In vivo analysis of subcellular dynamics in the zebrafish cerebellum provides new insights into centrosome positioning during vertebrate brain differentiation.
Abstract
The position of the centrosome ahead of the nucleus has been considered crucial for coordinating neuronal migration in most developmental situations. The proximity of the centrosome has also been correlated with the site of axonogenesis in certain differentiating neurons. Despite these positive correlations, accumulating experimental findings appear to negate a universal role of the centrosome in determining where an axon forms, or in leading the migration of neurons. To further examine this controversy in an in vivo setting, we have generated cell type–specific multi-cistronic gene expression to monitor subcellular dynamics in the developing zebrafish cerebellum. We show that migration of rhombic lip–derived neurons is characterized by a centrosome that does not persistently lead the nucleus, but which is instead regularly overtaken by the nucleus. In addition, axonogenesis is initiated during the onset of neuronal migration and occurs independently of centrosome proximity. These in vivo data reveal a new temporal orchestration of organelle dynamics and provide important insights into the variation in intracellular processes during vertebrate brain differentiation.
Introduction
Neuronal progenitors undergo a variety of developmental steps to form a functional brain. After proliferation, they migrate, differentiate terminally, and generate dendrites and axons to establish neuronal circuits. Cell behavior at each step is coordinated by the subcellular organelle dynamics occurring within the developing neurons. The centrosome in particular, through its function as a microtubule-organizing center (MTOC), has been proposed to act as a main organizer of polarized cell behaviors such as directed migration and axonogenesis (Higginbotham and Gleeson, 2007).
Within cells in a proliferating neuroepithelium, the centrosome localizes strictly to the apical (ventricular) side to maintain apico-basal polarity (Hinds and Ruffet, 1971; Shoukimas and Hinds, 1978). During both radial and tangential migration, the apical process of the immature neuron becomes disconnected from the proliferation zone and the cell body advances behind an extended membrane protrusion termed the leading process. Individual migratory steps of neurons are characterized by the forward movement of the nucleus—a process termed nucleokinesis—which can occur in a saltatory manner alternating with resting phases. In most analyzed neurons migrating by saltatory nucleokinesis, the centrosome is localized ahead of the nucleus to face toward the leading process, with the centrosome moving forward before the nucleus (Solecki et al., 2004; Tanaka et al., 2004; Bellion et al., 2005; Schaar and McConnell, 2005; Métin et al., 2006; Tsai et al., 2007). Due to these observations, a common model for saltatory nucleokinesis in migrating neurons—defined by the sequential subcellular events of a continuously leading centrosome followed by a trailing nucleus—attributes the centrosome with a permanently leading role in initiating and directing migration. (Tsai and Gleeson, 2005; Marín et al., 2006; Higginbotham and Gleeson, 2007; Métin et al., 2008).
Such an orientation of the centrosome in the direction of cell migration and ahead of the nucleus is not unique to neurons, but has been observed in many other cell types during migration, such as endothelial cells (Gotlieb et al., 1981), macrophages (Nemere et al., 1985), and fibroblasts (Kupfer et al., 1982; Schliwa et al., 1999). In non-neuronal migrating cells though, a correlation between migration and a leading centrosome is less consistent. For example, in fibroblasts migrating on grooved substrates or in collagen gels, the centrosome position is randomized with respect to the nucleus and the cell front (Schütze et al., 1991), whereas the centrosome in PtK cells lags behind the nucleus during wound-healing migration (Yvon et al., 2002). Similarly, a centrosome trailing the nucleus has been observed in cells of the migrating lateral line primordium in zebrafish embryos (Pouthas et al., 2008).
Reorientation of the centrosome can be stimulated by molecular interactions or gradients (Nemere et al., 1985; Renaud et al., 2008), electrical stimulation (Pu and Zhao, 2005; Zhao et al., 2006), or shear stress (Coan et al., 1993; Lee et al., 2005). This suggests that centrosome position is strongly influenced by the local molecular composition of the environment, but also by physical and physiological parameters such as morphogenetic constraints and electrical activity. Centrosome orientation may thus vary depending on the cell type, the tissue, and the developmental stage. Strikingly, it was recently shown that in radially migrating granule neurons of the developing cerebellum, the centrosome does not strictly lead migration during saltatory nucleokinesis, but it is often overtaken by the nucleus (Umeshima et al., 2007). This centrosomal behavior conflicts with the commonly used model of neuronal nucleokinesis; however, it was suggested that bidirectional movements of the nucleus may be the reason for the unusual temporary trailing of the centrosome in migrating granule neurons (Vallee et al., 2009). Clearly, further in vivo investigations are required to address a potential cell-type dependency of centrosome dynamics during neuronal migration.
An equally important role attributed to the centrosome is the determination of the site of axon outgrowth, as the centrosome is found in close proximity to the neurite that becomes the axon (Zmuda and Rivas, 1998; de Anda et al., 2005). In support, centrosome duplication through inhibition of cytokinesis resulted in two axons emerging adjacent to the centrosomes, whereas laser inactivation of the centrosome in cultured Drosophila neurons impaired axon formation (de Anda et al., 2005). In contrast though, axonogenesis is unaffected in DSas-4 mutant flies, which are unable to replicate centrioles and therefore lack functional centrosomes by the third instar larval stage (Basto et al., 2006). Additionally, it was recently reported that, at least after axon induction, continued axon outgrowth from hippocampal neurons occurs through centrosome-independent polymerization of microtubule fibers, and is in fact unaffected by laser ablation of the centrosome (Stiess et al., 2010). These controversial observations argue for differences between the cell culture and in vivo situation or suggest that organelle dynamics differ between neuronal cell types. Therefore, an in vivo investigation into the temporal relationship between centrosome dynamics and emerging axonogenesis is needed. Such an approach though, requires a cell type–specific multiple organelle labeling technology.
We have established efficient coactivation of multiple cell biological fluorescent reporter proteins expressed from Gal4-dependent multi-cistronic expression cassettes. In addition, we generated a stable transgenic zebrafish strain expressing the modified Gal4 transcriptional activator KalTA4 (Distel et al., 2009) under the control of regulatory elements from the zebrafish atonal1a gene and thus specifically in neuronal progenitors of the hindbrain rhombic lip. These neurons of the tegmental hindbrain nuclei (THN) have been shown to undergo long-distance migration along characteristic pathways through the developing zebrafish cerebellum and into the hindbrain tegmentum in an evolutionarily conserved manner (Köster and Fraser, 2001a; Volkmann et al., 2010). Expression of the multi-cistronic constructs in this transgenic Gal4 activator strain therefore allows the dynamics of the nucleus, the centrosome, and the emerging axon to be monitored simultaneously during THN migration in vivo. We demonstrate that the centrosome in migrating THN neurons is regularly overtaken by the nucleus and apparently does not lead migration by continuously advancing ahead of the nucleus. In addition, we reveal the temporal sequence of subcellular dynamics during THN neuron axonogenesis, which exclude an in vivo function of the centrosome in determining the site of axon outgrowth based on proximity to the centrosome. Thus, multicolor in vivo time-lapse imaging at subcellular resolution provides important insights into the dynamics of cellular processes and promises that the large fields of developmental genetics and cell biology can be merged in transparent zebrafish embryos into a field of vertebrate in vivo cell biology.
Results
Simultaneous subcellular intravital labeling in zebrafish cells
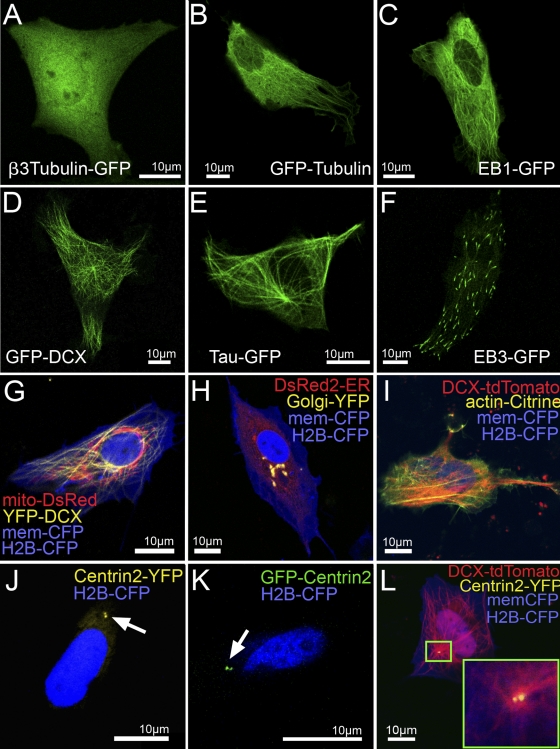
To fluorescently label subcellular structures in zebrafish, we first tested available fusion proteins in zebrafish Pac2 fibroblasts (Senghaas and Köster, 2009). Although β3-tubulin–GFP (mouse) was mostly localized to the cytoplasm (Fig. 1 A), GFP-tubulin (Fig. 1 B, mouse) and EB1-GFP (Fig. 1 C, mouse) each marked the microtubule network, although the specificity of labeling was dose dependent. In contrast, expression of GFP-DCX (Fig. 1 D, mouse) or Tau-GFP (Fig. 1 E, mouse) resulted in robust microtubule labeling, while EB3-GFP (Fig. 1 F, mouse) successfully marked microtubule plus-ends. These data indicate that subcellular labeling constructs cannot be easily transferred from one species to another, but require careful testing.
Figure 1.
Identification of subcellular markers for in vivo imaging of zebrafish cells. Images of zebrafish Pac2 fibroblasts transfected with pCS2+ constructs encoding fluorescently tagged markers for subcellular labeling 24 h after transfection. (A) β3-tubulin-GFP, (B) GFP-tubulin, (C) EB1-GFP, (D) GFP-DCX, (E) Tau-GFP, and (F) EB3-GFP. (G) mito-DsRed to label mitochondria in red, YFP-DCX to label microtubules in yellow, memCFP to label the cytoplasmic membrane in blue and H2B-CFP to label the nucleus in blue; (H) DsRed2-ER to label the ER in red, Golgi-YFP to label the Golgi apparatus in yellow, memCFP and H2B-CFP; (I) DCX-tdTomato to label microtubules in red, actin-Citrine to label the actin cytoskeleton in yellow, memCFP, H2B-CFP; (J) Centrin2-YFP to label the centrosome in yellow (arrow is indicating the two centrioles of the centrosome) and H2B-CFP; (K) GFP-Centrin2 to label the centrosome in green (arrow is indicating the two centrioles of the centrosome); and (L) DCX-tdTomato, Centrin2-YFP, memCFP, and H2B-CFP. The inset shows a higher magnification of the centrosome at the hub of the microtubule network. These data present a collection of subcellular-targeted fluorescent proteins tested for their specificity in zebrafish cells. “mem“ represents a membrane localization signal, which consists of a plamitylation and myristinylation sequence of the human Lck kinase.
Subsequently, we used a similar approach to achieve mitochondrial targeting in zebrafish fibroblasts (mitochondrial targeting sequence from subunit VIII of human cytochrome c oxidase; Fig. 1 G), as well as labeling of the endoplasmatic reticiulum (ER targeting sequence from calreticulin and ER retention sequence KDEL), the Golgi apparatus (N-terminal 81 aa of human β1-4 galactosyltransferase; Fig. 1 H), and the actin cytoskeleton (zebrafish cytoskeletal actin; Fig. 1 I). Finally, fluorescent protein fusions of the C terminus (Fig. 1 J) and the N terminus (Fig. 1 K) of a partial zebrafish centrin-2 cDNA were each used successfully to visualize the two centrioles of the centrosome at the hub of the microtubule network (Fig. 1 L, inset). To eventually allow for multiple combinations of subcellular markers, we generated fusion proteins with spectrally different fluorescent proteins for most of these subcellular markers (Table S1).
Simultaneous multicolor labeling
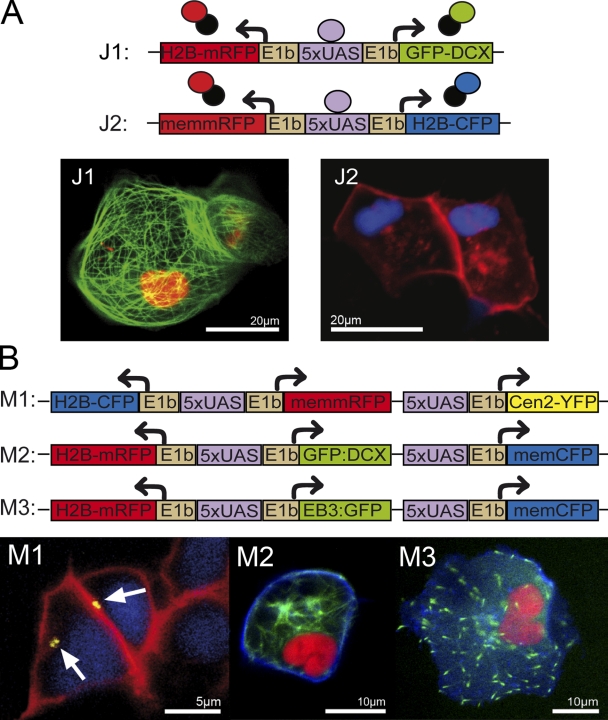
To mediate the simultaneous expression of two fluorescent subcellular markers by Gal4 transcriptional activators, we generated bidirectional Gal4-dependent effector constructs. These so-called Janus vectors carry a series of Gal4 binding sites (upstream activating sequences, UAS) flanked on both sides by E1b minimal promoters (Fig. 2 A; Paquet et al., 2009).
Figure 2.
Janus and Medusa Gal4 effector constructs for simultaneous expression of multiple subcellular labels. (A) Schematic representation of bidirectional Janus vectors J1 and J2. Upon binding of Gal4, two subcellular markers are expressed simultaneously (J1: H2B-mRFP labels the nucleus in red and GFP-DCX the microtubules in green; J2: memmRFP labels the membrane in red and H2B-CFP the nucleus in blue). (B) Schematic representation of Medusa vectors M1, M2, and M3. From each vector, the expression of three subcellular markers is activated in the presence of Gal4. M1 encodes H2B-CFP to label the nucleus in blue, memmRFP to mark the membrane in red, and Centrin2-YFP to label the centrioles of the centrosome in yellow. M2: H2B-mRFP to label the nucleus in red, GFP-DCX to label microtubules in green, and memCFP to label the membrane in blue. M3 codes for the same nuclear and membrane markers as M2, but contains EB3-GFP to label the plus-ends of microtubules. These data demonstrate that reliable coexpression of various transgenes can be achieved from Gal4-mediated multicistronic expression vectors. Images were obtained from living zebrafish embryos (24 hpf) coinjected at the one-cell stage with the respective Janus or Medusa vectors and a vector coding for Gal4. “mem“ represents a membrane localization signal, which consists of a plamitylation and myristinylation sequence of the human Lck kinase. Arrows in M1 indicate YFP-labeled centrosomes.
Two of these Janus vectors (Fig. 2 A; J1, J2) were used to evaluate the degree of coexpression of the two markers in vitro by cotransfection with a KalTA4-encoding expression vector (pCS-KalTA4GI) into Pac2 fibroblasts (not depicted) and in vivo by coinjection with pCS-KalTA4GI at the one-cell stage. Co-expression was found to be reliable, both in Pac2 cells and in embryos (ranging between 97–99%), indicating that bound KalTA4 is able to activate gene expression upstream and downstream, even when only a single UAS site is used (Fig. S1). Currently though, we cannot distinguish whether bidirectional activation occurs after KalTA4 is bound to UAS or whether bidirectional activation requires off and on events of KalTA4 binding to UAS.
To further evaluate if the position upstream or downstream of the UAS sites is favored by KalTA4 for activating transgene expression, we generated the Janus constructs mRFP:5xUAS:GFP and GFP:5xUAS:mRFP. Both vectors were cotransfected with pCS-KalTA4GI into zebrafish Pac2 fibroblasts and protein levels of GFP and mRFP were determined by Western blot analysis (n = 3). When the ratios of the expression levels were compared for both orientations, the position downstream of the UAS was found to be slightly more strongly activated (∼1.15 fold) than the position upstream of the UAS sites (unpublished data). Although this differential activation does not represent a marked difference, this information may be valuable when the dose of the expressed transgenes is of importance.
Next, we established so-called Medusa vectors containing additional UAS sites or Janus units for triple or quadruple transgene expression. This, for example, allows the nucleus, the cytoplasmic membrane, and the centrosome (M1) to be labeled simultaneously from a single Medusa expression construct. In addition, Medusa vectors labeling microtubule fibers (M2) or microtubule plus-ends (M3) together with the nucleus and the cell membrane were successfully expressed in zebrafish embryos (Fig. 2 B) and allow one to clearly observe microtubule dynamics in nondividing (Video 1) or dividing cells (Videos 2 and 3) in living zebrafish embryos. Furthermore, two Janus cassettes can be combined to achieve quadruple subcellular labeling (not depicted).
Generation of a rhombic lip–specific KalTA4 activator line
To express transgenes specifically in cells derived from the rhombic lip, we identified regulatory elements of the zebrafish atonal1a homologue and flanked a KalTA4 expression cassette (KalTA4GipA; Distel et al., 2009) with them. Subsequently, transgenic fish Tg(atoh1a:Gal4TA4)hzm2 were generated using I-SceI-meganuclease mRNA coinjection (Babaryka et al., 2009). When crossed to Tg(4xUAS:GFP)hzm3 carriers, the offspring showed the expected pattern of GFP expression throughout the rhombic lip (Fig. 3, B and C). Although some ectopic GFP expression domains in the retina and optic tectum (Fig. 3 C) could be observed, GFP fluorescence throughout the hindbrain and in sensory hair cells recapitulated the expression of endogenous atonal1a, as confirmed by coexpression analysis against atonal1a mRNA (Fig. 3, A and D) and anti-GFP immunohistochemistry (Fig. 3, B, E, and F).
Figure 3.
Characterization of Tg(atoh1a:Gal4TA4)hzm2 transgenic zebrafish. Lateral view of an offspring of Tg(atoh1a:Gal4TA4)hzm2 x Tg(4xUAS:GFP)hzm3 transgenic fish at 24 hpf. (A) Endogenous atonal1a expression in the rhombic lip as revealed by in situ hybridization (black). (B) Immunostaining for GFP after in situ hybridization for atonal1a on Tg(atoh1a:Gal4TA4)hzm2/Tg(4xUAS:GFP)hzm3 double-transgenic fish shows expression of GFP (green) in the rhombic lip in atonal1a-expressing cells (black). (C) In addition, some GFP-expressing cells can be found in the retina, the midbrain tegmentum, and the tectum of Tg(atoh1a:Gal4TA4)hzm2/Tg(4xUAS:GFP)hzm3 double-transgenic fish. (D) Enlargement of boxed area in A showing in situ hybridization for atonal1a in the hindbrain. (E) Immunostaining for GFP. (F) Overlay of D and E. These data show that KalTA4 expression in Tg(atoh1a:Gal4TA4)hzm2 embryos faithfully recapitulates rhombic lip expression of endogenous atonal1a. MHB, midbrain–hindbrain boundary; URL, upper rhombic lip.
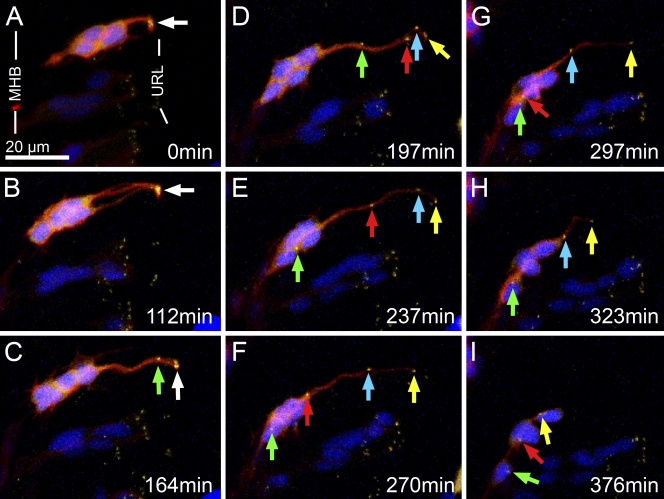
Confocal time-lapse recording of fluorescent URL-derived cells in embryos from crosses of Tg(atoh1a:Gal4TA4)hzm2 x Tg(shhb:Gal4TA4,5xUAS:mRFP)hzm4 (TG5xR; Distel et al., 2009), in which Gal4-expressing cells mosaically expressed mRFP, revealed that these cells divided at the URL starting at ∼24 hours postfertilization (hpf; Fig. 4 C), and subsequently moved radially to the MHB while still connected to the URL by a long trailing process (Fig. 4 D). They eventually retracted this trailing process and migrated ventrally in a tangential manner along the MHB (Fig. 4, E and F). These early URL-emigrating cells have recently been identified as neurons of tegmental hindbrain nuclei (Volkmann et al., 2010). Intriguingly, as has been described before (Köster and Fraser, 2001a), these THN neurons already begin to project axons along the MHB and into the midbrain before and during ventral migration (Fig. 4 F, white arrow; see Video 4).
Figure 4.
Time-lapse analysis of THN progenitor behavior. (A) Lateral view of the MHB region of a Tg(atoh1a:Gal4TA4)hzm2 x Tg(shhb:Gal4TA4,5xUAS:mRFP)hzm4 embryo at 24 hpf. URL-derived THN progenitors are labeled by mRFP expression. The boxed area is enlarged in B–F. (B) mRFP-expressing THN progenitors are connected to the apical surface by thin processes (arrow). (C) THN progenitors divide at the apical side (arrows indicate dividing cell). (D) During radial migration these cells maintain apical processes (arrows) that are retracted (E) once the nuclei reach the MHB. (F) Around the same time, axon-like processes become visible (arrow). These time-lapse data show that transgene expression mediated by Gal4 in Tg(atoh1a:Gal4TA4)hzm2 embryos reveals cell behavior consistent with that previously observed for URL-derived THN neurons (Köster and Fraser, 2001a; Volkmann et al., 2010). Images were taken from Video 4. MHB, midbrain–hindbrain boundary; URL, upper rhombic lip.
Subcellular analysis of THN neuronal progenitors during cell division
The establishment of subcellular markers and multi-cassette UAS-based vectors, in addition to the URL-specific KalTA4 expression in the transgenic strain Tg(atoh1a:Gal4TA4)hzm2, set up the possibility of doing in vivo cell biological experiments in a defined neuronal population. Tg(atoh1a:Gal4TA4)hzm2 embryos injected at the one-cell stage with the Medusa M1 construct showed expression of the subcellular markers in THN neuron progenitors from at least 22 hpf onwards. THN neuronal progenitors were observed to span the entire cerebellar primordium, being connected by endfeet-like processes to the apically positioned URL and basally located MHB (Fig. 5 A). THN progenitors underwent interkinetic nuclear movements (INM) and divided strictly at or close to the apical side, along the ventricle (Fig. 4 C, white arrows, and Fig. 5 C, yellow asterisk; n = 31 cells, 8 embryos). Mitotic events of these cells elsewhere in the cerebellar neuroepithelium were rarely observed, consistent with results from immunostainings against phosphorylated histone 3, an established M phase marker, which only labeled cells near the ventricle (Fig. S2). During INM phases, the centrosomes neither precede nor follow the movement of the nuclei, but remain stationary, being localized strictly to the apical membrane (Fig. 5, A–C, arrows; Video 5; n = 17, 4 independent embryos), as reported previously for mitotically active neuronal progenitors in polarized dividing neuroepithelia (Hinds and Ruffet, 1971; Chenn et al., 1998; Xie et al., 2007).
Figure 5.
In vivo subcellular imaging of INM and mitotic cleavages of THN progenitors. Lateral view of THN progenitors in the cerebellum of an ∼24-hpf Tg(atoh1a:Gal4TA4)hzm2 transgenic embryo injected with Medusa vector M1. Centrosomes are shown in yellow, cell nuclei in blue, and cellular membranes in red. (A) Centrosomes (arrows) were found to line the fourth ventricle at the apical side of the four THN progenitors undergoing INM between the midbrain–hindbrain boundary (MHB) and the fourth ventricle. Throughout INM, the centrosomes did not change their positions. Green asterisk demarcates a nucleus moving from apical to basal (A and B), while the corresponding centrosome (green arrow) stays at the apical side. The orange asterisk demarcates a nucleus that moves from basal to apical (A–C) to undergo a mitotic cleavage at the apical side (C). The corresponding centrosome (orange arrow) stays at the apical side, replicating to build the two spindle poles of the spindle apparatus (C, orange arrows). Thus, THN progenitors along the URL show characteristic INM behavior. Images are taken from Video 5. Note: some yellow-only labeling may suggest insufficient coexpression of transgenes from Medusa vectors. However, dependent on the z-level position of organelles and different intensities in expression levels, proper co-labeling can only be observed in cells of interest for which z-stacks were recorded. MHB, midbrain–hindbrain boundary; URL, upper rhombic lip.
THN neuronal progenitors maintain a mitotic organization during preparation for migration
When time-lapse recordings were performed over a prolonged period of time, we observed that THN progenitors appeared to change their behavior. THN progenitors are elongated and during INM the long axis of cells in the mid-cerebellum is oriented only ∼32 degrees off the apico-basal axis between the URL and MHB (Fig. 6, A–C; Fig. S3; 32.5 ± 4.6, n = 5 cells). As THN progenitors approach the MHB in preparation for ventral migration, however, the cells become more ventrally oriented, deviating now ∼65 degrees from the apico-basal axis of the cerebellum (Fig. 6 F; Fig. S3; 66.9 ± 6.9, n = 4 cells). Medusa vector labeling of the centrosome, the membrane, and the nucleus revealed that THN progenitors initiating migration leave behind a long trailing process that remained connected with the apical membrane. The centrosome remained stationary in the apical process, near the ventricular surface (Fig. 6, B and C), until shortly before the apical process began to detach from the germinal zone. If neuronal migration is considered to be an event whereby a neuron follows a leading process and moves from its birth place to its terminal site of later function without being connected to either position, then the final forward movement of bipolar THN progenitors toward the MHB does not represent neuronal migration. Rather, the apically positioned centrosome within a trailing process still in contact with the germinal zone is characteristic of proliferating neural progenitors and argues that the nuclear translocation toward the MHB in conjunction with a ventral turn of the cell actually represents an extended final step of INM.
Figure 6.
In vivo subcellular imaging of centrosome dynamics in THN progenitors. Lateral view of the cerebellar anlage of an ∼36-hpf Tg(atoh1a:Gal4TA4)hzm2 transgenic embryo injected with Medusa vector M1. Centrosomes (green arrow indicates the first centrosome, red arrow second, turquoise third, and yellow fourth; white arrows are shown when centrosomes are indistinguishable) are labeled in yellow, cell nuclei in blue, and cellular membranes in red. (A) Centrosomes (arrow) of the four THN progenitors were found to line the fourth ventricle. (B) Nuclei translocate basally toward the MHB, leaving behind a long trailing process containing the centrosome at its most apical part. This subcellular coordination argues that the final MHB-directed cell movement to initiate migration represents an extended final step of INM. (C–I) When nuclei reach the MHB, trailing processes containing the centrosomes at the most apical position are retracted, representing the initiation of THN neuron migration. Images are taken from Video 6. MHB, midbrain–hindbrain boundary; URL, upper rhombic lip.
Initiation of migration by THN neurons is accompanied by saltatory nuclear movements
When the cell soma approached the MHB, THN progenitors started to retract their trailing process, with the centrosome homing toward the soma and reaching the nucleus (n = 27 cells, 7 embryos) within ∼1.5 to 2 h (Fig. 6, C–I; Videos 6 and 7). Subsequently, THN neurons continued to migrate in a ventral direction to eventually reach their terminal positions in tegmental hindbrain areas.
To further quantify the subcellular processes during initiation of migration, we first performed a kymograph analysis of nuclear movements. THN progenitors connected with the URL via a trailing process showed a gradual pace of forward nuclear movement (see Fig. 8 A). In contrast, saltatory nucleokinesis alternating with resting phases was observed for nuclei of THN neurons migrating ventrally parallel to the MHB (see Fig. 8 B, white asterisk). When the distance of nuclear movement was plotted against time, a similar average pace of nucleokinesis could be determined for gradual (0.22 µm/min) and saltatory (0.21 µm/min) nucleokinesis. During the latter though, the nucleus only moved on average 1.17 µm (±0.7 µm) during resting phases, while suddenly progressing 7.54 µm (±1.43 µm) during saltatory steps (n = 8 saltatory steps, 3 independent videos), which is more than a nuclear diameter (6.63 ± 0.67 µm, 10 nuclei measured). Thus, migration of THN neurons disconnected from the URL is marked by a switch from gradual to saltatory nucleokinesis.
Figure 8.
Analysis of THN migratory movements. (A) Kymograph of a portion of Video 6 (229–390 min) showing the gradual movement of cell nuclei toward the MHB. Images have been rotated 45° and only the blue channel is shown in order to better visualize the nuclei. Each nucleus is labeled with a colored dot. Time between frames is 404.2 s. (B) Kymograph created from Video 8, showing two saltatory movements of the nucleus (asterisks) and the comparatively smooth forward migration of the centrosome during ventral migration. Images were rotated 45° and the time between frames is 522.6 s. The centrosome is ahead of the nucleus immediately before a nucleokinetic movement, but is overtaken when the nucleus jumps forward. (C) Graph of the cumulative migration distance (in any direction) for each nucleus in Video 6. Colors match the dots used for labeling nuclei in A. Tracking was done on 2D maximum projections with the Manual Tracking tool of ImageJ. The nuclei move at a gradual pace until they reach the MHB, at which point one nucleus undergoes a saltatory movement (red bar). The blue bar represents the region of the video shown in the kymograph in A. (D) Graph of the cumulative migration distance (in all directions) of the centrosome and nucleus in Video 8. During ventral migration, the cells undergo obvious nucleokinetic movements (red bars). The centrosome moves at a more consistent and gradual pace. (E) Pie chart showing the amount of time that the centrosome spends ahead of, lateral to, or behind the centroid of the nucleus (n = 4 cells, 3 embryos). (F) 3D graph showing the portion of time during which the centrosome and nucleus in Video 8 are each stationary, move in the direction of migration (forward), or move opposite to the direction of migration (backward). The movement of both organelles is predominantly in the direction of migration, but much of the forward centrosomal movement occurs while the nucleus is stationary.
The nucleus repeatedly surpasses the centrosome during nucleokinesis of THN neurons
A current model of saltatory nucleokinesis proposes that the centrosome permanently advances ahead of the nucleus, and that this organization is crucial for forward nuclear movement (Tsai and Gleeson, 2005; Métin et al., 2008). Indeed, during THN migration along the MHB, Medusa labeling revealed that the centrosome moved ahead of the nucleus (Fig. 6, F and G, red arrow; G–I, yellow arrowhead). Intriguingly, our time-lapse recordings showed that the centrosome did not remain strictly in front of the nucleus. Instead, with each forward migratory step, the nucleus passes the centrosome (Fig. 7, C–F; Fig. 8 B, white asterisks). Plotting of distance over time showed that the centrosome moved at a nearly constant pace, whereas the nucleus alternated between resting and sudden advances (Fig. 8 D). Quantification revealed that the centrosome most often trailed the nucleus and is ahead of it for only ∼35% of the time (Fig. 8 E). However, the centrosome repeatedly passed the nucleus during the preparatory phase of nuclear movement (Fig. 7, A–C, D, and E), when the centrosome seemed to indicate the direction of the next forward migratory step (n = 16 cells, 6 embryos; Video 8). Thus, when the nucleus is not stationary (72.22% ± 14.40% of time), the centrosome and the nucleus move in the same direction (70.9% ± 9.9) and rarely opposite (8.4% ± 4.5%) to one another (n = 4 ventrally migrating cells, 3 embryos), indicating the same directionality of their movements (for a detailed analysis of the direction of movements of the centrosome and the nuclear centroid in the ventrally migrating cell shown in Fig. 8 B, see Fig. 8 F).
Figure 7.
Subcellular imaging of saltatory nuclear movements in migrating THN progenitors. Lateral views of a region of the cerebellum of a 36-hpf Tg(atoh1a:Gal4TA4)hzm2 transgenic zebrafish embryo injected with Medusa vector M1. (A–C) According to the direction of migration, the centrosome (arrow) translocates in front of the nucleus (asterisk). (C and D) The nucleus then overtakes the centrosome in a rapid saltatory movement such that the centrosome locates posterior to the nucleus (D). Subesequently, the centrosome translocates once again ahead of the nucleus (E). In a second saltatory movement, the nucleus again overtakes the centrosome (F). These time-lapse data show that during saltatory nucleokinetic migration, THN neurons display iterative cycles of a centrosome leading and trailing the nucleus. Images are maximum projections of z-stacks. The time between images taken from Video 8 is indicated in the bottom right of each panel. MHB, midbrain–hindbrain boundary; URL, upper rhombic lip.
This iterative change in relative position of the nucleus and the centrosome to one another and the mostly trailing centrosome differ from the commonly used saltatory nucleokinetic model of a permanently leading centrosome during all steps of neuronal migration. Thus, either zebrafish THN neurons follow a different, cell population–specific form of saltatory nucleokinetic migration or saltatory nucleokinesis is less strictly orchestrated than previously thought, perhaps requiring a forward position of the centrosome only during a certain time window just before nuclear movement.
Axonogenesis of THN neurons occurs at the MHB
THN neurons approaching the MHB begin to extend from their leading edge a longer axon-like cellular process (Fig. 9 A, white asterisk; for axon projection into the midbrain see also final image sequence of Video 4). To verify the axonal identity of these emerging leading process structures, we performed immunohistochemistry against the axonal-specific marker acetylated tubulin. Acetylation of lysine40 of the α-tubulin subunit to stabilize microtubules is a key characteristic of the proximal axon and is important for axon outgrowth (Hammond et al., 2008; Witte and Bradke, 2008; Conde and Cáceres, 2009). Double-transgenic Tg(atoh1a:Gal4TA4)hzm2 x Tg(4xUAS:GFP)hzm3 embryos were raised until 40–42 hpf, by which time many GFP-expressing THN neurons had reached the MHB (Fig. 9 B). Subsequent fluorescent immunohistochemistry against GFP and acetylated α-tubulin showed that indeed GFP-expressing THN neurons at the MHB already possessed acetylated α-tubulin–positive processes (Fig. 9 C, white arrows), which likely emerged from the leading processes observed in Medusa-labeled cells (Fig. 9 A, white asterisk). These data strongly suggest that THN neurons initiate axon formation at the beginning of migration.
Figure 9.
THN progenitors initiate axonogenesis from their leading process independent of centrosome proximity. (A) Lateral view of the cerebellar anlage of an ∼42-hpf Tg(atoh1a:Gal4TA4)hzm2 transgenic zebrafish embryo injected with Medusa vector M1. An axon-like protrusion (white asterisk) has formed at the time when the centrosome (white arrow) is still homing toward the soma. (B) Lateral view of a Tg(atoh1a:Gal4TA4)hzm2 x Tg(4xUAS:GFP)hzm3 transgenic zebrafish embryo at 42 hpf. GFP-expressing cells are visualized by anti-GFP immunostaining (green) and acetylated microtubules by anti-acetylated tubulin immunostaining (red). (C) Enlargement of boxed area in B. Arrows indicate acetylated microtubules present in GFP-expressing THN progenitors, indicating the presence of axons by 42 hpf. (D–F) Lateral view of the cerebellum of a 40-hpf Tg(atoh1a:Gal4TA4)hzm2 transgenic zebrafish embryo injected with Medusa vector M1. (D) A THN progenitor (white asterisk) extends a process, the presumptive axon with a growth cone–like structure (red arrow), while the centrosome (white arrow) starts to translocate toward the soma. (E and F) The axon-like process elongates while the centrosome is moving toward the soma and is still far removed from the site of axonogenesis. These findings suggest that the site of axon formation in THN neurons is independent of a proximally positioned centrosome. Images in D–F are taken from Video 9. MHB, midbrain–hindbrain boundary; URL, upper rhombic lip.
Interestingly, time-lapse imaging of subcellular Medusa-labeled THN neurons revealed the emergence of the axon-like protrusion, which subsequently developed a clear growth cone structure (Fig. 9, D–F; red arrow), at the time when trailing process retraction and centrosome repositioning from the apical germinal zone toward the cell soma were still occurring (n = 9 cells, 8 embryos; Fig. 9, D–F, white arrow; Video 9). This suggests that axonogenesis in THN neurons occurs in parallel to, or shortly after, proliferative INM is terminated.
Axonogenesis in THN neurons is not induced by proximity to the centrosome
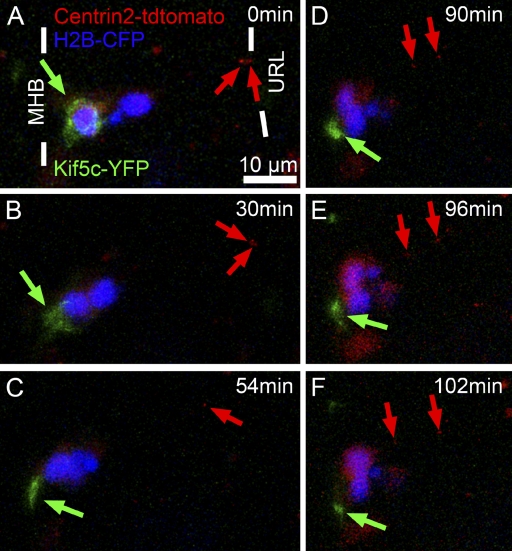
Our subcellular in vivo time-lapse studies of centrosome dynamics, together with the immunohistochemical analysis of axon formation, argue that axonogenesis in zebrafish THN neurons is initiated by a membrane protrusion far away from the microtubule-organizing centrosome. To directly resolve the temporal sequence of axonogenesis and centrosome dynamics in THN neurons in vivo, we made use of the reporter Kif5C560-YFP, which accumulates selectively in the forming axon very soon after axon specification (Jacobson et al., 2006; Reed et al., 2006). The Kif5C560-YFP axon reporter was expressed under UAS control in Tg(atoh1a:Gal4TA4)hzm2 embryos together with the Janus construct J8, demarcating the nucleus by blue and the centrosome by red fluorescence (see Table S1). Therefore, the emergence of THN axons could be visualized in real time relative to the position and movement of the centrosome within the same THN neuron. We purposely chose a coexpression strategy of two different vectors for this experiment, rather than generating a triple-cistron Medusa construct, in order to yield a high degree of mosaicism of transgene expression, allowing for observation of Kif5C560-YFP localization in a single cell of a Janus-labeled group or cluster of THN neurons. We confirmed by immunohistochemistry that such single Kif5C560-YFP–positive processes coexpressed axon-specific acetylated tubulin (unpublished data).
Time-lapse sequences starting at 36 hpf were recorded from THN neurons that had reached the MHB, but showed a fluctuating cytoplasmic distribution of Kif5C560-YFP (Fig. 10 A, green arrow) and had their centrosomes (Fig. 10 A, red arrows) still positioned in the apical URL, indicating that these THN neurons had not yet initiated axonogenesis. Strikingly, Kif5C560-YFP fluorescence soon accumulated in front of the nucleus and close to the MHB (Fig. 10, B and C; green arrow), while the centrosome remained stationary in the URL and far away from the emerging axon (Fig. 10 B, red arrows). Only when axonogenesis was well under way and the outgrowing axon extended ventrally along the MHB did the centrosome begin to detach from the proliferation zone and move toward the nucleus (Fig. 10, D–F; see also Video 10). This temporal sequence of axonogenesis occurring significantly before centrosome movements toward the cell soma and leading edge demonstrates that in vivo proximity to the centrosome is not relevant for axon determination from the leading process of THN neurons. Interestingly, reorientation of the THN neuron from an apico-basal to a dorso-ventral orientation (compare position of nuclei in Fig. 10, C and F) also preceded the arrival of the centrosome in the cell soma (Fig. 10, C–F). This strongly suggests that repolarization during turning behavior of THN neurons is similarly not mediated by repositioning of the centrosome.
Figure 10.
In vivo imaging of axonogenesis. Lateral view of the cerebellar anlage of a 36-hpf Tg(atoh1a:Gal4TA4)hzm2 transgenic zebrafish embryo coinjected with Janus vector J8 (marking nucleus in blue and centrosome in red, red arrows) and 5xUAS:Kif5c-YFP (emerging axons labeled with yellow fluorescence, here shown in green). (A) Due to coinjection of two vectors, only the more anteriorly located cell expresses the Kif5c-YFP fusion protein. Kif5c-YFP is initially distributed throughout the soma of the cell (green arrow), while the centrosomes of both cells are localized at the apical side (red arrows). (B–D) Kif5c-YFP localizes to a protrusion, the later axon, in the front of the cell, at the time when the centrosome is homing toward the soma. (E and F) Kif5c-YFP localizes to a growth cone–like structure of the emerging axon, while the centrosome has not reached the soma. This temporal sequence of axonogenesis and centrosome dynamics reveals that a proximal position of the centrosome is not required for selecting the site of axon formation in THN neurons in vivo. Images are taken from Video 10. MHB, midbrain-hindbrain boundary; URL, upper rhombic lip.
Discussion
In this study, we have expanded the use of Gal4 combinatorial genetics in zebrafish for cell biological characterization by establishing simultaneous expression of multiple subcellular markers in a cell type–specific manner. Enhancer and gene trap screens have already generated a multitude of zebrafish Gal4 activator strains (Davison et al., 2007; Scott et al., 2007; Asakawa et al., 2008; Distel et al., 2009) that could be used for cell type–specific cell biological analysis. The presented collection of subcellular fluorescent reporters and their cassette-like assembly in Janus or Medusa vectors will facilitate Gal4-mediated in vivo cell biology in zebrafish, but also in other vertebrates such as chick embryos (Fig. S4). It has to be noted though, that obtaining high contrast for specific subcellular structures sometimes requires the careful adjustment of the expression levels. For example, strong expression levels for centrosome labeling will lead to saturation effects, with excess fluorescent protein accumulating in the cytoplasm. Low expression levels instead will only sparsely label the centrosome, making it too dim for fast image acquisition. The Gal4 system allows one to fine-tune expression levels by either adjusting the number of UAS sites or by selecting a Gal4 activator with the appropriate transactivation potential (Distel et al., 2009). By this means, optimal labeling results can be achieved.
Medusa labeling showed that once THN neurons have become postmitotic, expressing markers such as PSA-NCAM or acetylated tubulin, and have initiated axonogenesis (Rieger et al., 2008), the apical centrosome-containing process detaches from the germinal URL and moves together with the centrosome toward the MHB-positioned cell somata to initiate neuronal migration. In a common model for tangentially migrating neurons, the centrosome typically moves significantly ahead of the nucleus, establishes an axonal swelling, and organizes the microtubule skeleton. This is followed by the saltatory translocation of the nucleus toward the displaced centrosome (Bellion et al., 2005; Schaar and McConnell, 2005; Métin et al., 2008). Also in zebrafish, tangentially migrating facial branchiomotor neurons display such a leading centrosome, which reorients during migratory turns. Failure to maintain the proper apico-basal positioning of the centrosome is accompanied by migration in ectopic directions, suggesting a role for the centrosome in regulating directional migration (Grant and Moens, 2010). In this respect the ventral migration of THN neurons along the MHB displays an unexpected subcellular orchestration. Only during the preparation for nucleokinesis is the centrosome positioned ahead of the nucleus, being then overtaken during the saltatory forward translocation of the nucleus. For most of the time, the centrosome trailed the nucleus, although it moved at a constant pace even during resting periods of the nucleus. Thereby an iterative cycle of centrosome–nucleus leapfrogging is created.
Interestingly, the role of the centrosome in permanently leading the nucleus of radially migrating neurons has also been called into question recently. In radially migrating cerebellar granule neurons, it was shown that the centrosome is similarly overtaken by the nucleus during nucleokinetic forward movements (Umeshima et al., 2007). In these neurons though, the centrosome advances ahead of the nucleus less regularly than in THN neurons and only during long resting phases. Notably, radial migration occurs along oriented glia fibers, which provide additional cues for the directionality of migration, whereas tangential migration of neurons occurs independently of a guiding glial meshwork. Therefore, tangential THN neuron migration may depend more strongly on a leading centrosome preparing forward migration during each migratory step, whereas radially migrating neurons may not need this consistent directional information from the centrosome. On the other hand, the centrosome in radially migrating cortical neurons does remain strictly ahead of the nucleus (Tsai et al., 2007). Taken together, these findings argue that, like in non-neuronal cells, the exact subcellular orchestration of saltatory nucleokinetic migration is strongly context dependent, varying with the neuronal cell type, local environment, and morphogenetic constraints.
A key role for the centrosome, and in particular its positioning, has been postulated for the induction of axonogenesis in several neuronal populations (Lefcort and Bentley, 1989; Zmuda and Rivas, 1998; Arimura and Kaibuchi, 2007). Findings in cultured hippocampal neurons and cortical neurons in vivo have established that the neurite nearest to the centrosome is selected to become the axon (de Anda et al., 2005, 2010). Subsequent axon extension though, does not require a functional centrosome (Stiess et al., 2010). Our in vivo time-lapse studies show that axonogenesis in THN neurons is initiated during the onset of migration and occurs clearly distant from the centrosome. These observations thus differ from hippocampal and cortical neurons, and THN neurons instead behave like retinal ganglion cells in the zebrafish retina, which initiate an axon from the basal process while retraction of the centrosome-containing apical process is still underway (Zolessi et al., 2006). Thus, in retinal ganglion cells and migratory THN neurons, the position of the centrosome does not predict the site of axonogenesis. Our findings argue that the cellular mechanisms of axonogenesis cannot be generalized and are, similar to migration, dependent on the cell type, morphogenetic constraints, and the makeup of the extracellular environment. For example, a strong influence on cerebellar granule neuron axonogenesis and centrosome positioning is exerted by the composition of the ECM and associated signaling molecules (Gupta et al., 2010), which can vary significantly among neuronal tissues. In THN neurons, cellular and molecular events occurring in the leading process itself may predispose the leading process to later axon formation. These events are probably initiated already during the extended interkinetic nuclear movement step of THN progenitors toward the MHB. Thus, the different molecular and cellular dynamics in the apical and leading processes, as well as their temporal orchestration, need to be further characterized in vivo in different cell types in order to better understand the course of neuronal development from birth to terminal differentiation. The cell type–specific multicolor labeling of individual neuronal cells, as established here, is a promising step in this direction.
Materials and methods
Maintenance of fish
Zebrafish strains were raised and maintained at 27°C in a manufactured fish facility (Aqua Schwarz GmbH, Göttingen, Germany) with circulating and constantly filtered water at 800–1,000 μS salinity (Kimmel et al., 1995; Westerfield, 1995). In the Tg(shhb:Gal4TA4,5xUAS:mRFP)hzm4 strain (TG5xR), the notochord-specific shhb (formerly twhh) promoter element drives the expression of GalTA4, which in turn activates expression of the red fluorescent protein mRFP under control of five UAS sites and the EIb basal promoter (Babaryka et al., 2009; Distel et al., 2009).
Construction of vectors
S14: #801 pCS GFP-DCX.
The ORF encoding GFP-DCX was isolated from the pEGFP-C2DCX vector (a kind gift of Fiona Francis, Institut Cochin, Université Paris Descartes, Paris, France) by Eco47III–SalI digest and cloned into StuI–XhoI-digested pCS2+ (Rupp et al., 1994).
U15: #699 pSK14xUAS:H2B-mRFP.
The ORF encoding H2B-mRFP was isolated from pCS-H2B-mRFP (a kind gift of Sean Megason, Harvard Medical School, Boston, MA) by Asp718 (Klenow blunted)–NsiI digest and cloned behind the E1b promoter of the XhoI (Klenow blunted)–NsiI-digested pSK14xUASE1b vector (Köster and Fraser, 2001b).
U16: #709 pSKH2B-mRFP:5xUASE1b.
The E1b minimal promoter and the ORF encoding H2B-mRFP were isolated from U15 by XbaI–NotI (Klenow blunted) digest and inserted into the SmaI site of the pSK5xUASE1b vector (Distel et al., 2009).
J1: #828 pSK-H2B-RFP:5xUAS:GFP-DCX.
The ORF encoding GFP-DCX was isolated from S14 by ClaI–Asp718 digest and cloned into ClaI–Asp718-digested U16.
U17: #627 pSK14xUAS:H2B-GFP.
H2B-GFP was isolated from pCSH2B-GFP SalI (Klenow blunted)–ApaI digest and cloned behind the E1b promoter of the XhoI (Klenow blunted)–ApaI-digested pSK14xUASE1b vector (Köster and Fraser, 2001b).
U18: #665 pSKH2B-GFP:5xUASE1b.
The E1b minimal promoter and the ORF encoding H2B-GFP were isolated from U17 by XbaI–NotI (Klenow blunted) digest and inserted into the SmaI site of the pSK5xUASE1b vector (Distel et al., 2009).
J11: #700 pSKH2B-GFP:5xUAS:memmRFP.
The ORF encoding memmRFP was isolated from pCSmemmRFP (a kind gift of Sean Megason) by XhoI (Klenow blunted)–ApaI digest and inserted into EcoRV–ApaI-digested U18. “mem“ represents a membrane localization signal, which consists of a plamitylation and myristinylation sequence of the human Lck kinase.
U2: #860 memmRFP:5xUAS.
The E1b minimal promoter and the ORF encoding memmRFP were isolated from J11 by XbaI–NotI (Klenow blunted) digest and inserted into the SmaI site of the pSK5xUASE1b vector (Distel et al., 2009).“mem“ represents a membrane localization signal, which consists of a plamitylation and myristinylation sequence of the human Lck kinase.
# 878 pH2B-CFP.
The ORF encoding H2B was isolated from pCSH2B-mRFP (a kind gift of Sean Megason) by XhoI–AgeI digest and inserted into XhoI–AgeI-digested pECFP-C1 (Takara Bio Inc.).
S5: # 895 pCSH2B-CFP.
The ORF encoding H2B-CFP was isolated from pH2B-CFP by NotI(Klenow blunted)–XhoI digest and cloned into XbaI(Klenow blunted)–XhoI-digested pCS2+.
J2: #939 pSKmemmRFP:5xUAS:H2B-CFP.
The ORF encoding H2B-CFP was isolated from S5 by XhoI–NotI (Klenow blunted) digest and inserted into Asp718-digested (Klenow blunted) U2.
#766 pCRIICentrin2.
The ORF of zebrafish centrin2 (acc. nr.: EU183505) was cloned by RT-PCR to generate C-terminal fusions using total RNA from adult brain and the following primers: HindIII centrin-up: 5′-TTAAGCTTATGGCGTCCGGCTTCAGGAA-3′; centrin-low BamHI: 5′-TAGGATCCCCGTACAGATTGGTTTTCTTCA-3′. The fragment was subcloned into the pCRII-Topo vector (Invitrogen) and sequenced.
#767 pCRIICentrin2STOP.
The ORF of zebrafish centrin2 (acc. nr.: EU183505) was amplified by RT-PCR to generate N-terminal fusions using total RNA from adult brain and the following primers: BamHI centrin-up: 5′-TTGGATCCATGGCGTCCGGCTTCAGGAA-3′; centrin-low XbaI: 5′-TTTCTAGATCAGTACAGATTGGTTTTCTTC-3′. The fragment was subcloned into the pCRII-Topo vector (Invitrogen) and sequenced.
#769 pCentrin2-YFP.
The ORF encoding Centrin2 was isolated by BamHI–HindIII digest from pCRII-Centrin2 and cloned into BamHI–HindIII-digested pEYFP-N1 (Takara Bio Inc.).
S3: #783 pEGFP-Centrin2.
The ORF encoding Centrin2 was isolated by BamHI–EcoRI digest from pCRII-Centrin2Stop and cloned into BglII–EcoRI-digested pEGFP-C1 (Takara Bio Inc.).
S2: #848 pCSCentrin2-YFP.
The ORF encoding Centrin2-YFP was isolated from pCentrin2-YFP by NotI (Klenow blunted)–XhoI digest and inserted into SnaBI–XhoI-digested pCS2+.
U5: #996 pSK5xUAS:Centrin2-YFP.
The ORF encoding Centrin2-YFP was isolated from pCSCentrin2-YFP by EcoRI–ApaI digest and inserted into EcoRI–ApaI-digested pSK5xUAS (Distel et al., 2009).
M1: #1595 pSKmemmRFP:5xUAS:H2B-CFP:5xUAS:Centrin2-YFP.
The 5xUAS:Centrin2-YFP cassette was isolated from U5 by SpeI–Asp718 digest (Klenow blunted) and inserted into SpeI-digested (Klenow blunted) J2.
S6: #938 pCSmemCFP.
mRFP of pCSmemmRFP (a kind gift of Sean Megason) was removed by SnaBI–AgeI digest and replaced with CFP from NotI (Klenow blunted)–AgeI-digested pECFP-1 (Takara Bio Inc.). “mem“ represents a membrane localization signal, which consists of a plamitylation and myristinylation sequence of the human Lck kinase.
U6: #997 pSK5xUAS:memCFP.
The ORF encoding memCFP was isolated from S6 by ClaI–ApaI digest and inserted into ClaI–ApaI-digested pSK5xUAS.
M2: #998 pSKH2B-mRFP:5xUAS:GFP-DCX-5xUAS:memCFP.
The 5xUAS:memCFP cassette was isolated from U6 by NotI (Klenow blunted) and inserted into Asp718-digested (Klenow blunted) J1.
S10: #771 pCSEB3-GFP.
The ORF encoding EB3-GFP was isolated from pEB3-GFP (a kind gift of Anna Akhmanova, Erasmus Medical Center, Rotterdam, Netherlands) by NotI (Klenow blunted)–SalI digest and inserted into XbaI(Klenow blunted)–XhoI-digested pCS2+.
J4: #780 pSKH2B-mRFP:5xUAS:EB3-GFP.
The ORF encoding EB3-GFP was isolated from S10 by StuI–Asp718 digest and inserted into EcoRV–Asp718-digested U16.
M3: #999 pSKH2B-mRFP:5xUAS:EB3-GFP-5xUAS:memCFP.
The 5xUAS:memCFP cassette was isolated from U6 by NotI (Klenow blunted) and inserted into Asp718-digested (Klenow blunted) J4.
#868 pCStdTomato.
The ORF encoding tdTomato was isolated from pRSETtdTomato (a kind gift of Roger Tsien, University of California, San Diego, La Jolla, CA) by BamHI–EcoRI digest and inserted into BamHI–EcoRI-digested pCS2+.
S1: #879 pCSCentrin2-tdtomato.
The ORF encoding Centrin2 was isolated from pCRII-Centrin2 by BamHI–HindIII digest and inserted into BamHI–HindIII-digested pCStdTomato.
#1532 pSKE1B5xUASE1b.
The E1b5xUASE1b cassette was isolated from U2 by EcoRI digest and inserted into EcoRI-digested pBSK.
U9: #2022 pSKH2B-CFP:5xUASE1b.
The ORF encoding H2B-CFP was isolated from pCSH2B-CFP by Asp718 digest and inserted into SmaI-digested pSKE1b5xUASE1b.
J8: #2146 pSKH2B-CFP:5xUAS:Centrin2-tdTomato.
The ORF encoding Centrin2-tdTomato was isolated from S1 by HindIII–Asp718 digest and inserted into HindIII–Asp718-digested U9.
pSC-BKif5c-YFP.
Kif5c-YFP was PCR amplified from Kif5c-YFP (a kind gift of Gary Banker, Oregon Health and Science University, Portland, OR) and a Kozak sequence was added using primers Kif5cEcoHinforKo: 5′-AAAGAATTCAAGCTTCCACCATGGCAGATCCAGCCGAATGCAGCATC-3′; and Venus4: 5′-TACTCGAGTTACTTGTACAGCTCGTCCAT-3′ and subcloned into pSC-B (Agilent Technologies).
S20: # 2328pCSKif5c-YFP.
The ORF encoding Kif5c-YFP was isolated from pSC-BKif5c-YFP by EcoRI (Klenow blunted)–HindIII digest and inserted into HindIII–SnaBI-digested pCS2+.
U14: #2329 pSK5xUAS:Kif5c-YFP.
The ORF encoding Kif5c-YFP was isolated from pCSKif5c-YFP by HindIII–Asp718 digest and inserted into HindIII–Asp718-digested pSK5xUAS
Further cloning strategies are available upon request.
Generation of atoh1a:KalTA4GI transgenic zebrafish
To express transgenes in rhombic lip–derived cells, KalTA4 was placed between up- and downstream regulatory elements of zebrafish atonal 1a. A 2950-bp fragment upstream of atoh1a was amplified from a BAC (RZPD, CH211-247L22) using primers #394: 5′-GCGGTCGACAATGGGACTGTATGGATGTTTCCC-3′ and #396: 5′-TGCGGATCCTCTGTTGGTTTGTGCTTTTGGGAG-3′. Likewise, a 5900-bp fragment downstream of atoh1a was amplified by using primers #395: 5′-ATAGCGGCCGCTTCTCGCCTCACTCGCACTTCA-3′ and #397: 5′-GCGCCGCGGAGCTTTGGGTTTAGTTCGGTAAGACTG-3′ for 1–3250 bp and #398: 5′-GACGGAGACCGCAGGTTTATTTCTCACAGAAG-3′ and #399: 5′-ATACCGCGGGCTATCTTGGTTACATTGATATGC-3′ for 3250–5900 bp and joining these fragments after subcloning by SacII digest. KalTA4GIpA (Distel et al., 2009) was inserted between the 5′ and 3′ fragments. This construct was flanked with I-Sce-I recognition sites and injected into one-cell stage zebrafish embryos together with mRNA coding for I-Sce-I at the one-cell stage (Babaryka et al., 2009). Injected zebrafish embryos were raised to adulthood and tested for successful integration by mating to Tg(shhb:Gal4TA4,5xUAS:mRFP)hzm4 or Tg(4xUAS:GFP)hzm3 transgenic fish (Babaryka et al., 2009; Distel et al., 2009). Tg(atoh1a:Gal4TA4)hzm2 carriers showing fluorescence expression in the rhombic lip were maintained up to the F4 generation.
Microinjection
Zebrafish embryos were injected with expression plasmids (25 ng/µl each, 1.5 nL) at the one-cell stage. Raised embryos were screened for expression right before microscopy analysis.
Microscopy
For image recording, embryos were dechorionated and embedded in 1.2% ultra low melting agarose/30% Danieau (Distel and Köster, 2007). Images of living embryos and of transfected cells were recorded using a confocal microscope (LSM 510; Carl Zeiss, Inc.) and LSM software (Carl Zeiss, Inc.). Images of in situ hybridizations were recorded using an Axioplan2 microscope equipped with an AxioCam HRc and Axiovision 4.5 software (all from Carl Zeiss, Inc.). Images in Fig. 1 and Fig. 2 B (M2, M3) were recorded using a 63x Plan-Apochromat oil immersion objective (NA 1.4); in Fig. 2, A and B (M1), Fig. 3, D–F, Figs. 5, 6, 7, 9, and 10 using a 40x C-Apochromat water immersion objective (NA 1.2); and in Fig. 3, A–C and Fig. 4 A using a 20x EC Plan-Neofluar objective (NA 0.5). Images in Figs. 4–7, Fig. 9, D–F, and Fig. 10 represent maximum intensity projections taken from respective videos of time-lapse analysis.
Quantification of subcellular dynamics
Migration angle analysis.
In a lateral view of the cerebellum, the apico-basal axis was determined by drawing a horizontal line from the URL to the MHB, perpendicular to the MHB. The nuclear centroids of THN progenitors were tracked using the Manual Tracking tool of ImageJ software (NIH, Bethesda, MD). The angle between the migration path of the respective cell and the apico-basal axis of the cerebellum was determined in THN progenitors undergoing INM and neurons initiating ventral migration using Adobe Photoshop CS3.
Centrosome and nuclear centroid tracking.
Centrosome and nuclear centroid tracking over time and measurements of migration distances and migration velocities of the respective organelles were performed on maximum projections of the respective z-stacks using the Manual Tracking tool of ImageJ software.
Generation of kymographs.
The kymographs in Fig. 8, A and B were generated manually using Adobe Photoshop CS4. 2D maximum projections shown in Videos 6 and 8 were rotated 45 degrees counterclockwise and regions containing the nuclei (Video 6, Fig. 8 A) or containing the nucleus and the centrosome (Video 8, Fig. 8 B) were extracted for each time point. Cropped regions were assembled in one image starting with the earliest time point on the left.
Determination of the position of the centrosome with respect to the nuclear centroid and migration direction.
A 0.4-µm-thick line was drawn through the nuclear centroid and orthogonal to the direction of cell migration, dividing the cell into two halves. The position of the centrosome was then scored manually to be either in the anterior half (anterior to the nuclear centroid), in the posterior half (posterior to the nuclear centroid), or on the line (scored as lateral to the nuclear centroid). Cells were analyzed during ventral migration, starting at the time point when the centrosome had reached the soma of the cell and ending when the cell no longer showed apparent migration. The pie chart in Fig. 8 was generated in Microsoft Excel.
Determination of directions of centrosomal and nuclear movements.
To determine the direction of centrosomal and nuclear movements during ventral migration, images were rotated where necessary so that cells migrated approximately along the y axis. Afterward, the y position of the respective cell organelle was determined using ImageJ (Measure tool or Manual Tracking plug-in). Subsequent y positions were substracted (and corrected for tissue growth if neccessary) to yield the net movement. Movements less than 0.5 µm were scored as stationary. Greater movements were scored as in the direction of migration (forward) or as opposite to the direction of migration (backward). The 3D bar graph of the respective movement combinations was generated in Microsoft Excel.
Transfection
Zebrafish Pac2 fibroblasts were maintained in Leibovitz L15 medium supplemented with 1x l-glutamine (Invitrogen) and 10% fetal bovine serum. Pac2 cells were transfected using the Effectene transfection kit (QIAGEN) or the Nanofectin transfection kit (PAA; Senghaas and Köster, 2009).
Immunohistochemistry
GFP-expressing offspring of Tg(atoh1a:Gal4TA4)hzm2 and Tg(4xUAS:GFP)hzm3 transgenic carriers (Distel et al., 2009) were fixed in 4% PFA at 36 and 42 hpf for 12 h and transferred into 100% MeOH. Embryos were then transferred to acetone (−20°C), incubated at −20°C for 7 min, and then incubated in H2O at room temperature for 1 h. Subsequently, embryos were washed with PBS/0.1% Tween (PTW) twice for 5 min each, after which embryos were incubated in 10 mM sodium citrate, pH 6.0/0.1% Tween for 15 min at 100°C for antigen retrieval. Subsequently, embryos were washed twice in PTW for 5 min each and blocked in 10% goat serum in PTW for 1 h at room temperature. Embryos were incubated with the primary antibodies chicken anti-GFP (1:500; Aves, catalog no. 1020) and mouse anti-acetylated tubulin (1:500; Sigma-Aldrich, catalog no. T6793) at 4°C overnight. After several washes in PTW, embryos were incubated with the secondary antibodies anti–mouse Alexa546 (1:100; Invitrogen) and anti–chicken FITC (1:100; Jackson ImmunoResearch Laboratories, Inc., catalog no. 703 095 155) overnight at 4°C. Nuclei were stained using DAPI (1 µg/µl; Roche, catalog no. 10236276001).
Online supplemental material
Online supplemental material is provided with this manuscript, including a table of expression constructs and their identifier numbers, which should be used for construct ordering. In addition, figures showing expression from a Janus construct using a single UAS-site (Fig. S1), anti-PH3 immunohistochemistry for detecting proliferating cells in the developing cerebellum (Fig. S2), a quantification of the ventral turning angle of migrating THN neurons (Fig. S3), and triple-cistronic Medusa vector expression in the developing chick tegementum (Fig. S4) are provided. Finally, the supplemental material contains all of the videos and their respective legends referred to in this manuscript. Video 1 visualizes MT dynamics in a zebrafish keratinocyte, Videos 2 and 3 visualize MT dynamics in cells of a gastrulating zebrafish embryo, Video 4 shows a time-lapse recording of THN neuron migration, Video 5 shows nucleus and centrosome dynamics during interkinetic nuclear movements of THN progenitors, Videos 6 and 7 show trailing process retraction and centrosome dynamics in THN neurons preparing to migrate ventrally, Video 8 shows centrosome dynamics in a ventrally migrating THN neuron, Video 9 shows a time-lapse recording of an emerging axon of a THN neuron, and Video 10 shows the localization of the axon specific marker Kif5c-YFP and the centrosome during THN axonogenesis.
Acknowledgments
We are grateful for excellent fish husbandry and technical assistance by Petra Hammerl, Enrico Kühn, and Christiane Lach. We thank Karsten Boldt for help with Western blot quantification and Damian Refojo for carefully reading and commenting on the manuscript. We thank Fiona Francis, Alexander Reugels, Jürgen Wehland, Anna Akhmanova, Sean Megason, Timm Schroeder, and Roger Tsien for generously providing us with different expression vectors.
M. Distel, J.C. Hocking, K. Volkmann, and R.W. Köster designed the experiments and performed the data analysis; M. Distel, J.C. Hocking, and K. Volkmann performed the experiments; M. Distel, J.C. Hocking, and R.W. Köster wrote the manuscript.
This work was generously supported by a BioFuture Award Grant of the German Ministry of Education and Research (BMBF 0311889), the German Research Association (DFG, KO1949/3-1), a Fellowship of the Studienstiftung des deutschen Volkes (M. Distel), and a postdoctoral fellowship from the Natural Sciences and Engineering Research Council (NSERC) of Canada (J.C. Hocking).
Footnotes
Abbreviations used in this paper:
- hpf
- hours postfertilization
- INM
- interkinetic nuclear movement
- MHB
- midbrain–hindbrain boundary
- MTOC
- microtubule-organizing center
- THN
- tegmental hindbrain nuclei
- UAS
- upstream activating sequence
- URL
- upper rhombic lip
References
- Arimura N., Kaibuchi K. 2007. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat. Rev. Neurosci. 8:194–205 10.1038/nrn2056 [DOI] [PubMed] [Google Scholar]
- Asakawa K., Suster M.L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M., Kawakami K. 2008. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA. 105:1255–1260 10.1073/pnas.0704963105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaryka A., Kühn E., Köster R.W. 2009. In vivo synthesis of meganuclease for generating transgenic zebrafish Danio rerio. J. Fish Biol. 74:452–457 10.1111/j.1095-8649.2008.02075.x [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. 2006. Flies without centrioles. Cell. 125:1375–1386 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bellion A., Baudoin J.P., Alvarez C., Bornens M., Métin C. 2005. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J. Neurosci. 25:5691–5699 10.1523/JNEUROSCI.1030-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A., Zhang Y.A., Chang B.T., McConnell S.K. 1998. Intrinsic polarity of mammalian neuroepithelial cells. Mol. Cell. Neurosci. 11:183–193 10.1006/mcne.1998.0680 [DOI] [PubMed] [Google Scholar]
- Coan D.E., Wechezak A.R., Viggers R.F., Sauvage L.R. 1993. Effect of shear stress upon localization of the Golgi apparatus and microtubule organizing center in isolated cultured endothelial cells. J. Cell Sci. 104:1145–1153 [DOI] [PubMed] [Google Scholar]
- Conde C., Cáceres A. 2009. Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 10:319–332 10.1038/nrn2631 [DOI] [PubMed] [Google Scholar]
- Davison J.M., Akitake C.M., Goll M.G., Rhee J.M., Gosse N., Baier H., Halpern M.E., Leach S.D., Parsons M.J. 2007. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304:811–824 10.1016/j.ydbio.2007.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda F.C., Pollarolo G., Da Silva J.S., Camoletto P.G., Feiguin F., Dotti C.G. 2005. Centrosome localization determines neuronal polarity. Nature. 436:704–708 10.1038/nature03811 [DOI] [PubMed] [Google Scholar]
- de Anda F.C., Meletis K., Ge X., Rei D., Tsai L.-H. 2010. Centrosome motility is essential for initial axon formation in the neocortex. J. Neurosci. 30:10391–10406 10.1523/JNEUROSCI.0381-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M., Köster R.W. 2007. In vivo time-lapse imaging of zebrafish embryonic development. Cold Spring Harbor Protocols. 10.1101/pdb.prot4816 [DOI] [PubMed] [Google Scholar]
- Distel M., Wullimann M.F., Köster R.W. 2009. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl. Acad. Sci. USA. 106:13365–13370 10.1073/pnas.0903060106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotlieb A.I., May L.M., Subrahmanyan L., Kalnins V.I. 1981. Distribution of microtubule organizing centers in migrating sheets of endothelial cells. J. Cell Biol. 91:589–594 10.1083/jcb.91.2.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P.K., Moens C.B. 2010. The neuroepithelial basement membrane serves as a boundary and a substrate for neuron migration in the zebrafish hindbrain. Neural Dev. 5:9 10.1186/1749-8104-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.K., Meiri K.F., Mahfooz K., Bharti U., Mani S. 2010. Coordination between extrinsic extracellular matrix cues and intrinsic responses to orient the centrosome in polarizing cerebellar granule neurons. J. Neurosci. 30:2755–2766 10.1523/JNEUROSCI.4218-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.W., Cai D., Verhey K.J. 2008. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 20:71–76 10.1016/j.ceb.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H.R., Gleeson J.G. 2007. The centrosome in neuronal development. Trends Neurosci. 30:276–283 10.1016/j.tins.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Hinds J.W., Ruffet T.L. 1971. Cell proliferation in the neural tube: an electron microscopic and Golgi analysis in the mouse cerebral vesicle. Zeitschrift für Zellforschung. 115:226–264 10.1007/BF00391127 [DOI] [PubMed] [Google Scholar]
- Jacobson C., Schnapp B., Banker G.A. 2006. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 49:797–804 10.1016/j.neuron.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310 [DOI] [PubMed] [Google Scholar]
- Köster R.W., Fraser S.E. 2001a. Direct imaging of in vivo neuronal migration in the developing cerebellum. Curr. Biol. 11:1858–1863 10.1016/S0960-9822(01)00585-1 [DOI] [PubMed] [Google Scholar]
- Köster R.W., Fraser S.E. 2001b. Tracing transgene expression in living zebrafish embryos. Dev. Biol. 233:329–346 10.1006/dbio.2001.0242 [DOI] [PubMed] [Google Scholar]
- Kupfer A., Louvard D., Singer S.J. 1982. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc. Natl. Acad. Sci. USA. 79:2603–2607 10.1073/pnas.79.8.2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Chang M.I., Tseng Y., Wirtz D. 2005. Cdc42 mediates nucleus movement and MTOC polarization in Swiss 3T3 fibroblasts under mechanical shear stress. Mol. Biol. Cell. 16:871–880 10.1091/mbc.E03-12-0910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefcort F., Bentley D. 1989. Organization of cytoskeletal elements and organelles preceding growth cone emergence from an identified neuron in situ. J. Cell Biol. 108:1737–1749 10.1083/jcb.108.5.1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O., Valdeolmillos M., Moya F. 2006. Neurons in motion: same principles for different shapes? Trends Neurosci. 29:655–661 10.1016/j.tins.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Métin C., Baudoin J.P., Rakić S., Parnavelas J.G. 2006. Cell and molecular mechanisms involved in the migration of cortical interneurons. Eur. J. Neurosci. 23:894–900 10.1111/j.1460-9568.2006.04630.x [DOI] [PubMed] [Google Scholar]
- Métin C., Vallee R.B., Rakic P., Bhide P.G. 2008. Modes and mishaps of neuronal migration in the mammalian brain. J. Neurosci. 28:11746–11752 10.1523/JNEUROSCI.3860-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemere I., Kupfer A., Singer S.J. 1985. Reorientation of the Golgi apparatus and the microtubule-organizing center inside macrophages subjected to a chemotactic gradient. Cell Motil. 5:17–29 10.1002/cm.970050103 [DOI] [PubMed] [Google Scholar]
- Paquet D., Bhat R., Sydow A., Mandelkow E.M., Berg S., Hellberg S., Fälting J., Distel M., Köster R.W., Schmid B., Haass C. 2009. A zebrafish model of tauopathy allows in vivo imaging of neuronal cell death and drug evaluation. J. Clin. Invest. 119:1382–1395 10.1172/JCI37537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouthas F., Girard P., Lecaudey V., Ly T.B., Gilmour D., Boulin C., Pepperkok R., Reynaud E.G. 2008. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J. Cell Sci. 121:2406–2414 10.1242/jcs.026849 [DOI] [PubMed] [Google Scholar]
- Pu J., Zhao M. 2005. Golgi polarization in a strong electric field. J. Cell Sci. 118:1117–1128 10.1242/jcs.01646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed N.A., Cai D., Blasius T.L., Jih G.T., Meyhofer E., Gaertig J., Verhey K.J. 2006. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16:2166–2172 10.1016/j.cub.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Renaud J., Kerjan G., Sumita I., Zagar Y., Georget V., Kim D., Fouquet C., Suda K., Sanbo M., Suto F., et al. 2008. Plexin-A2 and its ligand, Sema6A, control nucleus-centrosome coupling in migrating granule cells. Nat. Neurosci. 11:440–449 10.1038/nn2064 [DOI] [PubMed] [Google Scholar]
- Rieger S., Volkmann K., Köster R.W. 2008. Polysialyltransferase expression is linked to neuronal migration in the developing and adult zebrafish. Dev. Dyn. 237:276–285 10.1002/dvdy.21410 [DOI] [PubMed] [Google Scholar]
- Rupp R.A.W., Snider L., Weintraub H. 1994. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8:1311–1323 10.1101/gad.8.11.1311 [DOI] [PubMed] [Google Scholar]
- Schaar B.T., McConnell S.K. 2005. Cytoskeletal coordination during neuronal migration. Proc. Natl. Acad. Sci. USA. 102:13652–13657 10.1073/pnas.0506008102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M., Euteneuer U., Gräf R., Ueda M. 1999. Centrosomes, microtubules and cell migration. Biochem. Soc. Symp. 65:223–231 [PubMed] [Google Scholar]
- Schütze K., Maniotis A., Schliwa M. 1991. The position of the microtubule-organizing center in directionally migrating fibroblasts depends on the nature of the substratum. Proc. Natl. Acad. Sci. USA. 88:8367–8371 10.1073/pnas.88.19.8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E.K., Mason L., Arrenberg A.B., Ziv L., Gosse N.J., Xiao T., Chi N.C., Asakawa K., Kawakami K., Baier H. 2007. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat. Methods. 4:323–326 [DOI] [PubMed] [Google Scholar]
- Senghaas N., Köster R.W. 2009. Culturing and transfecting Pac2 zebrafish fibroblast cells. Cold Spring Harbor Protocols. 10.1101/pdb.prot5235 [DOI] [PubMed] [Google Scholar]
- Shoukimas G.M., Hinds J.W. 1978. The development of the cerebral cortex in the embryonic mouse: an electron microscopic serial section analysis. J. Comp. Neurol. 179:795–830 10.1002/cne.901790407 [DOI] [PubMed] [Google Scholar]
- Solecki D.J., Model L., Gaetz J., Kapoor T.M., Hatten M.E. 2004. Par6alpha signaling controls glial-guided neuronal migration. Nat. Neurosci. 7:1195–1203 10.1038/nn1332 [DOI] [PubMed] [Google Scholar]
- Stiess M., Maghelli N., Kapitein L.C., Gomis-Rüth S., Wilsch-Bräuninger M., Hoogenraad C.C., Tolić-Nørrelykke I.M., Bradke F. 2010. Axon extension occurs independently of centrosomal microtubule nucleation. Science. 327:704–707 10.1126/science.1182179 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Serneo F.F., Higgins C., Gambello M.J., Wynshaw-Boris A., Gleeson J.G. 2004. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J. Cell Biol. 165:709–721 10.1083/jcb.200309025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai L.H., Gleeson J.G. 2005. Nucleokinesis in neuronal migration. Neuron. 46:383–388 10.1016/j.neuron.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Tsai J.W., Bremner K.H., Vallee R.B. 2007. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 10:970–979 10.1038/nn1934 [DOI] [PubMed] [Google Scholar]
- Umeshima H., Hirano T., Kengaku M. 2007. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc. Natl. Acad. Sci. USA. 104:16182–16187 10.1073/pnas.0708047104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee R.B., Seale G.E., Tsai J.W. 2009. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 19:347–355 10.1016/j.tcb.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann K., Chen Y.-Y., Harris M.P., Wullimann M.F., Köster R.W. 2010. The zebrafish cerebellar upper rhombic lip generates tegmental hindbrain nuclei by long-distance migration in an evolutionary conserved manner. J. Comp. Neurol. 518:2794–2817 [DOI] [PubMed] [Google Scholar]
- Westerfield M. 1995. The Zebrafish Book. University of Oregon Press, Eugene, OR: 385 pp [Google Scholar]
- Witte H., Bradke F. 2008. The role of the cytoskeleton during neuronal polarization. Curr. Opin. Neurobiol. 18:479–487 10.1016/j.conb.2008.09.019 [DOI] [PubMed] [Google Scholar]
- Xie Z., Moy L.Y., Sanada K., Zhou Y., Buchman J.J., Tsai L.H. 2007. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 56:79–93 10.1016/j.neuron.2007.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvon A.-M.C., Walker J.W., Danowski B., Fagerstrom C., Khodjakov A., Wadsworth P. 2002. Centrosome reorientation in wound-edge cells is cell type specific. Mol. Biol. Cell. 13:1871–1880 10.1091/mbc.01-11-0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Song B., Pu J., Wada T., Reid B., Tai G., Wang F., Guo A., Walczysko P., Gu Y., et al. 2006. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature. 442:457–460 10.1038/nature04925 [DOI] [PubMed] [Google Scholar]
- Zmuda J.F., Rivas R.J. 1998. The Golgi apparatus and the centrosome are localized to the sites of newly emerging axons in cerebellar granule neurons in vitro. Cell Motil. Cytoskeleton. 41:18–38 [DOI] [PubMed] [Google Scholar]
- Zolessi F.R., Poggi L., Wilkinson C.J., Chien C.B., Harris W.A. 2006. Polarization and orientation of retinal ganglion cells in vivo. Neural Dev. 1:2 10.1186/1749-8104-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]