Degradation of Plk1 during mitotic exit allows recruitment of the Cep55 abscission factor and timely cytokinesis.
Abstract
Cytokinesis requires a membrane-remodeling and fission event termed abscission that occurs after chromosome segregation, cleavage furrow formation, and contraction have completed. In this study, we show how abscission factor recruitment is controlled by the Polo-like kinase 1 (Plk1). At the metaphase–anaphase transition, Plk1 initiates cleavage furrow formation and is then progressively degraded during mitotic exit. During this period, Plk1 phosphorylates the abscission factor Cep55 in trans and prevents its untimely recruitment to the anaphase spindle. A Plk1 phosphorylation site mutant of Cep55 is prematurely recruited to the anaphase spindle and fails to support abscission. Endogenous Cep55 behaves similarly after Plk1 inhibition by the drugs BI2536 or GW842862. Only once Plk1 is degraded can Cep55 target to the midbody and promote abscission. Blocking Plk1 degradation leads to elevated levels of Plk1 at the midbody and the failure of Cep55 recruitment. Thus, Plk1 activity negatively regulates Cep55 to ensure orderly abscission factor recruitment and ensures that this occurs only once cell contraction has completed.
Introduction
Polo-like kinase 1 (Plk1) is a conserved protein kinase controlling many of the key events of mitosis and cytokinesis (Barr et al., 2004). In prometaphase and metaphase, it is found on centrosomes and kinetochores where it promotes formation of a bipolar mitotic spindle (Lane and Nigg, 1996; Liu and Erikson, 2002; Sumara et al., 2004). Subsequently, it relocates to the central spindle in anaphase and activates Rho-GTPase regulators, initiating cleavage furrow formation and cell contractility (Golsteyn et al., 1995; Brennan et al., 2007; Burkard et al., 2007, 2009; Petronczki et al., 2007; Santamaria et al., 2007; Wolfe et al., 2009). A conserved phosphopeptide-binding domain, the Polo-box domain, is responsible for this complex pattern of spatial and temporal control (Cheng et al., 2003; Elia et al., 2003a,b). Through this domain, Polo kinases bind to phosphorylated partners containing a conserved motif (Elia et al., 2003a). This leads to concentration at particular sites and increased levels of kinase activity. According to the prevailing model, Cdk1–cyclin B creates these docking sites in prophase and metaphase (Cheng et al., 2003; Elia et al., 2003a,b), whereas in anaphase, self-priming by Plk1 itself predominates (Neef et al., 2007; Burkard et al., 2009). In anaphase, Plk1 self-primes a docking site at the C terminus of the anaphase spindle and microtubule-associated protein PRC1 on T602 and is then recruited to the central spindle (Neef et al., 2007).
Consistent with these mitosis-specific functions, Plk1 is slowly degraded as cells exit mitosis and is essentially absent by the time cells undergo cytokinesis (Golsteyn et al., 1994, 1995; Lindon and Pines, 2004). The anaphase-promoting complex/cyclosome–Cdh1 ubiquitin ligase is required for Plk1 degradation (Lindon and Pines, 2004). Anaphase-promoting complex/cyclosome–Cdh1 recognizes Plk1 as a target for ubiquitylation through a conserved destruction box (D-box) motif and, thus, earmarks it for proteasomal degradation (Lindon and Pines, 2004). However, although degradation of Plk1 as animal cells exit mitosis is clearly important in the control of mitotic exit and cytokinesis (Lindon and Pines, 2004), the mechanistic detail of its function at this time remains unclear.
Cytokinesis is terminated by a membrane-remodeling and fission event termed abscission that is mediated by the ESCRT-III membrane–remodeling proteins (Carlton and Martin-Serrano, 2007), which is an ancestral part of the cytokinesis machinery shared with Archaea (Samson et al., 2008). In dividing mammalian cells, ESCRT-III is specifically nucleated on the surface of the preassembled midbody by the Cep55 adaptor protein. Cep55 directly interacts with the central MKlp1 component of the midbody and the ESCRT proteins ALIX and TSG101 (Fabbro et al., 2005; Martinez-Garay et al., 2006; Zhao et al., 2006; Carlton and Martin-Serrano, 2007; Morita et al., 2007; Carlton et al., 2008; Lee et al., 2008). This series of events, whereby abscission components are recruited only after midbody formation when the plasma membrane is pulled down to a narrow 0.5-µm-diameter tube of membrane connecting the two daughter cells, may be caused by the limitations of the membrane-remodeling properties of ESCRT-III (Wollert et al., 2009). At earlier times, when the plasma membrane is less closely apposed and has a larger radius of curvature, ESCRT-III cannot promote the membrane-remodeling event leading to abscission. In addition, other membrane delivery and microtubule-remodeling events have to occur before abscission can be triggered (Kouranti et al., 2006; Barr and Gruneberg, 2007; Simon et al., 2008). Thus, timing is critical for efficient abscission, yet it remains unclear how this is achieved.
Results and discussion
Identification of Plk1-sensitive midbody components
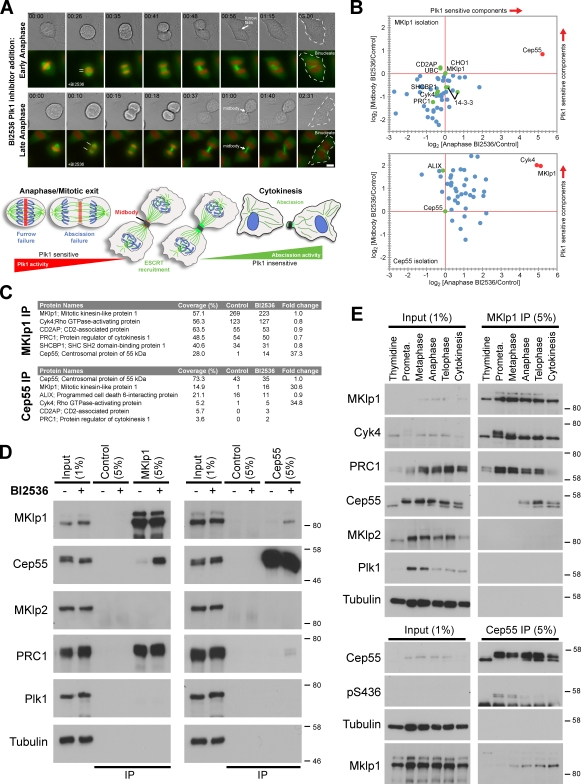
Previous studies have indicated that Plk1 has unexplained functions in the late stages of cytokinesis (Lindon and Pines, 2004; Santamaria et al., 2007). To confirm this idea, the rapid-acting Plk1 inhibitor BI2536 was used (Lénárt et al., 2007; Steegmaier et al., 2007). When Plk1 was inhibited as chromosome segregation initiated in early anaphase, cleavage furrow formation failed (Fig. 1 A, top). However, although furrow formation was not obviously perturbed when BI2536 was added as chromosomes reached the maximum point of segregation later in anaphase, cytokinesis ultimately failed and the cells became binucleate (Fig. 1 A, bottom). These observations suggest that Plk1 may function as the control of abscission.
Figure 1.
Plk1 phosphorylation negatively regulates the interaction of Cep55 with the midbody component MKlp1. (A, top) HeLa cells stably expressing EGFP-tagged α-tubulin (green) and mCherry-tagged histone H2B (red) were treated with 1 µM BI256. Still images from videos are indicated with arrows to mark the position of the midbody and intracellular bridge. Bar, 10 µm. (bottom) The model depicts the decay in Plk1 levels and activity in anaphase while chromosome segregation by the microtubule spindle is taking place and the outcomes of inhibiting Plk1 in early or late anaphase. Lines at the second time point indicate the extent of chromosome segregation. Dotted lines in the last time point mark the outline of the binucleate cells. (B) HeLa cells in anaphase or undergoing cytokinesis were treated with DMSO (control) or 1 µM BI2536 for 25 min. Cep55 and MKlp1 complexes were isolated with sheep antibodies and analyzed by quantitative nano liquid chromatography MS/MS. The ratio of identified proteins in BI2536 and control conditions was plotted for anaphase versus midbody stages. Green dots indicate midbody proteins, red dots mark proteins increased after Plk1 inhibition in anaphase, and blue dots indicate other proteins. (C) The response of midbody proteins to Plk1 inhibition is summarized in the table. The number of peptides found is listed in the control and BI2536 columns. Fold change in abundance based on the summed intensities of the peptides forming each protein group was calculated using MaxQuant. IP, immunoprecipitation. (D) The same samples were Western blotted. (E) Western blotting of MKlp1 and Cep55 complexes isolated from synchronized HeLa cells at the different stages of mitosis. Western blot markers are given in kilodaltons.
To identify central spindle and midbody proteins whose levels alter according to the activity of Plk1, MKlp1 complexes were isolated from early anaphase cells treated with solvent control or BI2536 to inhibit Plk1 and then analyzed by mass spectrometry (MS). To give a second dimension to the analysis, this was repeated for cells at a later stage where midbodies had formed. MKlp1 is predominantly in a complex with the Rho family GAP Cyk4 and PRC1, and these proteins cluster together about the center of the plot (Fig. 1 B). Plk1 inhibition with BI2536 shifts Cep55 away from this cluster, corresponding to an increase of >37-fold (Fig. 1, B–D), indicating that it is a Plk1-sensitive component of MKlp1 complexes. Reciprocal isolation of Cep55 was performed to corroborate these observations. This showed that Cep55 was found predominantly in a complex with the ESCRT protein ALIX in control cells (Fig. 1 C). After BI2536 treatment, >30-fold increases in the amounts of MKlp1 and Cyk4 were observed (Fig. 1, B–D). MKlp1 and Cyk4 were the only components altered by Plk1 inhibition in this way, which is consistent with these proteins forming a heterooligomeric complex (Mishima et al., 2002). Other midbody components such as the 14–3-3 proteins (Douglas et al., 2010) were not altered by Plk1 inhibition (Fig. 1 B). To investigate whether the Cep55–MKlp1 interaction occurs in normal cytokinesis in the absence of Plk1 inhibitors, MKlp1 complexes were isolated throughout mitosis and cytokinesis. Cep55 was only precipitated together with MKlp1 from anaphase onwards when phosphorylation on Cep55 at the conserved Plk1 site at S436 was lost (Fig. 1 E). Therefore, Plk1 phosphorylation may negatively regulate the interaction of Cep55 with the midbody component MKlp1 in anaphase cells and could provide an inhibitory signal preventing premature abscission factor recruitment.
Central spindle–targeted Plk1 is a negative regulator of Cep55 recruitment
To provide further evidence that Plk1 is a specific regulator of Cep55, cells were treated with specific chemical inhibitors to Cdk1 (flavopiridol; Potapova et al., 2006), aurora B (ZM447439; Ditchfield et al., 2003), and Plk1 (BI2536 or GW843682; Lansing et al., 2007; Lénárt et al., 2007). In control cells, Cep55 showed the expected pattern of late recruitment to the midbody in cytokinesis (Fig. 2 A, Fig. S1 A, and Video 1). Strikingly, Plk1 inhibition with BI2536 or GW843682 resulted in premature recruitment of Cep55 to the central spindle in anaphase and telophase (Fig. 2 A; and Fig. S1, A–C). The Cep55 partner and ESCRT-III component ALIX was also prematurely recruited to the central spindle in a Cep55-dependent manner after Plk1 inhibition with BI2536 (Fig. S2, A and B). Premature recruitment of Cep55 to the anaphase spindle was not seen with Cdk1 and aurora B inhibitors; although after these treatments cells showed chromosome segregation errors, they still formed Cep55-positive midbodies during cytokinesis (Fig. 2 A; and Fig. S1, D and E).
Figure 2.
Plk1 phosphorylation negatively regulates central spindle recruitment of Cep55. (A) HeLa cells treated with DMSO, 1 µM BI2536, 5 µM flavopiridol, or 1 µM ZM447439 for 25 min were stained for DNA with DAPI and mouse anti–α-tubulin, rabbit anti-Cep55 (green), and goat anti-Plk1 (red). (B) HeLa cells transiently expressing EGFP-PRC1 or the Plk1-binding defective ST602AA mutant were transfected with control or PRC1 3′ untranslated region (UTR) siRNAs for 36 h. Cells were stained for DNA with DAPI, sheep anti-Cep55 (false-colored green in the merge), and goat anti-Plk1 (red). PRC1 was visualized using EGFP fluorescence. (C and D) Cep55 (C) and MKlp1 (D) complexes were isolated using sheep antibodies from mitotic HeLa cells treated with DMSO (control), 1 µM BI2536, or 1 µM ZM447439 for 25 min and then Western blotted. The asterisks indicate cross-reactivity to the heavy chain of the antibody used for immunoprecipitation. Western blot markers are given in kilodaltons. Bars, 10 µm.
In anaphase, Plk1 self-primes its own binding to the anaphase spindle microtubule-associated protein PRC1 at T602 (Neef et al., 2007). As predicted by this model, when cells are treated with the Plk1 inhibitor BI2536 for 5 min, PRC1 T602 phosphorylation is lost, and Plk1 is absent from the central spindle in anaphase and telophase (Figs. 2 A and S3). The absence of Plk1 correlated with the appearance of Cep55 at the central spindle under these conditions (Fig. 2 A). Furthermore, cells expressing only a Plk1-binding defective ST602AA mutant of PRC1 were unable to recruit Plk1 to the central spindle in anaphase yet showed premature recruitment of Cep55 (Fig. 2 B). These observations provide further support both for the model that PRC1 is a major structural partner for Plk1 at the central spindle and the role of Plk1 as a negative regulator of Cep55 recruitment.
Plk1 phosphorylates Cep55 on S436 within a C-terminal region containing the binding site for MKlp1 (Fabbro et al., 2005; Zhao et al., 2006). Cep55 complexes isolated from early anaphase cells treated with solvent control, BI2536, or ZM447439 were therefore Western blotted with Cep55 pS436 antibodies. This revealed that endogenous Cep55 phosphorylation at S436 is sensitive to Plk1 but not aurora B inhibition and that MKlp1 is only present when S436 phosphorylation is absent (Fig. 2 C). These same lysates were used for the isolation of MKlp1 complexes. This revealed that the Cep55 present in MKlp1 complexes was not phosphorylated at S436 (Fig. 2, compare C and D). To test the functional consequences of this phosphorylation by Plk1, the conserved Plk1 sites in Cep55 were mutated, and stable doxycycline-inducible cell lines produced expressing EGFP-tagged forms of the wild-type and mutant forms of Cep55. The N-terminal T46A, S47A double mutant behaved as the wild-type protein and targeted to the midbody only late in cytokinesis (Fig. 3 A). This is the same behavior seen with Cep55 antibodies (Fig. 2 A). In contrast, the S436A mutant was present at the central spindle from early anaphase (Fig. 3 A). This recapitulates the behavior of wild-type Cep55 when Plk1 is inhibited (Fig. 2 A). Live cell imaging also showed that Cep55 S436A was prematurely recruited to the central spindle and then accumulated to higher levels than the wild-type protein at the midbody (Fig. 3 B). Typically, Cep55 was recruited 60 min after the onset of anaphase, whereas Cep55 S436A was visible on the central spindle after 5–10 min (Fig. 3 B, bar graph). These cells remained arrested at the midbody stage for many hours (Fig. 3 B, insets). Measurements of Cep55 levels at the central spindle and midbody confirmed that Cep55 S436A recruitment occurs with more rapid kinetics and accumulates to higher levels than the wild-type protein (Fig. 3 C). These observations support the proposal that Plk1 is a negative regulator of Cep55 recruitment.
Figure 3.
Cep55 S436A is prematurely recruited to the central spindle in anaphase. (A) HeLa cells transiently expressing EGFP-tagged wild-type, TS46, 47AA, and S436A mutant Cep55 were stained for DNA with DAPI and mouse anti–α-tubulin. Bar, 10 µm. (B, left) HeLa cells stably expressing doxycycline-inducible mCherry-Cep55 or the S436A mutant of Cep55 were imaged as they exited mitosis. Insets in phase-contrast images show enlargements of the midbody region (arrows). Times are shown in hours and minutes from the onset of anaphase. Bar, 5 µm. (right) The time of Cep55 recruitment from the start of anaphase was measured in wild-type and S436A-expressing cells and is plotted as a bar graph (n = 10). Error bars indicate SD. (C) The fluorescence intensity of Cep55 was measured for the full 3D central spindle and midbody volume using the quantitation and tracking software and is plotted against time for both wild-type Cep55 and the S436A mutant–expressing cells. Arrows indicate the point of metaphase to anaphase transition obtained by inspection of the chromosomes in phase-contrast images. All timings are set relative to this point.
Plk1 degradation is required for Cep55 recruitment to the midbody
A key aspect of the model proposed in this study is that Plk1 degradation acts as a timer for Cep55 recruitment. Accordingly, preventing Plk1 degradation with the proteasomal inhibitor MG132 resulted in increased levels of Plk1 at MKlp1 midbody structures after 120-min treatment compared with control cells (Fig. 4 A). As expected, Cep55 was absent from these Plk1-positive midbody structures (Fig. 4 A). To investigate this in more detail, inducible stable cells lines expressing EGFP-tagged Cep55 and either wild-type or D-box mutant mCherry-tagged Plk1 were created. Live cell imaging confirmed that there is an inverse correlation between Plk1 and Cep55 levels at the midbody during cytokinesis (Fig. 4, B [left] and D). Furthermore, MG132 reduces Plk1 degradation and, thus, prevents recruitment of Cep55 to the midbody. Inhibition of this MG132-stabilized Plk1 triggers immediate recruitment of Cep55 to the midbody (Fig. 4, B [right] and D), supporting the idea that Plk1 is a key factor stabilized by MG132 addition. Cep55 recruitment to the midbody was also delayed in cells expressing the Plk1 D-box mutant compared with controls expressing wild-type Plk1 (Fig. 4, C and E). This was matched by a pronounced delay in cytokinesis in Plk1 D-box–expressing cells (Fig. 4 C, bar graph). No changes in the timing of chromosome segregation or furrowing were observed (Fig. 4 C, bar graph). These findings are consistent with the idea that destruction of Plk1 provides a mechanism for controlling the recruitment of Cep55 and thus the timing of abscission.
Figure 4.
Plk1 destruction controls the timing of Cep55 recruitment to the midbody. (A) HeLa cells were mock treated or treated with 100 µM MG132 for 120 min and stained for DNA with DAPI and mouse anti–α-tubulin, rabbit anti-Cep55 or MKlp1 (green), and goat anti-Plk1 (red). Bars, 10 µm. The number of Plk1-positive and Cep55-negative midbody structures in the presence and absence of 100 µM MG132 was counted for 120 cells in each of three independent experiments. (B and C) HeLa cells stably expressing mCherry-Cep55 and either doxycycline-inducible EGFP-tagged wild-type Plk1 or the D-box mutant of Plk1 were imaged every minute with a spinning-disk confocal microscope. At the times indicated (arrows), 100 µM MG132 or 1 µM BI2536 was added. Fluorescence intensity of Plk1 and Cep55 was measured for the full central spindle and midbody volume and is plotted against time. A bar graph shows the times spent in the different stages of mitosis for 15 cells in three independent experiments. (D and E) Still images of Cep55 behavior in control, MG132, and BI2536 (D) cells and wild-type and D-box mutant Plk1 (E) cells are shown. Times are shown in hours:minutes. Error bars indicate SD. Bar, 5 µm.
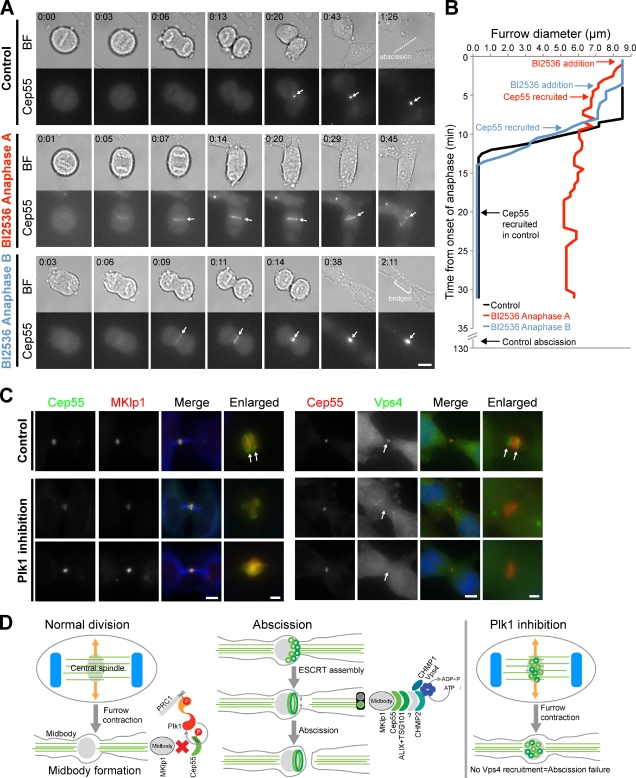
Premature Cep55 recruitment results in aberrant midbody structures
Live cell imaging of EGFP-Cep55 cell lines revealed that furrowing and midbody formation only fail to occur when Plk1 was inhibited in anaphase A (Fig. 5 A). Once chromosome segregation has occurred, cells treated with BI2536 in anaphase B completed furrowing and formed a midbody with timings similar to control cells (Fig. 5 B) but remained connected by a bridge of cytoplasm and did not complete cytokinesis (Fig. 5 A). Importantly, rapid and untimely recruitment of Cep55 to the central spindle was observed irrespective of the time of Plk1 inhibition in anaphase A or B (Fig. 5, A and B). Control cells treated with the solvent DMSO alone showed normal timing of Cep55 recruitment only once furrowing was completed (Fig. 5, A and B). These findings suggest that after Plk1 inhibition or upon expression of a Cep55 phosphorylation mutant, cytokinesis fails because of misregulation of a late-acting component of the abscission machinery. To test this idea, the detailed localization of midbody and ESCRT-III components was investigated. When Plk1 was inhibited in anaphase using BI2536, midbodies had an abnormal architecture (Fig. 5 C). Instead of forming two concentric rings on either side of the MKlp1-positive midbody structure, Cep55 either formed multilobed structures or a single large structure overlapping with MKlp1 (Fig. 5 C). Furthermore, although positive for the ESCRT-III component ALIX, these midbodies (Fig. S2 B) were functionally abnormal and did not recruit the Vps4 AAA-ATPase required for the final step of abscission (Fig. 5 C). These observations support the hypothesis that Plk1 is acting as an important regulator of abscission by preventing Cep55 recruitment to the central spindle earlier in anaphase and telophase. Interference with the Plk1 control mechanism leads to Cep55 recruitment before midbody formation, which perturbs normal midbody architecture and leads to the failure of the Vps4-dependent abscission process (Fig. 5 D).
Figure 5.
Plk1 is required for furrow formation and abscission. (A) HeLa cells expressing mCherry-Cep55 were imaged as they exited mitosis after treatment with either DMSO or 1 µM BI2536. Fluorescence (Cep55) and brightfield (BF) still images from the live cell imaging shown in the bottom panels. BI2536 was added as chromosome segregation started in anaphase A or after chromosome segregation, but before furrowing in anaphase B. A dotted line marks abscission in the controls samples. Arrows indicate Cep55 recruitment to the central spindle or midbody. Times are shown in hours:minutes from the start of anaphase, defined as the time point before chromosome segregation was first detected. (B) The graph shows furrow diameter from the onset of anaphase measure in minutes. Arrows show the time of Cep55 recruitment and abscission, if this occurred. (C) HeLa cells treated with DMSO or 1 µM BI2536 for 25 min were stained for DNA with DAPI, mouse anti–α-tubulin (blue), rabbit anti-Cep55 (green), and sheep anti-MKlp1 (red). (right) HeLa cells expressing Vps4-EGFP and Cep55-mCherry were treated with DMSO or BI2536 for 25 min before fixation. Arrows mark the rings of Cep55 and Vps4 flanking the midbody position. (D) A model summarizing how the timely recruitment of Cep55 may be important for proper ESCRT function at the midbody. Bars: (main panels) 5 µm; (enlargements) 1 µm.
Plk1 is a negative regulator of abscission factors in human cells
In this study, we have provided evidence showing that Plk1 is a negative regulator of abscission factors. By phosphorylating Cep55 at the conserved S436, Plk1 prevents it from interacting with the central spindle and midbody protein MKlp1 in anaphase cells. Proteomic analysis indicates that Plk1 does not regulate the interaction of Cep55 with other ESCRT and midbody components. Only once the midbody has formed and Plk1 levels are reduced does Cep55 recruitment occur in two concentric rings on either side of the midbody core. This timing is critical because premature recruitment of Cep55 leads to abnormal midbody architecture and abscission failure. Critically, the final component of the ESCRT machinery Vps4 does not recognize these abnormal midbodies. This suggests a simple mechanism for the timing of abscission relative to furrowing and provides a reason why the previously described degradation properties of Plk1 are important for its function during exit from mitosis (Lindon and Pines, 2004). When Plk1 is active at the central spindle, it initiates and maintains the cleavage furrow while simultaneously preventing abscission factor recruitment. Plk1 is then degraded by the proteasome and falls to a level unable to sustain this inhibition of abscission. Previous work has shown that once cells have started anaphase, inhibiting proteasome-mediated degradation with the drug MG132 causes abscission to fail and cells to become binucleate (Straight et al., 2003). This is consistent with both the data presented in this study and observations that Plk1 degradation is necessary for orderly mitotic exit (Lindon and Pines, 2004). A further aspect of the Plk1 regulation pathway may involve the peptidylprolyl isomerase Pin1, although this is unlikely to be cell essential (van der Horst and Khanna, 2009; van der Horst et al., 2009). Pin1 is not an essential gene, whereas cytokinesis and abscission are essential processes needed for the production of new cells during early embryonic development and in the adult. It therefore seems unlikely that Pin1 regulation is critical for Cep55 function. However, Pin1 may play a role in controlling the level of the Plk1-phosphorylated form of Cep55 (van der Horst and Khanna, 2009; van der Horst et al., 2009). A model in which Pin1 fine tunes Cep55 levels before abscission could reconcile these observations. In the absence of Pin1, excess Cep55 might accumulate and interfere with the abscission machinery under some conditions. Consistent with this idea, the Cep55 S436A Plk1 phosphorylation-defective mutant progressively accumulates at the midbody and appears to block cytokinesis.
Cep55 has been reported to be an essential regulator of cytokinesis controlled by Cdk1 and Plk1, and the findings in this study are broadly in agreement with these previous studies (Fabbro et al., 2005; Martinez-Garay et al., 2006; Zhao et al., 2006). However, in these studies, Plk1 was suggested to be a positive regulator of Cep55 function (Fabbro et al., 2005; Zhao et al., 2006). In contrast, we show that S436 phosphorylation by Plk1 negatively regulates Cep55 recruitment to the central spindle in anaphase. Consistent with this view, experiments using two independent antibodies and tagged proteins in inducible stable cell lines show that Cep55 is absent from the central spindle in anaphase, whereas Plk1 is lost from the midbody in cells undergoing cytokinesis. Plk1 has been reported to dock with Cep55 via two Cdk1/ERK phosphorylation sites at S425 and S428, but this is not consistent with an anaphase function for this interaction. As previously shown, Plk1 self-primes its own docking sites in anaphase, and Plk1 inhibition results in a loss of Plk1 targeting in anaphase (Neef et al., 2007; Burkard et al., 2009). A further concern is that although these sites conform with a general Cdk1/ERK phosphorylation consensus, they do not match the Plk1-docking consensus based on a series of biochemical studies and x-ray crystal structures, which mandates that a serine precedes the phosphorylated docking site residue (Cheng et al., 2003; Elia et al., 2003a,b). Proteomic analysis of anaphase Cep55 complexes also failed to reveal any Plk1, and these two proteins are recruited at different times to the central spindle and midbody as cells exit mitosis. Thus, Cep55 is not a docking partner for Plk1 and is therefore phosphorylated in trans by the PRC1–Plk1 complex at the central spindle.
Materials and methods
Reagents and antibodies
General laboratory chemicals were obtained from Sigma-Aldrich and Thermo Fisher Scientific. Antibodies to Cep55 (1–222 aa), Cep55 pS436 (peptide antigen, ALNEpSLVE; sheep only), MKlp1 (24–146 aa), Vps4 (1–129 aa), and MKlp2 (63–193 aa) were raised in sheep and rabbit using peptides or hexahistidine-tagged human proteins expressed in and purified from bacteria. Specific antibodies were purified using the antigens conjugated to Affigel-15, eluted with 0.2 M glycine, pH 2.8, then dialyzed against PBS before storage at −80°C. Rabbit antibodies to Plk1, astrin, PRC1, PRC1 pT602, and MKlp1 pS911 have been described previously (Neef et al., 2003, 2006, 2007; Thein et al., 2007). Commercially available antibodies were used to α-tubulin (mouse DM1A; Sigma-Aldrich), Plk1 (mouse SC-17783; Santa Cruz Biotechnology, Inc.), aurora B (mouse AIM1; BD), and cyclin B1 (mouse GNS3; Millipore). Kinase inhibitors were obtained from Sigma-Aldrich (5 mM flavopiridol 1,000× stock and 1 mM GW843862 1,000× stock), Tocris Bioscience (100 mM ZM447439 10,000× stock), and Axon Medchem (1 mM BI2536 1,000× stock).
Molecular biology
Human Cep55, ALIX, MKlp1, MKlp2, and Vps4 were amplified by PCR from Image clones (Source Bioscience) using KOD polymerase (Takara Bio Inc.). Mammalian expression constructs for Cep55, ALIX, Vps4, α-tubulin, and histone H2B were made using pcDNA4/TO and pcDNA5/FRT/TO vectors (Invitrogen) modified to encode the mCherry or EGFP reading frames. Bacterial expression constructs were made in pQE32 (QIAGEN). Mutagenesis was performed using the QuikChange method (Agilent Technologies). Primers were obtained from Metabion GmbH. Table S1 lists the siRNA duplexes used in this study.
Cell culture
HeLa cells were cultured in growth medium (DME containing 10% bovine calf serum) at 37°C and 5% CO2. For plasmid transfection and siRNA transfection, LT1 (Mirus Bio LLC) and Oligofectamine (Invitrogen) were used, respectively. Stable HeLa cell lines with single copies of the desired transgene were created using the T-Rex doxycycline-inducible Flp-In system (Invitrogen). HeLa cells stably expressing mCherry–histone H2B and EGFP–α-tubulin were provided by K. Zeng (University of Liverpool, Liverpool, England, UK). For synchronization, cells were treated for 18 h with 2 mM thymidine and washed three times in PBS and twice with growth medium.
Protein complex purification
Three 15-cm dishes of synchronized HeLa S3 cells per condition were released for 3 h at 37°C, then 100 ng/ml nocodazole was added and incubated further for 16 h. Mitotic cells were collected by shake off, and nocodazole was removed by washing three times with prewarmed PBS and twice with growth medium. At each time point, cells were collected and washed three times with ice-cold PBS. Cell pellets were resuspended in lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% IGEPAL, 0.1% sodium deoxycholate, 40 mM β-glycerophosphate, 10 mM NaF, 0.3 mM Na vanadate, 100 nM okadaic acid, and protease inhibitor cocktail [Sigma-Aldrich]), left for 15 min on ice, and clarified by centrifugation at 20,000 gav for 20 min at 4°C. Protein complexes were isolated from 3 mg of cell lysate using 3 µg sheep antibodies against either mCherry, EGFP, Cep55, or Mklp1 bound to 20 µl protein G–Sepharose by incubation for 2 h at 4°C. Isolated complexes were washed three times with lysis buffer then twice with 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% IGEPAL. Samples were analyzed by SDS-PAGE, MS, and Western blotting. Sample preparation for MS was performed as described previously (Yoshimura et al., 2010). MaxQuant and Mascot (Matrix Science) were used to compile and search the raw data against the human International Protein Index database. Protein group and peptide lists were sorted and analyzed in Excel (Microsoft) and MaxQuant (Cox and Mann, 2008). MS and MS/MS spectra were inspected using Xcalibur Qualbrowser (Thermo Fisher Scientific).
Live and fixed cell microscopy
Fixed cells on glass slides were imaged using a 60× 1.35 NA oil immersion objective on a standard upright laboratory microscope with a camera (CoolSNAP HQ2; Roper Industries) under the control of MetaMorph software (version 7.5; MDS Analytical Technologies). A staging system was used to identify the different phases of mitosis and cytokinesis based on the DNA and spindle morphology and extent of chromosome alignment and separation (Fig. 2 B). For live cell imaging, cells were plated in 2-cm dishes with a cover glass window in the bottom. Imaging was performed at 37°C in 5% CO2 using an inverted microscope with a 60× 1.42 NA oil immersion objective, an electron multiplying charge-coupled device camera (QuantEM 512; Roper Industries), an XY motorized z stage (Piezo; Applied Scientific Instrumentation), and a lamp (Lambda DG4; Sutter Instrument Co.) under the control of MetaMorph software or a spinning-disk confocal system (Ultraview Vox; PerkinElmer). Image stacks of 25–35 planes spaced 0.5–0.7 µm were taken at one to four stage positions every minute for 2–12 h. Exposure times were 10–33 ms for EGFP- or mCherry-tagged histone H2B, Cep55, Plk1, and tubulin using 1% of full lamp intensity or 3% laser power. A brightfield reference image stack was collected using 10-ms exposures. Maximum intensity projection images of the fluorescent channels were cropped in ImageJ (National Institutes of Health) and placed into Illustrator (CS3; Adobe) to produce the figures. Intensity measurements were performed on the full 3D dataset using the volume quantitation and object tracking tools of Volocity (version 5; PerkinElmer).
Online supplemental material
Figs. S1–S3 show additional experiments relating to Plk1 regulation of Cep55 and the ESCRT protein ALIX. Video 1 shows the timing of Cep55 recruitment to the midbody late during cytokinesis in living cells. Table S1 lists the siRNA duplexes used in the study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008108/DC1.
Acknowledgments
We thank Dr. Ulrike Gruneberg and our other colleagues for much helpful discussion and advice during the course of this work.
This work was supported by the Cancer Research UK Programme (grant C20079/A9473 to F.A. Barr). The North West Cancer Research Fund generously provided support for equipment used in this study.
Footnotes
Abbreviations used in this paper:
- D-box
- destruction box
- MS
- mass spectrometry
- Plk1
- Polo-like kinase 1
References
- Barr F.A., Gruneberg U. 2007. Cytokinesis: placing and making the final cut. Cell. 131:847–860 10.1016/j.cell.2007.11.011 [DOI] [PubMed] [Google Scholar]
- Barr F.A., Silljé H.H., Nigg E.A. 2004. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5:429–440 10.1038/nrm1401 [DOI] [PubMed] [Google Scholar]
- Brennan I.M., Peters U., Kapoor T.M., Straight A.F. 2007. Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS One. 2:e409 10.1371/journal.pone.0000409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard M.E., Randall C.L., Larochelle S., Zhang C., Shokat K.M., Fisher R.P., Jallepalli P.V. 2007. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 104:4383–4388 10.1073/pnas.0701140104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard M.E., Maciejowski J., Rodriguez-Bravo V., Repka M., Lowery D.M., Clauser K.R., Zhang C., Shokat K.M., Carr S.A., Yaffe M.B., Jallepalli P.V. 2009. Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 7:e1000111 10.1371/journal.pbio.1000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.G., Martin-Serrano J. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 316:1908–1912 10.1126/science.1143422 [DOI] [PubMed] [Google Scholar]
- Carlton J.G., Agromayor M., Martin-Serrano J. 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. USA. 105:10541–10546 10.1073/pnas.0802008105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K.Y., Lowe E.D., Sinclair J., Nigg E.A., Johnson L.N. 2003. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 22:5757–5768 10.1093/emboj/cdg558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas M.E., Davies T., Joseph N., Mishima M. 2010. Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr. Biol. 20:927–933 10.1016/j.cub.2010.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A.E., Cantley L.C., Yaffe M.B. 2003a. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 299:1228–1231 10.1126/science.1079079 [DOI] [PubMed] [Google Scholar]
- Elia A.E., Rellos P., Haire L.F., Chao J.W., Ivins F.J., Hoepker K., Mohammad D., Cantley L.C., Smerdon S.J., Yaffe M.B. 2003b. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 115:83–95 10.1016/S0092-8674(03)00725-6 [DOI] [PubMed] [Google Scholar]
- Fabbro M., Zhou B.B., Takahashi M., Sarcevic B., Lal P., Graham M.E., Gabrielli B.G., Robinson P.J., Nigg E.A., Ono Y., Khanna K.K. 2005. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell. 9:477–488 10.1016/j.devcel.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Golsteyn R.M., Schultz S.J., Bartek J., Ziemiecki A., Ried T., Nigg E.A. 1994. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J. Cell Sci. 107:1509–1517 [DOI] [PubMed] [Google Scholar]
- Golsteyn R.M., Mundt K.E., Fry A.M., Nigg E.A. 1995. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol. 129:1617–1628 10.1083/jcb.129.6.1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouranti I., Sachse M., Arouche N., Goud B., Echard A. 2006. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol. 16:1719–1725 10.1016/j.cub.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Lane H.A., Nigg E.A. 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 135:1701–1713 10.1083/jcb.135.6.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansing T.J., McConnell R.T., Duckett D.R., Spehar G.M., Knick V.B., Hassler D.F., Noro N., Furuta M., Emmitte K.A., Gilmer T.M., et al. 2007. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol. Cancer Ther. 6:450–459 10.1158/1535-7163.MCT-06-0543 [DOI] [PubMed] [Google Scholar]
- Lee H.H., Elia N., Ghirlando R., Lippincott-Schwartz J., Hurley J.H. 2008. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 322:576–580 10.1126/science.1162042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lénárt P., Petronczki M., Steegmaier M., Di Fiore B., Lipp J.J., Hoffmann M., Rettig W.J., Kraut N., Peters J.M. 2007. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 17:304–315 10.1016/j.cub.2006.12.046 [DOI] [PubMed] [Google Scholar]
- Lindon C., Pines J. 2004. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J. Cell Biol. 164:233–241 10.1083/jcb.200309035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Erikson R.L. 2002. Activation of Cdc2/cyclin B and inhibition of centrosome amplification in cells depleted of Plk1 by siRNA. Proc. Natl. Acad. Sci. USA. 99:8672–8676 10.1073/pnas.132269599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garay I., Rustom A., Gerdes H.H., Kutsche K. 2006. The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics. 87:243–253 10.1016/j.ygeno.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Mishima M., Kaitna S., Glotzer M. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2:41–54 10.1016/S1534-5807(01)00110-1 [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.Y., Morham S.G., Gygi S.P., Rodesch C.K., Sundquist W.I. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227 10.1038/sj.emboj.7601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R., Preisinger C., Sutcliffe J., Kopajtich R., Nigg E.A., Mayer T.U., Barr F.A. 2003. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162:863–875 10.1083/jcb.200306009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef R., Klein U.R., Kopajtich R., Barr F.A. 2006. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr. Biol. 16:301–307 10.1016/j.cub.2005.12.030 [DOI] [PubMed] [Google Scholar]
- Neef R., Gruneberg U., Kopajtich R., Li X., Nigg E.A., Sillje H., Barr F.A. 2007. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat. Cell Biol. 9:436–444 10.1038/ncb1557 [DOI] [PubMed] [Google Scholar]
- Petronczki M., Glotzer M., Kraut N., Peters J.M. 2007. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev. Cell. 12:713–725 10.1016/j.devcel.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Potapova T.A., Daum J.R., Pittman B.D., Hudson J.R., Jones T.N., Satinover D.L., Stukenberg P.T., Gorbsky G.J. 2006. The reversibility of mitotic exit in vertebrate cells. Nature. 440:954–958 10.1038/nature04652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.Y., Obita T., Freund S.M., Williams R.L., Bell S.D. 2008. A role for the ESCRT system in cell division in archaea. Science. 322:1710–1713 10.1126/science.1165322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria A., Neef R., Eberspächer U., Eis K., Husemann M., Mumberg D., Prechtl S., Schulze V., Siemeister G., Wortmann L., et al. 2007. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol. Biol. Cell. 18:4024–4036 10.1091/mbc.E07-05-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G.C., Schonteich E., Wu C.C., Piekny A., Ekiert D., Yu X., Gould G.W., Glotzer M., Prekeris R. 2008. Sequential Cyk-4 binding to ECT2 and FIP3 regulates cleavage furrow ingression and abscission during cytokinesis. EMBO J. 27:1791–1803 10.1038/emboj.2008.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M., Hoffmann M., Baum A., Lénárt P., Petronczki M., Krssák M., Gürtler U., Garin-Chesa P., Lieb S., Quant J., et al. 2007. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 17:316–322 10.1016/j.cub.2006.12.037 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Cheung A., Limouze J., Chen I., Westwood N.J., Sellers J.R., Mitchison T.J. 2003. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 299:1743–1747 10.1126/science.1081412 [DOI] [PubMed] [Google Scholar]
- Sumara I., Giménez-Abián J.F., Gerlich D., Hirota T., Kraft C., de la Torre C., Ellenberg J., Peters J.M. 2004. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 14:1712–1722 10.1016/j.cub.2004.09.049 [DOI] [PubMed] [Google Scholar]
- Thein K.H., Kleylein-Sohn J., Nigg E.A., Gruneberg U. 2007. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J. Cell Biol. 178:345–354 10.1083/jcb.200701163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A., Khanna K.K. 2009. The peptidyl-prolyl isomerase Pin1 regulates cytokinesis through Cep55. Cancer Res. 69:6651–6659 10.1158/0008-5472.CAN-09-0825 [DOI] [PubMed] [Google Scholar]
- van der Horst A., Simmons J., Khanna K.K. 2009. Cep55 stabilization is required for normal execution of cytokinesis. Cell Cycle. 8:3742–3749 10.4161/cc.8.22.10047 [DOI] [PubMed] [Google Scholar]
- Wolfe B.A., Takaki T., Petronczki M., Glotzer M. 2009. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 7:e1000110 10.1371/journal.pbio.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J.H. 2009. Membrane scission by the ESCRT-III complex. Nature. 458:172–177 10.1038/nature07836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura S., Gerondopoulos A., Linford A., Rigden D.J., Barr F.A. 2010. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J. Cell Biol. 191:367–381 10.1083/jcb.201008051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.M., Seki A., Fang G. 2006. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell. 17:3881–3896 10.1091/mbc.E06-01-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]