFP-Tub1 during mitotic exit. Cell shape is outlined in white. Orange arrows track shrinking spindle halves, whereas the yellow arrows indicate a growing spindle-half. (left) Graphs represent the normalized lengths of depolymerizing spindle halves after the spindle had broken. Each line represents an individual spindle-half that was selected at random. For the cdh1Δ mutant, the shrinkage of four nonhyperstable and four hyperstable spindle halves is charted. Bar, 5 µm. (D) Tetrad dissection of spores resulting from a cross between kip3Δ and cdh1Δ mutants. Offspring possessing both mutations are indicated with yellow boxes. (E) Histograms representing the persistence times of hyperstable spindle halves observed after spindle breakage in cdh1Δ and cdh1Δ kip3Δ mutants (n = 30 for both). (bottom) A series of fluorescence images depicting a cdh1Δ kip3Δ cell after spindle breakage in which the hyperstable half-spindle persisted for at least 70 min.
Table I.
Depolymerization kinetics of spindle halves after onset of spindle disassembly
| Genotype | Recovery frequency | Shrinkage rate | n | P-value |
| F(rec) (s−1) | R(hs) (µm × s−1) | |||
| 23°C | ||||
| Wild type | 0.0050 | 0.069 ± 0.006 | 42 | NA |
| cdh1Δ nonhyperstable | 0.0054 | 0.051 ± 0.007 | 21 | 0.085 |
| cdh1Δ hyperstable | 0.0046 | 0.013 ± 0.004 | 16 | <0.001 |
| doc1Δ nonhyperstable | 0.0064 | 0.044 ± 0.006 | 25 | 0.013 |
| doc1Δ hyperstable | 0.0016 | 0.004 ± 0.002 | 16 | <0.001 |
| dbf2Δ nonhyperstable | 0.0079 | 0.029 ± 0.004 | 19 | <0.001 |
| dbf2Δ hyperstable | 0.002 | 0.012 ± 0.002 | 16 | <0.001 |
| kip3Δ | 0.0102 | 0.044 ± 0.004 | 39 | 0.002 |
| dcc1Δ | 0.0078 | 0.043 ± 0.004 | 40 | 0.003 |
| ctf8Δ | 0.0082 | 0.041 ± 0.005 | 39 | 0.008 |
| she1Δ | 0.0053 | 0.048 ± 0.004 | 45 | 0.008 |
| mcm21Δ | 0.0059 | 0.060 ± 0.005 | 42 | 0.31 |
| 37°C | ||||
| Wild type | 0.0028 | 0.090 ± 0.007 | 40 | NA |
| ipl1-321 | 0.0045 | 0.052 ± 0.004 | 36 | <0.001 |
| kip3Δ | 0.0125 | 0.061 ± 0.009 | 19 | 0.014 |
| ipl1-321 kip3Δ | 0.0076 | 0.042 ± 0.010 | 17 | <0.001 |
NA, not applicable. Cells with indicated the genotypes expressing GFP-Tub1 were imaged every 10 s, and the lengths of the spindle halves were measured at each time point. A recovery event was scored each time a spindle-half switched from shrinkage to growth. The shrinkage rate (R(hs)) represents the slope of the half-spindle lengths when plotted versus time (mean ± SEM). P-values were calculated using a model I analysis of variance test. Half-spindle shrinkage rates for each mutant were directly compared with the wild-type rates.
In contrast, spindle halves in kip3Δ cells alternated between shrinkage and growth, suggesting that Kip3 is necessary for sustained ipMT depolymerization during spindle disassembly (Fig. 1 C). Consistently, the frequency of spindle-half recovery was twofold higher in kip3Δ cells (F(rec) = 0.0102 s−1) relative to wild-type or cdh1Δ cells (Table I). We never observed hyperstable spindle halves in kip3Δ mutant cells (n = 39). These differences in spindle morphology and dynamics suggest that Kip3 and APCCdh1 play different roles during spindle disassembly.
If APCCdh1 and Kip3 operate via different mechanisms, we would expect combinatorial depletion of Cdh1 and Kip3 to produce synergistic defects in spindle disassembly and possibly cell proliferation. Indeed, tetrad analysis revealed a synthetic genetic interaction between kip3Δ and cdh1Δ null alleles (Fig. 1 D). Furthermore, spindles in cdh1Δ kip3Δ double mutants were longer (maximum spindle length = 9.00 ± 1.7 µm; n = 20) and more often fishhook shaped (95%) than in either single mutant (Fig. S1 A). Addition of the kip3Δ mutation also prolonged the persistence time of the hyperstable half-spindles seen in cdh1Δ cells. In cdh1Δ kip3Δ cells, only ∼6% of stable spindles halves disassembled in under 15 min, whereas ∼70% lasted >55 min (Fig. 1 E). These results indicate that Kip3 is necessary to disassemble the persistent half-spindle structures left when APCCdh1 activity is compromised.
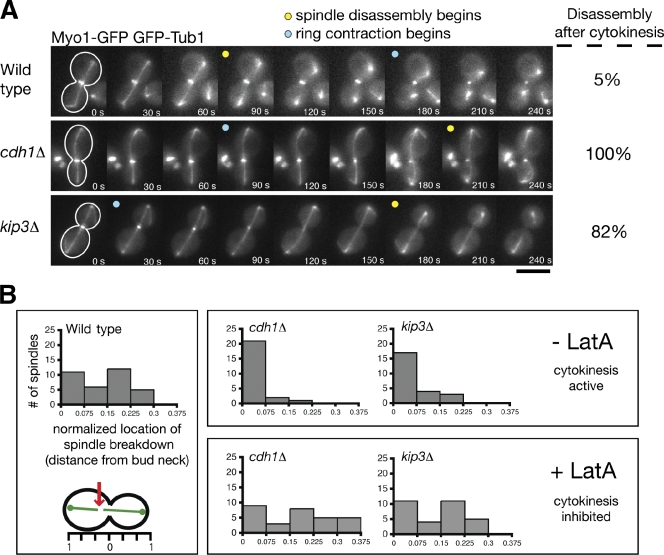
We were surprised that spindles broke apart in cdh1Δ kip3Δ double mutants because inactivation of the MEN, which is upstream of spindle disassembly, arrests cells in late anaphase with intact spindles (Visintin and Amon, 2001). Because the MEN initiates cytokinesis in addition to spindle disassembly (Luca et al., 2001), we considered the possibility that actinomyosin ring contraction might shear the mitotic spindle when spindle disassembly is inhibited. To test this hypothesis, we simultaneously visualized spindles and cytokinetic rings in cells expressing GFP-Tub1 and the actinomyosin ring marker Myo1-GFP. In 90 ± 5% (mean ± SD; n = 3 experiments; 20 events/experiment) of wild-type cells, the mitotic spindle disassembled before cytokinetic ring contraction, indicating that cytokinesis is not required for spindle disassembly (Fig. 2 A and Video 1; VerPlank and Li, 2005). However, in 100% of cdh1Δ cells and 82 ± 10.4% of kip3Δ cells (n = 3 experiments), the spindle disassembled only after the cytokinetic ring contracted (Fig. 2 A and Videos 2 and 3). In addition, we noticed that the initial location of spindle-half separation during spindle disassembly was widely distributed within wild-type cells, suggesting a lack of connection between cytokinetic ring contraction and spindle breakdown (Fig. 2 B). However, spindle breakage occurred exclusively at the bud neck in kip3Δ and cdh1Δ mutants. When cdh1Δ and kip3Δ cells were treated with 250 µM latrunculin A, a potent inhibitor of actinomyosin ring contraction (Ayscough et al., 1997), spindle breakage was no longer restricted to the bud neck region, which is similar to wild-type cells (Fig. 2 B). These results show that in cells lacking Kip3 or APCCdh1, the spindle breaks as a result of cytokinetic ring contraction. Thus, it is likely that cytokinesis is not ordinarily required for spindle disassembly but can break the spindle when disassembly is delayed. Even after the spindle breaks apart in this cytokinesis-dependent manner, Kip3 and APCCdh1 activities are still required to fully disassemble the mitotic spindle halves.
Figure 2.
Cytokinetic ring contraction breaks the mitotic spindle when normal disassembly mechanisms are impaired. (A) Time-lapse images of cells expressing GFP-Tub1 and the cytokinetic ring marker Myo1-GFP during mitotic exit. Yellow dots indicate when the spindle breaks, whereas blue dots indicate when the cytokinetic ring has begun contracting. The frequency of spindle disassembly events that occurred after ring contraction is indicated to the right of each image series (n = 3 separate experiments each analyzing 20 disassembly events for each strain; mean ± SD). Note that cytokinetic ring contraction appears to break the spindle in kip3Δ and cdh1Δ cells but not in wild-type cells. Bar, 5 µm. (B) Cells were monitored as in Fig. 1 A, and the location of breakdown relative to the bud neck was recorded and normalized with the length of the cell (n = 34, 24, and 24 for wild-type, cdh1Δ, and kip3Δ strains, respectively). All numbers represent absolute values. kip3Δ and cdh1Δ cells were treated with 250 µM latrunculin A to inhibit actinomyosin ring contraction (n = 30 for each strain).
Synthetic lethal screens identify novel spindle disassembly factors
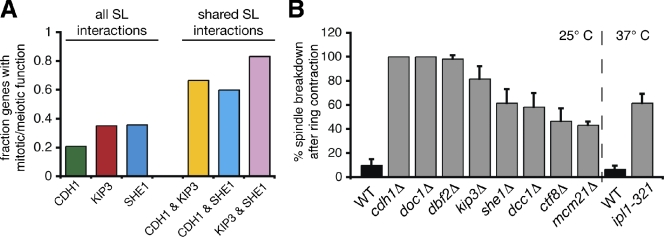
Because kip3Δ and cdh1Δ null alleles showed a synthetic genetic interaction, we hypothesized that other mutants displaying synthetic genetic interactions with these alleles might also be defective in spindle disassembly. Therefore, we searched published synthetic interactions for KIP3 and CDH1 (Costanzo et al., 2010). Among the interesting genes identified was SHE1, which encodes a spindle and bud neck–localized protein with no previously known role in spindle disassembly (Wong et al., 2007; Woodruff et al., 2009). Spindles in cells lacking She1 frequently disassembled after cytokinetic ring contraction (61.7 ± 11.5%; n = 3 experiments; Fig. 3 B), which is similar to the phenotype seen in kip3Δ and cdh1Δ mutants. These results demonstrate that She1 is important for spindle disassembly and validate use of synthetic lethal profiles to identify novel disassembly factors.
Figure 3.
Synthetic lethal screens identify additional genes important for spindle disassembly. (A) Fraction of the genes found in the CDH1, KIP3, and SHE1 synthetic interaction profiles that have previously described roles in mitosis or meiosis. SL, synthetic lethal. (B) Frequency in which spindles disassembled after initiation of cytokinetic ring contraction in the indicated strains (n = 3 experiments each analyzing 20 disassembly events for each strain; mean ± SD). Analysis was performed similar to the experiments described in Fig. 2 A. Analysis of ipl1-321 mutants was performed at 37°C and compared with wild-type (WT) cells cultured at the same temperature.
We found that analysis of genetic interactions shared by CDH1, KIP3, and SHE1 provided an approximately two- to threefold enrichment for genes encoding proteins with previously described mitotic or meiotic functions (Fig. 3 A and Fig. S2). Importantly, ∼44% of these proteins had been previously implicated in spindle disassembly (Fig. S2; Juang et al., 1997; Vizeacoumar et al., 2010). Therefore, genes showing synthetic interactions with more than one of the genes CDH1, KIP3, and SHE1 are strong candidates for novel spindle disassembly factors. For the remainder of our study, we largely focused on such genes, listed in Fig. S2.
In addition to further analyzing the effects of mutations in CDH1, KIP3, and SHE1, we also characterized the effects of mutations in six additional genes: DOC1, DBF2, IPL1, MCM21, DCC1, and CTF8. Mutant alleles of all nine genes caused delays in spindle disassembly (Fig. 3 B). Doc1 is a subunit of the APC (Hwang and Murray, 1997), and Dbf2 is a downstream component of the MEN that indirectly activates APCCdh1 (Visintin et al., 1998). Ipl1 is the essential Aurora B kinase in yeast implicated previously in spindle disassembly (Buvelot et al., 2003), and Mcm21 is a component of the COMA kinetochore complex shown to affect Ipl1 localization (Vizeacoumar et al., 2010). Finally, Dcc1 and Ctf8 are components of the seven-protein alternative replication factor C (A-RFC) complex that regulates sister chromatid cohesion (Mayer et al., 2001). Like She1, Dcc1 and Ctf8 have not previously been linked to spindle disassembly.
Real-time analysis reveals distinct roles for spindle disassembly factors in regulating ipMT depolymerization
Our results up to this point suggest that multiple pathways orchestrate spindle disassembly, and they identify a diverse set of proteins governing this process. As a first step toward elucidating the mechanisms underlying spindle disassembly and the complexity of the pathways controlling this process, we analyzed the effects of depleting Doc1, Dbf2, She1, Ctf8, Dcc1, Mcm21, and Ipl1 on ipMT depolymerization. We used time-lapse fluorescence microscopy to follow shrinking ipMT plus ends after disassembly onset, which is similar to our experiments in Fig. 1 (A–C).
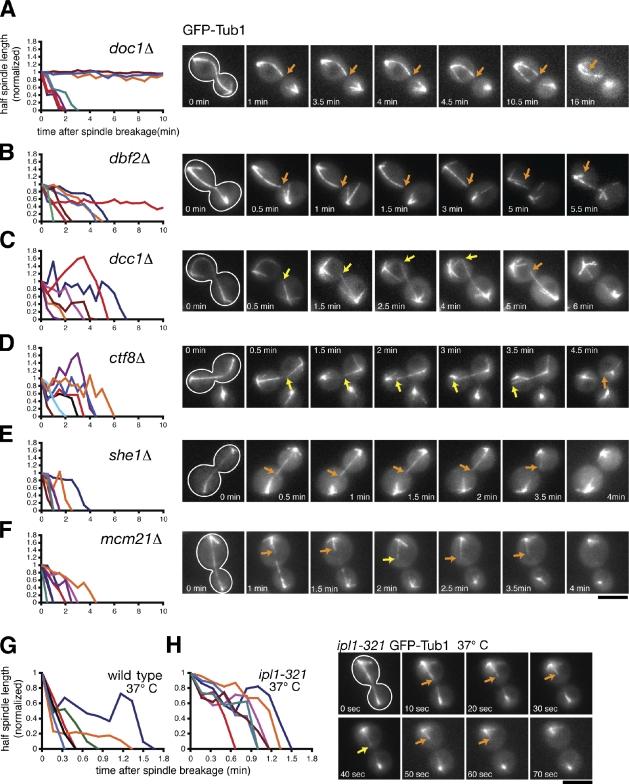
We observed hyperstable half-spindles in doc1Δ and dbf2Δ cells (R(hs) = 0.004 ± 0.002 µm × s−1 and 0.012 ± 0.002 µm × s−1, respectively) similar to the phenotype we described for cdh1Δ cells (Fig. 4, A and B; and Table I). This result is consistent with the fact that all three mutations compromise APCCdh1 activity. Surprisingly, nonhyperstable spindle halves disassembled more slowly and underwent more recoveries in dbf2Δ mutants (R(hs) = 0.029 ± 0.004 µm × s−1; F(rec) = 0.0079 s−1) than in cdh1Δ mutants (R(hs) = 0.051 ± 0.007 µm × s−1; F(rec) = 0.0054 s−1) or doc1Δ mutants (R(hs) = 0.044 ± 0.006 µm × s−1; F(rec) = 0.0064 s−1; Table I). This result suggests that Dbf2 might play a role in spindle disassembly in addition to activating APCCdh1.
Figure 4.
Dynamics of ipMT depolymerization in spindle disassembly mutants. (A–F) Cells expressing the spindle marker GFP-Tub1 were analyzed by time-lapse microscopy during mitotic exit similar to the experiments described in Fig. 1 (A–C). Cells were incubated at 23°C. The cell periphery is outlined in white. Orange arrows track shrinking spindle halves, whereas yellow arrows track growing spindle halves. (left) Line graphs represent the normalized lengths of individual depolymerizing spindle halves after the spindle had broken. (G and H) Comparison of spindle disassembly in wild-type and ipl1-321 cells incubated at 37°C. Bars, 5 µm.
We did not observe hyperstable half-spindles in dcc1Δ, ctf8Δ, she1Δ, mcm21Δ, or ipl1-321 cells (n > 40 breakdown events/strain; Fig. 4, C–H; unpublished data). Instead, dcc1Δ and ctf8Δ cells displayed spindle halves that alternated between periods of shrinkage and sustained growth, which is similar to the phenotype we observed in kip3Δ cells (Fig. 1 C). In fact, half-spindle shrinkage rates and recovery frequencies were elevated 1.5–2-fold in kip3Δ, dcc1Δ, and ctf8Δ cells (Table I). Strikingly, spindle halves in dcc1Δ and ctf8Δ mutants occasionally grew beyond their initial length, sometimes reaching the opposing cell cortex (Fig. 4, C and D). These results suggest that the conserved A-RFC complex is required to inhibit ipMT growth at the end of anaphase.
Interestingly, depletion of She1 slowed half-spindle shrinkage rates (R(hs) = 0.048 ± 0.004 µm × s−1) but did not dramatically alter half-spindle recovery frequencies (F(rec) = 0.0053 s−1; Fig. 4 E and Table I). However, depletion of Mcm21 dampened half-spindle shrinkage rates and elevated half-spindle recovery frequencies only modestly compared with wild-type cells (R(hs) = 0.060 ± 0.005 µm × s−1; F(rec) = 0.0059 s−1; Fig. 4 F and Table I), indicating that Mcm21 plays a minimal role in regulating ipMT dynamics during spindle disassembly.
Finally, we analyzed the effects of inactivating Ipl1 on ipMT depolymerization by incubating ipl1-321 temperature-sensitive cells at the nonpermissive temperature (37°C) for 2 h and performing live cell fluorescence microscopy. On average, spindle halves shrank more slowly and underwent more recoveries in ipl1-321 cells than in wild-type cells incubated at 37°C (ipl1-321: R(hs) = 0.052 ± 0.004 µm × s−1; F(rec) = 0.0045 s−1; wild type: R(hs) = 0.090 ± 0.007 µm × s−1; F(rec) = 0.0028 s−1; Fig. 4, G and H; and Table I). These results suggest that Ipl1 mediates spindle disassembly by enhancing ipMT depolymerization and preventing ipMT recovery.
The aforementioned data indicate that regulating ipMT depolymerization and recovery is crucial for efficient spindle disassembly. Given the range of distinct phenotypes we observed and, most importantly, that mutations in proteins operating in the same pathway produced similar phenotypes, we conclude that real-time imaging of spindle-half depolymerization is a powerful method for categorizing spindle disassembly factors.
Ipl1 coordinates inhibition of the MT growth factor Bim1 and activation of the spindle-destabilizing factor She1
Sustained ipMT depolymerization at the end of mitosis could occur either by enhancing MT shrinkage, inhibiting MT growth, or both. It is likely that Kip3, an ATP-dependent MT depolymerase, acts by the former mechanism. In contrast, our results suggest that APCCdh1 accelerates depolymerization of ipMTs by removing and degrading spindle-stabilizing proteins like Ase1 and Cin8. Additional activities may be required, considering that other proteins that are not APCCdh1 substrates (e.g., Bim1, Stu1, and Stu2) contribute to spindle stability and length (Severin et al., 2001; Yin et al., 2002; Gardner et al., 2008). It is possible that removal of these proteins from the midzone could suppress MT growth to encourage spindle disassembly. This hypothesis is supported by previous work showing that Ipl1 phosphorylation regulates Bim1 association with MTs in vitro (Zimniak et al., 2009). Thus, we decided to test whether Ipl1-mediated removal of Bim1 from spindle MTs facilitates spindle disassembly in vivo.
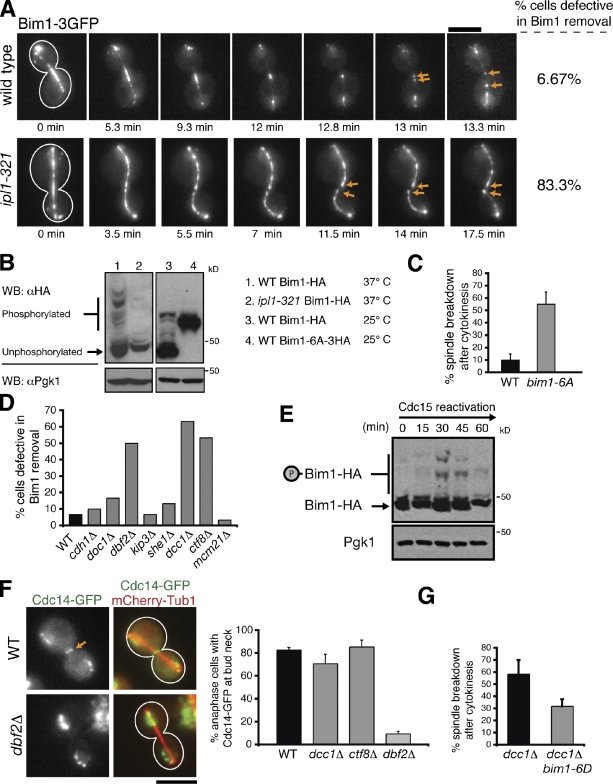
Consistent with previous findings (Zimniak et al., 2009), we observed Bim1-3GFP localization at the spindle midzone that decreased dramatically as cells progressed through anaphase (Fig. 5 A), suggesting that Bim1 is unloaded from spindle MTs during spindle disassembly. This localization pattern was observed in 28/30 wild-type cells. However, in 25/30 ipl1-321 cells, Bim1-3GFP continued to decorate the entire length of the spindle and did not concentrate to the midzone during mitotic exit, indicative of a defect in Bim1 removal (Fig. 5 A). We observed a dramatic decrease in Bim1-HA phosphorylation when either Ipl1 was inactivated or when serine/threonine to alanine mutations were made within the six Ipl1 phosphorylation sites in Bim1 (Bim1-6A-3HA; Fig. 5 B; Zimniak et al., 2009). Strikingly, cells expressing Bim1-6A displayed delays in spindle disassembly reminiscent of the phenotype seen in ipl1-321 mutant cells (Fig. 5 C). These results strongly suggest that Ipl1 phosphorylation and consequent dissociation of Bim1 from ipMTs are required for efficient spindle disassembly.
Figure 5.
The MEN and the A-RFC complex regulate Ipl1-mediated removal of Bim1 from the spindle midzone. (A) Time-lapse fluorescence images of Bim1-3GFP localization in wild-type and ipl1-321 cells incubated at 37°C. Arrows indicate Bim1-3GFP decoration of the plus ends of spindle halves after disassembly onset. (B) Protein extracts isolated from asynchronous populations of the indicated strains were separated by SDS-PAGE supplemented with 30 µM Phos-tag acrylamide. The tagged phosphomutant version of Bim1 (Bim1-6A-3HA) contained a larger C-terminal epitope and a longer linker than Bim1-HA; thus, Bim1-6A-3HA migrates slower than Bim1-HA. The presence of a single Bim1-6A-3HA band indicates the absence of phosphorylation. Pgk1 was used as a loading control. WB, Western blotting. (C) Frequency in which spindles disassembled after initiation of cytokinetic ring contraction in the indicated cells. (D) Bim1-3GFP localization just before spindle disassembly in the indicated strains (n = 30 for each). Only dbf2Δ, ctf8Δ, and dcc1Δ mutants displayed pronounced defects in Bim1 removal. (E) cdc15-2 cells expressing Bim1-HA were synchronized in late anaphase after a 3-h incubation at 37°C. These cells were then released by lowering the temperature to 23°C, and cells were harvested every 15 min. Protein extracts were separated by SDS-PAGE supplemented with 30 µM Phos-tag acrylamide. Pgk1 was used as a loading control. (F) In wild-type (WT), dcc1Δ, and ctf8Δ cells, Cdc14-GFP was exported from the nucleus and localized to the bud neck (arrow) during late anaphase. However, Cdc14-GFP export was defective in dbf2Δ cells (n = 75). (G) Expression of bim1-6D alleviates the spindle disassembly defects in dcc1Δ cells. (C and G) n = 3 experiments, with 20 events per experiment. Error bars indicate mean ± SD. Bars, 5 µm.
Next, we analyzed the localization of Bim1 in other spindle disassembly mutants. We reasoned that mutations in any upstream activators of Ipl1 should also prevent Bim1 removal from the midzone. Bim1-3GFP removal from the spindle midzone occurred normally in cdh1Δ, doc1Δ, kip3Δ, she1Δ, and mcm21Δ mutants but not in dbf2Δ, dcc1Δ, and ctf8Δ mutants (Fig. 5 D). These results suggest that Dbf2 and the A-RFC complex (Dcc1, Ctf8, and others) regulate Ipl1 phosphorylation of Bim1 and that APCCdh1, Kip3, She1, and Mcm21 do not operate upstream of Ipl1. The persistence of Bim1 on ipMTs could explain why spindle halves recover more frequently in ipl1-321, dbf2Δ, dcc1Δ, and ctf8Δ mutants than in wild-type cells (Table I).
That ipl1-321 and dbf2Δ mutants share a similar phenotype indicates that Ipl1 phosphorylation of Bim1 occurs downstream of the MEN. Indeed, the timing of Bim1 phosphorylation is consistent with this conclusion. We synchronized cdc15-2 cells in late anaphase and observed phosphorylated forms of Bim1-HA only after Cdc15 was reactivated (Fig. 5 E). We then asked whether the A-RFC complex indirectly activates Ipl1 by stimulating MEN activity. However, during late anaphase, although nuclear export of Cdc14-GFP was inhibited in dbf2Δ cells (Vizeacoumar et al., 2010), Cdc14-GFP relocalization was normal in dcc1Δ and ctf8Δ mutants (Fig. 5 F), suggesting that the A-RFC complex does not regulate the MEN. This result raises the possibility that the A-RFC complex might operate in parallel to the MEN and Ipl1 to mediate Bim1 removal. However, expression of Bim1-6D, which mimics Bim1 when fully phosphorylated by Ipl1 (Zimniak et al., 2009), partially rescued the spindle disassembly defects seen in dcc1Δ mutants (Fig. 5 G). Thus, the A-RFC complex might represent a novel cofactor that directly regulates the cell cycle activity and/or substrate specificity of Ipl1 while operating either downstream of or in parallel to the MEN.
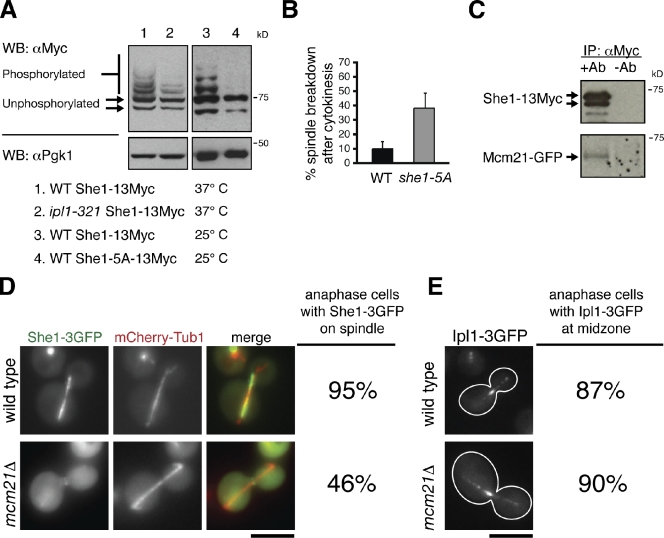
It is possible that Bim1 is not the sole target of Ipl1 during mitotic exit. Likely, Ipl1 not only deactivates spindle-stabilizing factors, like Bim1, but activates spindle-destabilizing factors as well. She1 interacts with the Ipl1 complex component Sli15 (yeast INCENP homologue) in yeast two-hybrid assays, making it an excellent candidate for an Ipl1 target (Wong et al., 2007). Furthermore, She1 likely destabilizes the mitotic spindle, considering that mitotic spindles often rely on cytokinesis to initiate disassembly when She1 is mutated (Fig. 3 B). She1-13Myc migrated as multiple bands on Phos-tag poly-acrylamide gels (Fig. 6 A). All but the two fastest migrating bands disappeared upon phosphatase treatment; thus, the slower migrating bands are phosphorylated species (Fig. S3 A). We observed a dramatic decrease in She1-13Myc phosphorylation when either Ipl1 was inactivated or when serine/threonine to alanine mutations were made in the five Ipl1 consensus sites in She1 (She1-5A-13Myc; Fig. 6 A). Finally, spindle disassembly was delayed relative to cytokinetic ring contraction in cells expressing She1-5A (Fig. 6 B). These findings suggest that Ipl1 mediates spindle disassembly by directly phosphorylating She1.
Figure 6.
Ipl1 and Mcm21 mediate spindle disassembly by regulating She1 activity. (A) Protein extracts isolated from asynchronous populations of the indicated strains were separated by SDS-PAGE supplemented with 20 µM Phos-tag acrylamide. Detection of Pgk1 was used as a loading control. WB, Western blotting. (B) Frequency in which spindles disassembled after initiation of cytokinetic ring contraction in the indicated strains (n = 3 experiments each analyzing 20 disassembly events for each strain; mean ± SD). (A and B) WT, wild type. (C) Protein lysate from cells expressing She1-13Myc and Mcm21-GFP was passed over a column conjugated either to anti-Myc antibody (+Ab) or nothing (−Ab). Mcm21-GFP copurified with She1-13Myc only when anti-Myc antibody was conjugated to the column. IP, immunoprecipitation. (D) She1-3GFP localization in wild-type and mcm21Δ cells expressing the spindle marker mCherry-Tub1 during late anaphase (n = 80 for both strains). (E) Ipl1-3GFP localization in wild-type and mcm21Δ cells during late anaphase (n = 40 for both strains). Bars, 5 µm.
Because she1Δ and mcm21Δ mutants share similar phenotypes (delays in spindle disassembly without any substantial defects in half-spindle recovery frequencies, midzone protein degradation, or Bim1 localization), She1 and Mcm21 might operate in the same pathway. In support of this hypothesis, both proteins localize to the kinetochore (Fig. S3, B and C; Vizeacoumar et al., 2010), and Mcm21-GFP copurified with She1-13Myc isolated from wild-type yeast extracts (Fig. 6 C). Depletion of Mcm21 reduced She1-3GFP accumulation on the spindle (Fig. 6 D). Surprisingly, in contrast with previous findings (Vizeacoumar et al., 2010), mutation of Mcm21 did not affect Ipl1-3GFP localization to the spindle midzone (Fig. 6 E). This result suggests that Mcm21 does not operate upstream of Ipl1, which is consistent with our observations that mcm21Δ and ipl1-321 mutants have distinct spindle disassembly phenotypes (Figs. 4 and 5 and Table I). Instead, Mcm21 and likely other components of the COMA kinetochore complex mediate spindle disassembly by binding to She1 and recruiting it to the spindle.
Kip3 operates independently of Ipl1 to regulate spindle length and promote sustained depolymerization of ipMTs
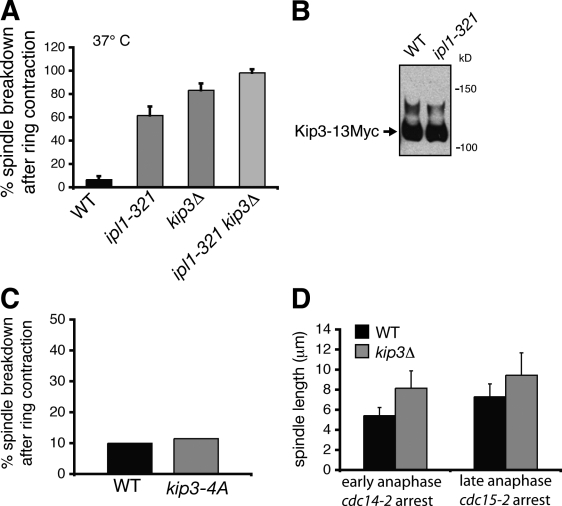
Lastly, we considered the possibility that Ipl1 might also mediate spindle disassembly by regulating Kip3 via phosphorylation. However, we found that the spindle disassembly defects observed in ipl1-321 cells were intensified upon additional depletion of Kip3. The frequency of spindles disassembling after cyotkinetic ring contraction, half-spindle shrinkage rates, and half-spindle recovery frequencies were higher in kip3Δ ipl1-321 mutants than in single ipl1-321 mutants (Fig. 7 A and Table I). These results suggest that Ipl1 and Kip3 perform distinct functions during spindle disassembly. When we used Phos-tag SDS-PAGE to analyze protein extracts from wild-type and ipl1-321 cells expressing Kip3-13Myc, we could not detect any difference in band migration pattern (Fig. 7 B), suggesting that Ipl1 does not phosphorylate Kip3 in vivo. We considered the possibility that our inability to detect Ipl1-dependent band migration shifts for Kip3-13Myc may have been because of insensitivity of our detection method. However, we obtained the same result when we affixed alternative tags to Kip3 (GFP and 3HA) and strengthened the concentration of Phos-tag acrylamide and the primary antibody used for detection by Western blotting (unpublished data). We still cannot completely rule out the possibility that Ipl1 phosphorylates Kip3 in a manner that does not affect the mobility of Kip3 on an acrylamide gel. Nevertheless, our conclusion that Ipl1 does not mediate spindle disassembly by phosphorylation-dependent activation of Kip3 is further supported by the fact that expression of a Kip3 construct containing serine/threonine to alanine mutations in all four putative Ipl1 consensus motifs (kip3-4A) did not compromise spindle disassembly (Fig. 7 C).
Figure 7.
Kip3 and Ipl1 operate in separate pathways during spindle disassembly. (A) Frequency in which spindles disassembled after initiation of cytokinetic ring contraction in wild-type (WT) cells and selected mutants (n = 3 separate experiments each analyzing 20 disassembly events for each strain). All cells were incubated at 37°C. (B) Analysis of Kip3-13Myc phosphorylation in wild-type and ipl1-321 strains. Protein extracts were separated by SDS-PAGE supplemented with 20 µM Phos-tag acrylamide then visualized by immunoblotting against the Myc epitope. (C) Quantification of delayed spindle disassembly in wild-type cells (n = 60) and in cells expressing an allele of KIP3 with all four Ipl1 consensus sites mutated (kip3-4A; n = 26). (D) Kip3 regulates mitotic spindle length throughout anaphase. Spindle length was measured in kip3Δ or wild-type cells expressing GFP-Tub1 arrested in early anaphase (cdc14-2) or late anaphase (cdc15-2; n = 60 for each strain). Error bars indicate mean ± SD.
The exclusion of Kip3 from the APCCdh1 and Ipl1 pathways, both of which seem to be activated by the MEN, raises questions about whether Kip3 activity is cell cycle regulated. We found that loss of Kip3 increased spindle length ∼1.5-fold in both early and late anaphase (Fig. 7 D), suggesting that Kip3-mediated depolymerization is not restricted to late mitosis, which is in contrast to APCCdh1-mediated destruction of midzone proteins or Ipl1-mediated inactivation of Bim1. Thus, although Ipl1, APCCdh1, and Kip3 are all important for disassembling the mitotic spindle, our findings indicate that Ipl1 and APCCdh1 are the major factors governing the transition from spindle growth to disassembly.
Discussion
Up to this point, understanding of spindle disassembly in any organism has been limited because this process has not been thoroughly analyzed in real time to fully dissect the mechanisms at play. Previous studies have relied on two methods to assess defects in spindle disassembly: (1) static analysis of late anaphase spindle morphology, looking for the accumulation of abnormally long or bent spindles, or (2) analysis of spindle persistence after the onset of anaphase B, looking for spindles that remain intact for longer periods of time than normal (Buvelot et al., 2003; Woodbury and Morgan, 2007; Vizeacoumar et al., 2010). Although these types of measurements can identify mutants that regulate spindle dynamics and integrity, they cannot distinguish between mutants that modify the rate of spindle elongation during anaphase and those that perturb spindle disassembly during telophase, and they are inadequate for distinguishing the various steps of spindle disassembly.
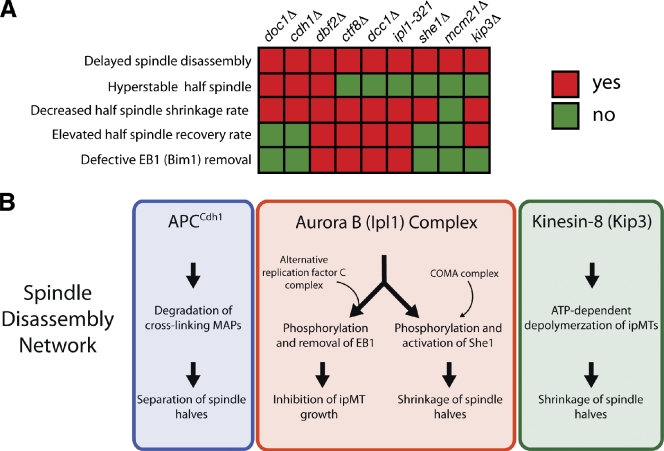
In this study, we combined genetic analysis with live cell fluorescence microscopy to identify proteins critical for spindle disassembly and to reveal their specific contributions to this process. We assessed in detail the effects of nine mutations (cdh1Δ, doc1Δ, dbf2Δ, kip3Δ, she1Δ, dcc1Δ, ctf8Δ, mcm21Δ, and ipl1-321) on spindle-half separation, ipMT depolymerization, and removal of MT-stabilizing proteins from the spindle. The phenotypes we observed for each mutant (Fig. 8 A) support a model wherein at least three pathways, which we call the spindle disassembly network, mediate spindle disassembly (Fig. 8 B).
Figure 8.
Multiple pathways regulating distinct subprocesses drive spindle disassembly. (A) Summary of phenotypic analysis for nine spindle disassembly mutants. Decreased half-spindle shrinkage rate indicates that the mean rate of half-spindle shrinkage was significantly lower than the wild-type rate (P < 0.05; Table I). Additionally, elevated half-spindle recovery rate indicates that the spindle-half recovery frequency was >1.5-fold higher than the wild-type frequency. (B) Multiple pathways model for spindle disassembly. In the first pathway (blue box), the APC loaded with the late anaphase cofactor Cdh1 (APCCdh1) degrades cross-linking MAPs (e.g., Ase1, Cin8, and others) to facilitate separation and destabilization of spindle halves. In the second pathway (red box), Aurora B (Ipl1 in yeast) phosphorylates the spindle stabilizer EB1 (Bim1 in yeast) and the spindle destabilizer She1. The ARF-C complex is needed for EB1 phosphorylation by Aurora B. The COMA kinetochore complex loads She1 onto the spindles. In the third pathway (green box), the kinesin-8 family member Kip3 actively depolymerizes spindle MTs and prevents their regrowth during mitotic exit. Inhibition of multiple pathways produces additive defects on spindle-half disassembly and cell fitness.
In the first pathway, the MEN stimulates association of the APC with the late anaphase cofactor Cdh1. This holoenzyme (APCCdh1) then ubiquitinates Ase1 (yeast Prc1 homologue), Cin8 (yeast BimC homologue), and other MT-cross-linking MT-associated proteins (MAPs; Hildebrandt and Hoyt, 2001; Woodbury and Morgan, 2007), thereby effecting their degradation and facilitating separation of the spindle halves.
In the next pathway, the Ipl1 complex (yeast Aurora B) arrests ipMT growth by inactivating the MT growth factor Bim1 (yeast EB1). Our findings suggest that the MEN activates this pathway, but the mechanism remains to be determined. We can exclude regulation of Ipl1 localization as a possible mechanism because we observed Ipl1-3GFP decoration of the midzone in dbf2Δ cells (Fig. S4). Interestingly, we also discovered that a novel factor uncovered in our screen, the highly conserved A-RFC complex (Dcc1, Ctf8, and others; Mayer et al., 2001), is necessary for Ipl1-mediated removal of Bim1. In the future, it will be important to determine how the A-RFC complex regulates Ipl1 activity.
In addition, Ipl1 destabilizes the spindle by activating She1, a novel disassembly factor identified in our screen, although the mechanism of She1 function remains to be determined. Likely, She1 does not regulate MEN, APC, or Kip3 activity, considering that Cdc14-GFP localization, Ase1-4GFP degradation, and Kip3-GFP localization are not detectably affected in she1Δ cells (Fig. S3, D–F). Because She1 localizes along the mitotic spindle, it is possible that She1 directly regulates MT dynamics. Finally, our data indicate that the kinesin-8 member Kip3 operates independently of the APCCdh1 and Ipl1 pathways to drive spindle MT depolymerization during spindle disassembly.
Why does spindle disassembly require coordination of multiple pathways that function via distinct mechanisms? The mitotic spindle is a highly complex structure assembled from MTs interlinked and stabilized by an assortment of MAPs. Thus, it seems logical that the diversity of stabilizing interactions must be reversed by a comparably diverse set of destabilizing mechanisms. Furthermore, the mitotic spindle is highly dynamic and irreversibly disassembled during a brief window of time (∼2 min in yeast; Maddox et al., 2000) in telophase. Deployment of multiple distinct destabilizing mechanisms may be necessary to achieve this switch-like transition. An alternative but not mutually exclusive explanation is that these multiple pathways may serve as a multitiered back-up system to ensure disassembly in the event that one pathway is compromised. This latter possibility is supported by our observation that disassembly of the hyperstable half-spindles in cdh1Δ cells was dramatically inhibited after additional mutation of Kip3. It is possible that Kip3-mediated depolymerization might help dissociate MT cross-linking proteins that remain on the spindle when APCCdh1 is inactive. This conclusion is further supported by the multitude of synthetic genetic interactions between null mutations that inhibit spindle disassembly (Fig. S2).
Our results demonstrate that yeast cells can accommodate the loss of one spindle disassembly pathway but not the loss of multiple pathways, thus highlighting the importance of spindle disassembly for cell cycle progression and overall fitness. We propose that the fitness defect seen when multiple spindle disassembly pathways are inactivated results from a G1 arrest and not a failure in mitotic exit. In kip3Δ cdh1Δ double mutants, cytokinetic ring contraction simultaneously divided the daughter cells and separated the spindle halves, yielding G1 cells with spindle remnants that persisted for >80 min, which is >40 times longer than in wild-type cells. During this time, these kip3Δ cdh1Δ double mutants remained arrested in G1, whereas their wild-type counterparts exited from G1 and formed new spindles (unpublished data). Our results suggest that cytokinesis can break the spindle when primary mechanisms are impaired, allowing entry into the next cell cycle. However, it seems that progression out of G1 is intimately linked with complete disassembly of preexisting spindle structures. Perhaps the inability to completely depolymerize spindle MTs prevents spindle pole body duplication and/or formation of a new spindle in the next cell cycle.
Considering the high degree of conservation among APCCdh1, Aurora B, kinesin-8, and the A-RFC complex, we expect our findings to have important implications for spindle disassembly in all eukaryotes. Despite the fact that direct, real-time analysis of spindle disassembly in metazoans has yet to be performed, there is evidence that the APC, Aurora B, and kinesin-8 might participate in this process in these organisms. In human cells, APCCdh1 concentrates to the central spindle and targets a variety of spindle-stabilizing proteins (e.g., Prc1, Polo kinase, and TPX2) for proteasome-mediated degradation during late mitosis (Peters, 2006). Based on our results, this activity might disrupt the central spindle and facilitate separation of spindle halves in human cells. Aurora B also concentrates to the central spindle in Caenorhabditis elegans (Severson et al., 2000) and human cells (Gruneberg et al., 2004), as in yeast, but whether Aurora B facilitates removal of MT-stabilizing proteins like EB1 during telophase in these organisms remains to be seen. Finally, similar to Kip3 in yeast, the kinesin-8 members Kif18A and Klp67A have been shown to regulate spindle size in human and Drosophila melanogaster cells, respectively (Goshima et al., 2005; Mayr et al., 2007). Thus, it is possible that sustained MT depolymerization driven by kinesin-8 represents a universal mechanism to mediate spindle disassembly.
Our goal for this study was to gain insight into how eukaryotic cells rapidly disassemble the mitotic spindle, an extremely complex structure, and to identify the proteins critical for this process. We conclude that mechanistically distinct pathways governed by APCCdh1, Aurora B, and kinesin-8 drive spindle disassembly in budding yeast. These pathways act in concert to break the proteinaceous attachments that hold overlapping ipMTs in a rigid structure, inhibit ipMT growth, and promote sustained ipMT depolymerization.
Materials and methods
Yeast strains
The yeast strains used in this study are derivatives of S288C and are listed in Table S1. doc1Δ, cdh1Δ, dbf2Δ, dcc1Δ, ctf8Δ, mcm21Δ, and kip3Δ null mutants originated from the Research Genetics Collection. The Ase1-4GFP and Cin8-4GFP strains were provided by E. Schiebel (ZMBH, Heidelberg, Germany). The phosphomutant alleles kip3-4A (S74A, T172A, S402A, and S792A) and she1-5A (T14A, S165A, S269A, T280A, and S325A) were generated using QuikChange site-directed mutagenesis. Ipl1 phosphorylation sites selected for mutation matched the (R/K)X(T/S)(I/L/V/T) consensus motif (Cheeseman et. al, 2002). Plasmids containing full-length kip3-4A and she1-5A genes were integrated at each gene’s endogenous locus in kip3Δ and she1Δ strains, respectively. The bim1-6A and bim1-6D strains were provided by S. Westermann (Research Institute of Molecular Pathology, Vienna, Austria), and the Kip3-GFP strain was provided by K. Bloom (University of North Carolina at Chapel Hill, Chapel Hill, NC).
Fluorescence microscopy
Live cell microscopy at room temperature was performed using a microscope (IX-71; Olympus), a 100× NA 1.4 objective, and a camera (Orca-ER; Hamamatsu Photonics). All microscopy at 37°C was performed using a microscope (IX-81; Olympus) equipped with a temperature-controlled enclosure (Precision Control Weather Station), a 100× NA 1.4 objective, and a camera (Orca-ER). Two-color images were obtained by sequential switching between RFP and GFP filter sets. For time-lapse microscopy of shrinking half-spindles, actinomyosin ring contraction, and Bim1-3GFP localization, images were collected at 10-s intervals with 300-ms exposures. Each image represents a maximum intensity projection from a z stack containing six planes 0.2 µm apart. All image processing was performed using MetaMorph software (MDS Analytical Technologies).
Synthetic lethal profile comparison
To examine the synthetic interactions for SHE1, we used the synthetic lethal and synthetic sick dataset described previously by Costanzo et al. (2010) and Tong et al. (2004). Although synthetic interactions between SHE1 and CDH1 and between SHE1 and DOC1 did not appear in these lists, we had previously identified such interactions through tetrad analysis. When comparing the synthetic interaction profiles of KIP3, CDH1, and SHE1, we used only the raw dataset with an intermediate cutoff applied (|ε| > 0.08, P < 0.05) Costanzo et al., 2010), plus several interactions we had identified in our laboratory.
Detection of protein phosphorylation
To detect phosphorylation-dependent mobility shifts of She1-13Myc, Kip3-13Myc, and Bim1-HA, whole cell extracts were loaded onto 6–8% SDS polyacrylamide gels containing either 20 µM or 30 µM Phos-tag acrylamide (Hiroshima University, Hiroshima, Japan) and 40 µM or 60 µM MnCl2, respectively. Membranes were then probed with 1:500 mouse anti-Myc (9E10; our laboratory) or 1:1,000 mouse anti-HA (12CA5; Roche).
Immunoprecipitation
Immunoprecipitation was performed essentially as described previously (Woodruff et al., 2009) with a few minor modifications. Lysates were incubated with mouse anti-Myc antibody for 1 h and then passed over protein G–Sepharose beads (GE Healthcare) to purify She1-13Myc. The column was washed with lysis buffer and eluted with SDS loading buffer.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was conducted essentially as described previously (Kang et al., 2001) with a few minor changes. Cells were fixed for 1 h in 1% formaldehyde. Whole cell extracts were passed over IgG Sepharose (GE Healthcare) to pull down She1-TAP and Mif2-TAP.
Chromosome spreads
Chromosome spreads were prepared as described previously (Cheeseman et al., 2001). Anti-Duo1 antibody (Hofmann et al., 1998) was used at a dilution of 1:2,000 to detect Duo1, and anti-GFP (rabbit; Torrey Pines Institute) was used at a dilution of 1:2,000 to detect She1-3GFP.
Online supplemental material
Fig. S1 describes spindle morphology in cells compromised for Kip3 and APCCdh1 activity. Fig. S2 depicts the pairwise comparison of previously described synthetic genetic interactions shared between CDH1, KIP3, and SHE1. Fig. S3 shows She1 phosphorylation and localization to the kinetochore and localization of Cdc14 and Kip3 in an she1Δ mutant. Fig. S4 shows Ipl1-3GFP localization in dbf2Δ cells. Videos 1–3 show spindle disassembly relative to cytokinetic ring contraction in wild-type, cdh1Δ, and kip3Δ cells, respectively. Table S1 lists the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006028/DC1.
Acknowledgments
We thank Stefan Westermann, Kerry Bloom, and Elmar Schiebel for strains and Charles Boone and Michael Costanzo for sharing data before publication. We also thank Doug Koshland for comments on the manuscript and Yutian Peng, Yidi Sun, Yuko Nakajima, and Randall Tyers for experimental help and suggestions.
This work was supported by the National Institutes of Health (grant GM 47842 to G. Barnes) and the National Science Foundation (graduate research fellowship to J.B. Woodruff).
Footnotes
Abbreviations used in this paper:
- APC
- anaphase-promoting complex
- A-RFC
- alternative replication factor C
- ipMT
- interpolar MT
- MAP
- MT-associated protein
- MEN
- mitotic exit network
- MT
- microtubule
References
- Ayscough K.R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D.G. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399–416 10.1083/jcb.137.2.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S., Tatsutani S.Y., Vermaak D., Biggins S. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329–339 10.1083/jcb.200209018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Enquist-Newman M., Müller-Reichert T., Drubin D.G., Barnes G. 2001. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 152:197–212 10.1083/jcb.152.1.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Anderson S., Jwa M., Green E.M., Kang J., Yates J.R., III, Chan C.S., Drubin D.G., Barnes G. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172 10.1016/S0092-8674(02)00973-X [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S., et al. 2010. The genetic landscape of a cell. Science. 327:425–431 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Haase J., Mythreye K., Molk J.N., Anderson M., Joglekar A.P., O’Toole E.T., Winey M., Salmon E.D., Odde D.J., Bloom K. 2008. The microtubule-based motor Kar3 and plus end–binding protein Bim1 provide structural support for the anaphase spindle. J. Cell Biol. 180:91–100 10.1083/jcb.200710164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. 2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10:9–20 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Wollman R., Stuurman N., Scholey J.M., Vale R.D. 2005. Length control of the metaphase spindle. Curr. Biol. 15:1979–1988 10.1016/j.cub.2005.09.054 [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Honda R., Nigg E.A., Barr F.A. 2004. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166:167–172 10.1083/jcb.200403084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.L., Jr, Carvalho P., Roof D.M., Pellman D. 2006. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8:913–923 10.1038/ncb1457 [DOI] [PubMed] [Google Scholar]
- Hildebrandt E.R., Hoyt M.A. 2001. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol. Biol. Cell. 12:3402–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C., Cheeseman I.M., Goode B.L., McDonald K.L., Barnes G., Drubin D.G. 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143:1029–1040 10.1083/jcb.143.4.1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.-Y., Sun Z.-W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F., et al. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 102:279–291 10.1016/S0092-8674(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Murray A.W. 1997. A novel yeast screen for mitotic arrest mutants identifies DOC1, a new gene involved in cyclin proteolysis. Mol. Biol. Cell. 8:1877–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang Y.L., Huang J., Peters J.M., McLaughlin M.E., Tai C.Y., Pellman D. 1997. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 275:1311–1314 10.1126/science.275.5304.1311 [DOI] [PubMed] [Google Scholar]
- Kang J., Cheeseman I.M., Kallstrom G., Velmurugan S., Barnes G., Chan C.S. 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)–related protein Sli15 during chromosome segregation. J. Cell Biol. 155:763–774 10.1083/jcb.200105029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale C.V., Frei S.B., Stern B.M., Biggins S. 2007. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev. Cell. 13:433–445 10.1016/j.devcel.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F.C., Mody M., Kurischko C., Roof D.M., Giddings T.H., Winey M. 2001. Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol. Cell. Biol. 21:6972–6983 10.1128/MCB.21.20.6972-6983.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Bloom K.S., Salmon E.D. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2:36–41 10.1038/71357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.L., Gygi S.P., Aebersold R., Hieter P. 2001. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol. Cell. 7:959–970 10.1016/S1097-2765(01)00254-4 [DOI] [PubMed] [Google Scholar]
- Mayr M.I., Hümmer S., Bormann J., Grüner T., Adio S., Woehlke G., Mayer T.U. 2007. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 17:488–498 10.1016/j.cub.2007.02.036 [DOI] [PubMed] [Google Scholar]
- Peters J.M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell. 9:931–943 10.1016/S1097-2765(02)00540-3 [DOI] [PubMed] [Google Scholar]
- Peters J.M. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7:644–656 10.1038/nrm1988 [DOI] [PubMed] [Google Scholar]
- Severin F., Habermann B., Huffaker T., Hyman T. 2001. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 153:435–442 10.1083/jcb.153.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson A.F., Hamill D.R., Carter J.C., Schumacher J., Bowerman B. 2000. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 10:1162–1171 10.1016/S0960-9822(00)00715-6 [DOI] [PubMed] [Google Scholar]
- Sullivan M., Morgan D.O. 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8:894–903 10.1038/nrm2276 [DOI] [PubMed] [Google Scholar]
- Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. 2004. Global mapping of the yeast genetic interaction network. Science. 303:808–813 10.1126/science.1091317 [DOI] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A.A., Tanaka T.U., Howard J. 2006. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8:957–962 10.1038/ncb1462 [DOI] [PubMed] [Google Scholar]
- VerPlank L., Li R. 2005. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol. Biol. Cell. 16:2529–2543 10.1091/mbc.E04-12-1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Amon A. 2001. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell. 12:2961–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., Amon A. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2:709–718 10.1016/S1097-2765(00)80286-5 [DOI] [PubMed] [Google Scholar]
- Vizeacoumar F.J., van Dyk N., S Vizeacoumar F., Cheung V., Li J., Sydorskyy Y., Case N., Li Z., Datti A., Nislow C., et al. 2010. Integrating high-throughput genetic interaction mapping and high-content screening to explore yeast spindle morphogenesis. J. Cell Biol. 188:69–81 10.1083/jcb.200909013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J., Nakajima Y., Westermann S., Shang C., Kang J.S., Goodner C., Houshmand P., Fields S., Chan C.S., Drubin D., et al. 2007. A protein interaction map of the mitotic spindle. Mol. Biol. Cell. 18:3800–3809 10.1091/mbc.E07-06-0536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury E.L., Morgan D.O. 2007. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat. Cell Biol. 9:106–112 10.1038/ncb1523 [DOI] [PubMed] [Google Scholar]
- Woodruff J.B., Drubin D.G., Barnes G. 2009. Dynein-driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment. Mol. Biol. Cell. 20:3003–3011 10.1091/mbc.E09-03-0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., You L., Pasqualone D., Kopski K.M., Huffaker T.C. 2002. Stu1p is physically associated with beta-tubulin and is required for structural integrity of the mitotic spindle. Mol. Biol. Cell. 13:1881–1892 10.1091/mbc.01-09-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K., Westermann S. 2009. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186:379–391 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]