Abstract
DNA chromosomal DSBs (double-strand breaks) are potentially hazardous DNA lesions, and their accurate repair is essential for the successful maintenance and propagation of genetic information. Two major pathways have evolved to repair DSBs: HR (homologous recombination) and NHEJ (non-homologous end-joining). Depending on the context in which the break is encountered, HR and NHEJ may either compete or co-operate to fix DSBs in eukaryotic cells. Defects in either pathway are strongly associated with human disease, including immunodeficiency and cancer predisposition. Here we review the current knowledge of how NHEJ and HR are controlled in somatic mammalian cells, and discuss the role of the chromatin context in regulating each pathway. We also review evidence for both co-operation and competition between the two pathways.
Keywords: double-strand break (DSB), double-strand break repair (DSB repair), homologous recombination, mammalian DNA repair, non-homologous end-joining (NHEJ)
INTRODUCTION
Chromosomal DSBs (double-strand breaks) can result from either endogenous or exogenous sources. Naturally occurring DSBs are generated spontaneously during DNA synthesis when the replication fork encounters a damaged template and during certain specialized cellular processes, including V(D)J recombination, class-switch recombination at the immunoglobulin heavy chain (IgH) locus and meiosis. In addition, exposure of cells to ionizing radiation (X-rays and gamma rays), UV light, topoisomerase poisons or radiomimetic drugs can produce DSBs and other types of DNA damage [1]. The ends of a DSB may contain additional chemical modifications, potentially requiring processing prior to the engagement of canonical DSB repair enzymes.
Failure to repair DSBs, or their misrepair, may result in cell death or chromosomal rearrangements, including deletions and translocations. This chromosomal instability can promote carcinogenesis and accelerate aging. Two major pathways have evolved to repair DSBs and thereby suppress genomic instability. Repair by HR (homologous recombination) can be error-free, but requires the presence of a homologous template, such as a sister chromatid (reviewed in [2,3]). The NHEJ (non-homologous end-joining) pathway joins the two ends of a DSB through a process largely independent of homology. Depending on the specific sequences and chemical modifications generated at the DSB, NHEJ may be precise or mutagenic (reviewed in [4]). Inherited defects in NHEJ account for approx. 15% of human severe combined immunodeficiency [4], whereas inherited defects in HR contribute to a variety of human cancers [5] (Table 1).
Table 1. Disease and cancer incidence associated with impaired DSB repair.
A summary of some human hereditary cancer predisposition syndromes that are known to be associated with germ-line mutation of individual DSB repair genes. The chromosome location is the location of the human gene on the chromosome. AD, autosomal dominant; AR, autosomal recessive; EBV, Epstein–Barr virus.
| Gene | Inheritance | Chromosome location | Disease association | Cancer predisposition |
|---|---|---|---|---|
| Ku70 | AR | 22q11-13 | T-cell lymphomas | |
| Ku80 | AR | 2q35 | Pro-B cell lymphomas | |
| Artemis | AR | 10p | Severe combined immunodeficiency | EBV-associated lymphomas |
| DNA ligase IV | AR | 13q22-24 | Ligase IV (LIG4) syndrome | Lymphoid malignancies |
| Cernunnos (XLF) | AR | 2q35 | Severe combined immunodeficiency | |
| BRCA1 | AD | 17q21 | Hereditary breast cancer | Breast and ovarian cancer |
| BRCA2 (FANCD1) | AD (AR) | 13q12.3 | Hereditary breast cancer (Fanconi anaemia) | Breast and ovarian cancer |
| Mre11 | AR | 11q21 | Ataxia telangiectasia-like disorder | |
| Rad50 | AR | 5q31 | Slightly elevated breast cancer risk | |
| Nbs1 | AR | 8q21 | Nijmegen breakage syndrome | Haematologic malignancies |
| ATM | AR | 11q22.3 | Ataxia telangiectasia | Haematologic malignancies |
| ATR | AR | 3q22-24 | Seckel syndrome | |
| FANCD2 | AR | 3p25.3 | Fanconi anaemia | Acute myeloid leukaemia, squamous cell carcinoma |
| BLM | AR | 15q26.1 | Bloom’s syndrome | Broad spectrum (leukaemia, lymphoma, carcinoma) |
| PALB2 (FANCN) | AR | 16p12 | Fanconi anaemia | Breast and ovarian cancer |
| BRIP1 (BACH1/FANCJ) | AR | 17q22 | Fanconi anaemia | Breast cancer |
| CHEK2 | AR | 22q12.1 | Li–Fraumeni syndrome 2 | Breast cancer, sarcomas, brain tumours |
| WRN | AR | 8p12-11.2 | Werner’s syndrome | Sarcomas |
NON-HOMOLOGOUS END-JOINING
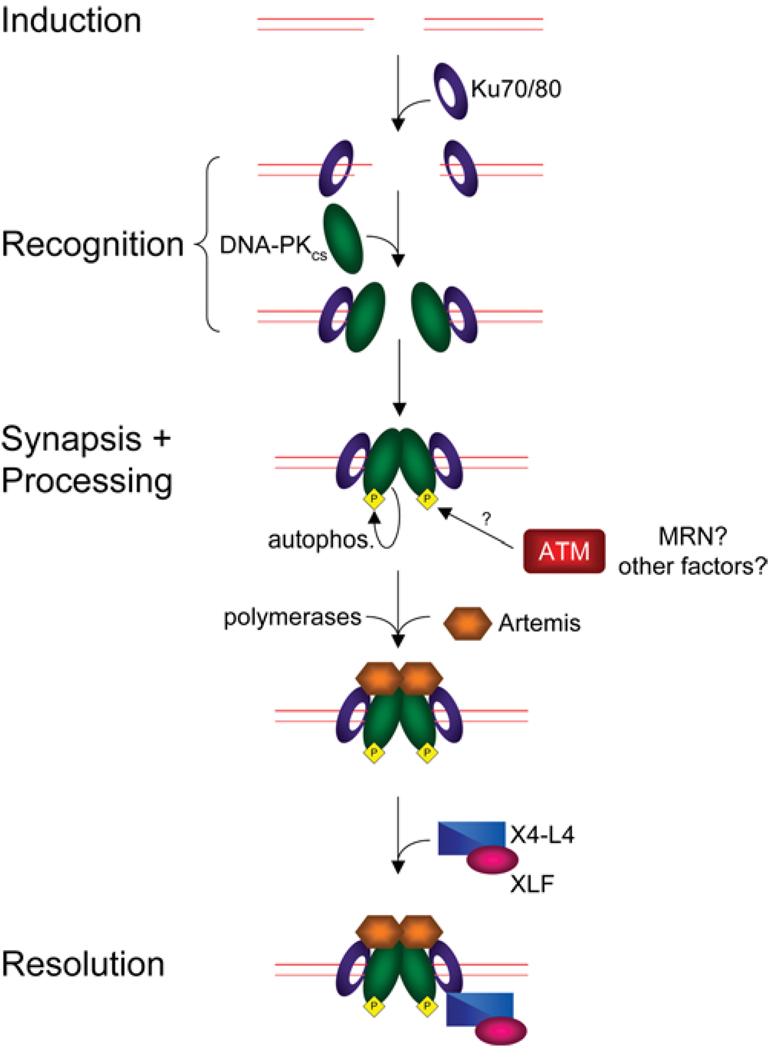
NHEJ is an efficient DSB repair pathway in multicellular eukaryotes such as mice and humans. NHEJ provides a mechanism for the repair of DSBs throughout the cell cycle, but is of particular importance during G0-, G1- and early S-phase of mitotic cells [6,7]. Briefly, the Ku70/Ku80 (Ku) protein binds with high affinity to DNA termini in a structure-specific manner and can promote end alignment of the two DNA ends [8,9]. The DNA-bound Ku heterodimer recruits DNA-PKcs (DNA-dependent protein kinase catalytic subunit), and activates its kinase function [10]. Together with the Artemis protein, DNA-PKcs can stimulate processing of the DNA ends [11]. Finally, the XRCC4 (X-ray repair complementing defective repair in Chinese hamster cells 4)–DNA ligase IV complex, which does not form a stable complex with DNA but interacts stably with the Ku–DNA complex, carries out the ligation step to complete repair [12] (Figure 1). A great deal is known about the process of NHEJ; however, due to space constraints we can only cover a small portion of the relevant literature.
Figure 1. NHEJ in mammalian cells.
Induction of a DSB forms DNA ends that are bound by the Ku heterodimer. Ku translocates inwards, allowing recruitment of DNA-PKcs to the DNA termini. The two DNA-PKcs molecules can then interact to tether the DSB ends together. Synapsis of DNA-PKcs triggers phosphorylation of DNA-PKcs [including autophosphorylation (autophos.)], altering the conformation and dynamics of DNA-PKcs. Phosphorylation of DNA-PKcs allows for recruitment of Artemis and other end-processing factors such as Pol X (DNA polymerare X) family members to generate the proper DNA ends required for ligation. Once the ends are processed, the X4-L4 complex, along with XLF, ligates the ends, repairing the break.
Recognition
NHEJ is initiated by the binding of a heterodimeric complex composed of Ku70 and Ku80 to both ends of the broken DNA molecule. Ku interacts with many proteins in vitro, including DNA-PKcs [13] and the XRCC4–DNA ligase IV complex [14]. Association of Ku with DNA ends may serve as a scaffold for the assembly of the NHEJ synapse. The Ku–DNA complex recruits DNA-PKcs , a 460 kDa member of the PIKKs (phosphoinositide 3-kinase-like family of protein kinases) [13]. Ku then moves inward on the DNA, allowing DNA-PKcs to contact DNA [15]. The association of DNA-PKcs with both DNA and Ku leads to activation of the serine/threonine kinase activity of DNA-PKcs [10]. Inward translocation of Ku also allows two DNA-PKcs molecules to interact across the DSB, forming a molecular ‘bridge’ or synapse between the two DNA ends [16].
In yeast, the MRX (MRE11-RAD50-XRS2) complex participates in both NHEJ and HR, and is one of the first complexes to interact with a DSB [17]. The orthologous MRN [Mre11–Rad50–Nbs1 (Nijmegen breakage syndrome 1)] complex in vertebrates has an established role in HR [18,19]. Recent evidence suggests that mammalian MRN is also involved in NHEJ [20,20a–20c]. The Rad50 protein contains a high-affinity DNA-binding domain and a two-cysteine Zn2+ -binding hook that may assist synapsis [21]. In yeast, this is in fact the case, as DSB ends remain associated after break induction only when the MRX complex is intact [22,23].
The Mre11 polypeptide exhibits an in vitro nuclease function, cleaning hairpin structures and 3′ single-strand overhangs at the ss (single-stranded)/ds (double-stranded) DNA junction as well as harbouring a 3′-to-5′ exonuclease activity [24,25]. This nucleolytic function of Mre11 is not essential in yeast, and the major function of the MRX complex in yeast NHEJ appears to be to tether DNA ends and recruit the ligase complex [24]. Interestingly, the nuclease function of Mre11 is essential in mammals, perhaps reflecting a critical role in HR [26]. Vertebrate DNA-PKcs may function in parallel with MRN as a DNA end-bridging factor for NHEJ [16,23]. This partial redundancy may explain why, in yeast, where no orthologues of DNA-PKcs have been identified, NHEJ is relatively inefficient.
DNA-PKcs can phosphorylate a number of substrates in vitro, including Ku70, Ku80, XRCC4, XLF, Artemis and DNA ligase IV [27–31]. However, it is not clear to what extent these phosphorylation events are required for NHEJ in vivo. Indeed, the best candidate substrate for DNA-PK is DNA-PKcs itself; a number of autophosphorylation sites in DNA-PKcs have been identified and in vivo phosphorylation of DNA-PKcs occurs in response to DNA damage [32–35]. The phosphorylation status of DNA-PKcs is known to influence its conformation and dynamics, probably serving to relieve the blockage of the ends by DNA-PKcs, thus allowing further processing of the DNA [36].
Processing
Since DSBs can occur with a variety of different ends, a number of processing enzymes may be required to repair breaks. Ends must be transformed to 5′-phosphorylated ligatable ends in order for repair to be completed. One key end-processing enzyme in mammalian NHEJ is Artemis, a member of the metallo-β-lactamase superfamily of enzymes, which may be recruited to DSBs through its ability to interact with DNA-PKcs [11,37]. Artemis possesses both a DNA-PKcs-independent 5′-to-3′ exonuclease activity and a DNA-PKcs-dependent endonuclease activity towards DNA-containing ds–ssDNA transitions and DNA hairpins, each of which might be important for processing of DNA termini during NHEJ [11,38]. Inactivation of Artemis results in radiation sensitivity; however, cells lacking Artemis do not have major defects in DSB repair, suggesting that only a subset of DNA lesions are repaired in an Artemis-dependent manner in vivo [39].
Processing of complex lesions might lead to the creation of DNA gaps that must be filled in by DNA polymerases to enable break repair. The DNA polymerase X family of polymerases, including polymerase μ, polymerase λ and terminal deoxyribonuc-leotidyltransferase, have been implicated in fulfilling this role during NHEJ (reviewed in [40]).
Resolution
NHEJ is completed by ligation of the DNA ends, a step that is carried out by X4–L4 (a complex containing XRCC4, DNA ligase IV and XLF) [12]. XRCC4 has no known enzymatic activity, but is required for both NHEJ and V(D)J recombination [41]. It forms a homodimer that acts as a scaffold, interacting with Ku [14] and DNA [42]. The ligase activity of DNA ligase IV is stimulated by both XRCC4 [43] and XLF [44]. DNA ligase IV can ligate blunt DNA ends as well as those with compatible overhangs [12]. X4–L4 has the unique ability to ligate one DNA strand independent of the other, which might allow processing enzymes to act concurrently with the ligation machinery [45]. In addition to this ability, the complex is able to ligate across gaps of several nucleotides and can ligate some incompatible DNA ends with short overhangs [46,47]. NHEJ occurs even in the absence of X4-L4, suggesting that another ligase can partially substitute for DNA ligase IV [48].
ALTERNATIVE PATHWAYS OF NHEJ
A loosely defined alternative end-joining pathway operates in the absence of classical NHEJ factors such as Ku, XRCC4 or DNA ligase IV. These repair events frequently involve small deletions and entail short stretches of homology at the break point [48–52]. This MMEJ (microhomology-mediated end-joining) pathway dominates during alternative end-joining. However, MMEJ and alternative end-joining are not synonymous, since error-free ligation can occur at low frequency in the absence of X4-L4 [51]. Furthermore, MMEJ accounts for a proportion of V(D)J recombination events in wild-type cells [53,54]. Notably, yeast lacking MRX reveal reduced repair by MMEJ, but the complete set of genes that participate in alternative NHEJ in mammalian cells is not yet known [49,52,55].
Analysis of immune development in mice lacking X4-L4 has shown that alternative end-joining is fairly robust [48,56,57]. MMEJ also appears to contribute to genomic instability in cancer. Translocation breakpoints in human cancers very often reveal microhomology, suggesting a role for MMEJ in translocation [58]. MMEJ may also facilitate chemotherapy resistance by genetic reversion in cells lacking wild-type BRCA2 (breast-cancer susceptibility gene 2)[59].In these cases, in-frame microhomologous deletions flanking the original mutation occurred in the resistant cells. The genetic requirements for MMEJ in cancer remain unclear.
SPECIALIZED FUNCTIONS OF NHEJ
The vertebrate immune system is characterized by intrinsic DSB production and repair as a mechanism of diversification of the B- and T-lymphocyte repertoire (reviewed in [60]). Core members of the NHEJ pathway perform direct roles in V(D)J recombination. For example, Ku-deficient cells [61,62] and DNA ligase IV-deficient cells [63] are defective in both coding and signal joint formation. Cells harbouring mutations in DNA-PKcs are severely impaired in their ability to form coding joints, but show little or no defect in signal joint formation [64–66]. Artemis is also implicated in the formation of coding joints, but not signal joints [67,68].
In contrast with V(D)J recombination, multiple DNA repair pathways are likely to be involved in CSR (class switch recombination), including base excision repair, mismatch repair and NHEJ [69]. DNA sequences located between S (switch) regions can be detected in the form of excised circularized DNA, consistent with the involvement of DSB intermediates in CSR [70,71]. Sequences from CSR junctions show little or no sequence homology between donor and acceptor S regions, and often contain short deletions or insertions of nucleotides, all of which are consistent with DSB repair by NHEJ [72]. Further evidence from knockout mice also suggests a role for NHEJ in CSR. DNA-PKcs-deficient mice have significantly reduced levels of serum Ig isotypes, and the only detectable isotype in Ku-deficient mice containing rearranged IgH and IgL genes is IgM [73–75].
NHEJ also plays a role in telomere biology (reviewed in [76]). The formation of dicentric chromosomes as a consequence of DNA end-joining is a hallmark of telomere dysfunction. NHEJ appears to play a central role in the formation of dicentric chromosomes in cells with telomere dysfunction, since fusion of uncapped telomeres is strictly DNA ligase IV-dependent [77]. In addition, Ku, DNA-PKcs and the MRN complex participate in multiple facets of normal telomere biology. All three components of the MRN complex bind telomeres, and disruption of the MRN complex leads to telomere length deregulation [78,79]. The Ku heterodimer and DNA-PKcs also play roles in the regulation of normal telomere length [78,80–83].
CHROMATIN RESPONSE IN NHEJ
Chromosomal DSBs in eukaryotes provoke a rapid and extensive response in chromatin flanking the break, highlighted by phosphorylation of histone H2AX in mammalian cells (γH2AX), on C-terminal Ser139. γH2AX facilitates repair of the break by either HR or NHEJ [84–88]. Phosphorylated H2AX is detected within 1 min of damage [89,90]. The H2AX phosphorylation site, Ser139, is a common recognition site for the PIKKs, and in principle, all three major PIKK members, ATM (ataxia telangiectasia mutated), ATR (ataxia telangiectasia mutated- and Rad3-related) and DNA-PKcs , have the potential to phosphorylate H2AX. There is evidence that each of these kinases can carry out this phosphorylation when the others are compromised, but ATM seems to be the main kinase associated with γ H2AX formation under normal physiological conditions [91–93].
The γH2AX mark around a DSB may extend more than 1 Mb from the break [89,90,93]. In Saccharomyces cerevisiae, γH2AX is present in a 40–50 kb region around an unrepairable DSB and the greatest enrichment of γ H2AX occurred 3–5 kb on either side of the break, with γH2AX absent in sequences 1–2 kb on both sides of the DSB [94]. In mammalian cells, γH2AX is bound by MDC1 (mediator of DNA damage checkpoint 1), which interacts constitutively with the MRN complex and thereby activates ATM [95–97]. The interaction of MDC1 and γH2AX has therefore been proposed to amplify the γH2AX signal [95,97,98]. However, recent chromatin immunoprecipitation analysis suggests a more nuanced picture, whereby MDC1 may reinforce an existing γH2AX signal, but the extent of spread of the signal is not dependent upon MDC1 [93]. This recent work raises the possibility that the signal that generates the γH2AX mark is diffusable.
A number of DNA damage response proteins, such as MDC1, the MRN complex, ATM, 53BP1 (p53-binding protein 1) and BRCA1/BARD1 [BRCA1-associated RING (really interesting new gene) domain 1], accumulate on γH2AX-containing chro-matin. MDC1 is a critical adaptor protein that directly interacts with γH2AX [95]. 53BP1, BRCA1 and MRN/ATM can also associate with DSBs in H2AX−/− cells, suggesting H2AX-independent roles at the DSB [99]. The recruitment of 53BP1 and BRCA1 to γ H2AX chromatin is indirect, requiring the activity of the E3 ubiquitin ligases RNF8 (RING finger protein 8) [100–103] and RNF168 [104,105].
γH2AX accumulates in an AID (activation-induced cytidine deaminase)-dependent manner at the IgH locus in cells undergoing switching [87], and B-cells from H2AX null mice reveal defects in CSR [85,86]. H2AX is not required for switch targeting, initial lesion formation or end-processing during CSR, suggesting that γH2AX affects the efficiency of repair itself [86,106]. However, V(D)J recombination appears to be unaffected by H2AX deletion [85].
Studies of CSR at the IgH locus and of the fusion of dysfunctional telomeres have revealed quantitative roles for H2AX, MDC1 and 53BP1 in ‘long-range’ NHEJ [107,108]. These defects are more severe in 53BP1-null mice than in H2AX- or MDC1-null mice, but less severe than that observed in cells lacking classical NHEJ [109]. 53BP1 localizes rapidly to DSBs and colocalizes with IR (ionizing radiation)-induced γH2AX nuclear foci, but can also accumulate in the absence of H2AX [99,110]. 53BP1-deficient mice are immunodeficient, predisposed to T-cell lymphomas, and reveal severely diminished CSR but normal V(D)J recombination [109,111–113]. 53BP1 has also been implicated inXRCC4-dependent NHEJ of a conventional DSB [114]. 53BP1 accumulation on γH2AX-containing chromatin is mediated by interaction of the 53BP1 tandem Tudor domain with the exposed constitutive chromatin mark, H4K20me2 (histone H4 dimethylated at Lys20) [115]. It is not yet clear whether the same 53BP1–H4K20me2 interaction mediates the H2AX-independent functions of 53BP1.
HOMOLOGOUS RECOMBINATION
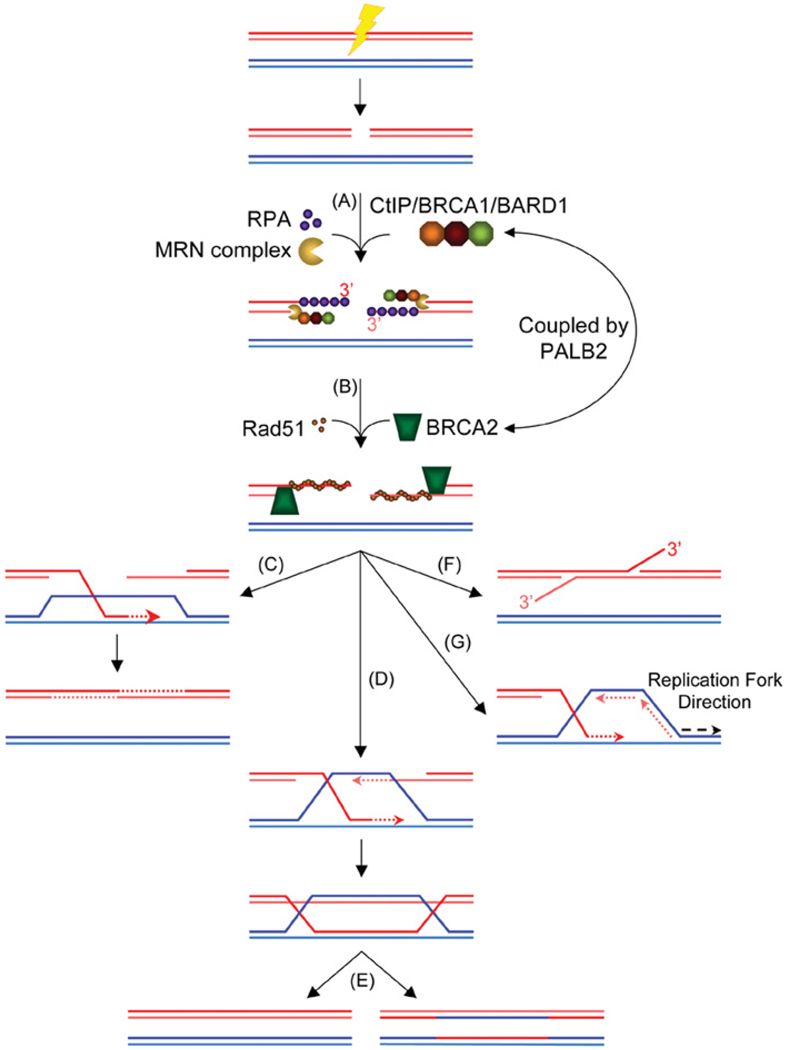
Several distinct mechanisms of ‘homology-directed repair’ have been identified. In yeast, these include HR, SSA (single-strand annealing) and BIR (break-induced replication; reviewed in [2]). Whereas HR potentially results in conservative repair of a DSB, both SSA and BIR are mutagenic pathways. Early steps in HR are the resection of the DNA ends to yield 3′-ssDNA overhangs (Figure 2, pathway A), followed by Rad51-mediated homologous DNA pairing and strand exchange (Figure 2, pathway B). In SSA, a DSB in or near one of two direct repeats leads to the annealing of complementary strands from each repeated sequence, yielding a homologous deletion (Figure 2, pathway F). In contrast with HR, BIR in yeast requires lagging-strand synthesis and appears to be mediated by formation of a replication fork (Figure 2, pathway G) [116]. Consequently, BIR can involve extensive copying from the donor, leading to non-reciprocal translocations and other types of genomic instability [117,118]. Although BIR has been invoked to explain some examples of genomic instability in mammalian cells, direct evidence for a mammalian BIR pathway is lacking.
Figure 2. Homology-directed repair in eukaryotic cells.
(A) Induction of a DSB is recognized by the MRN complex, which tethers the DNA ends together and participates in end processing. The CtIP–BRCA1–BARD1 complex co-operates with the MRN complex to aid in end resection. ssDNA is initially bound by the ssDNA-binding protein RPA to keep the ssDNA from forming secondary structures. BRCA1/BARD1 promotes accumulation of BRCA2 via PALB2. (B) BRCA2 catalyses the nucleation of Rad51 on to the free 5′ end of a dsDNA–ssDNA junction. Once the Rad51 filament is assembled it captures duplex DNA and searches for homology. (C) The SDSA model predicts that a migrating D loop fails to capture the second DNA end and, following extension, the invading strand is displaced and anneals with the resected second end. (D) The DSB repair model predicts that the second DNA end is captured by annealing to the extended D loop, forming two HJs. (E) The double HJ structure is then resolved to yield either crossover or non-crossover products. (F) The SSA pathway: a break near one of two direct repeat sequences leads to annealing of complementary strands from each repeated sequence. The product of this repair event contains a single copy of the repeat with a deletion of the intervening sequences. (G) BIR occurs when the 3′ end of the invading strand leads to the formation of a replication fork, potentially copying long tracts from the donor DNA molecule. Dotted arrows indicate new DNA synthesis.
Recognition
The MRN complex plays a critical role in the early DSB response. MRN complexes on adjacent DNA ends are thought to associate via Rad50 homodimerization to connect the DNA ends prior to repair [21,23,119]. In post-replicative repair, the broken ends may also be kept in close proximity with the neighbouring sister chromatid. Genetic studies in yeast link the cohesin complex (SMC1/3) and the related SMC5/6 complex to HR by maintaining close association of sister chromatids (reviewed in [120,121]). In addition to its role in tethering DNA ends, the MRN complex also recruits and activates the catalytic function of the ATM protein kinase through direct interaction of ATM and Nbs1 [122,123]. ATM phosphorylates numerous substrates in the DNA damage response, including histone H2AX, making H2AX phosphorylation an early marker in chromatin of DSB formation [93,124].
Processing
HR requires processing of the DSB to yield ssDNA containing a 3′-hydroxyl overhang. Genetic evidence in yeast suggests that this end processing involves the MRX complex, as deletions of MRE11, RAD50 and XRS2 slow down the rate of 5′-to-3′ exonuclease activity in vivo [125–127]. However, Mre11 possesses a 3′-to-5′ ATP-independent exonuclease activity, rather than the 5′-to-3′ exonuclease activity required for generation of ssDNA with a 3′-hydroxyl end [25,124a,124b].
Recent evidence in yeast paints a more complex picture of 5′-end resection. In S. cerevisiae, Sae2 interacts with the MRX complex, and these proteins collaborate to trim the DNA ends to an intermediate form [124c]. The DSB is then processed more extensively by either the 5′-to-3′ exonuclease activity of Exo1 or by the Sgs1 helicase in conjunction with an as-yet unidentified single-strand specific nuclease [124d,125]. Sae2, Exo1 and Sgs1 each have orthologues in mammalian cells {CtIP [CTBP (C-terminus-binding protein of adenovirus E1A)-interacting protein], Exo1 and BLM (Bloom’s syndrome protein) respectively}, suggesting a general mechanism for DSB processing in eukaryotes. Indeed, mammalian CtIP, in association with BRCA1, has been implicated in DSB end-processing [133,134].
ssDNA is rapidly bound by the ssDNA-binding protein RPA (replication protein A), which melts the DNA’s secondary structure [3]. However, the DNA strand invasion and homology search steps of HR require formation of a nucleoprotein filament composed of multimers of the Rad51 recombinase bound to ssDNA. Since RPA binds more avidly to ssDNA than Rad51, additional activities are required to load Rad51 on to RPA-coated ssDNA and to displace RPA. In mammalian cells, a critical mediator complex appears to include BRCA1/BARD1 and BRCA2(FANCD1)/DSS1, probably bridged by the PALB2 (partner and localizer of BRCA2) (FANCN) polypeptide [135,136]. Although each of these protein complexes is required for the formation of IR-induced Rad51 nuclear foci, the direct Rad51-loading function is provided by BRCA2, which interacts directly with Rad51 [137–139]. Studies on the Ustilago maydis BRCA2 orthologue Brh2, and a polypeptide harbouring critical functional domains of the human BRCA2 protein, have provided direct biochemical evidence of the Rad51-loading function of BRCA2 [140,141].
Many proteins involved in Rad51 function are products of hereditary cancer predisposition genes (Table 1), implying that failure to adequately regulate HR, and the consequent genomic instability, plays a causal role in cancer. The critical role of HR in suppressing genomic instability is reflected in the early embryonic lethality of mice lacking Rad51, BRCA1 or BRCA2. The abundance of chromatid-type errors observed in such mutant cells suggests that the major function of these genes is to control SCR (sister chromatid recombination) [142,143].
Resolution
The Rad51 nucleoprotein filament then captures duplex DNA and searches for homology. Studies using bacterial RecA indicate that the homology search probably occurs by way of random collisions between the nucleoprotein filament and the duplex DNA, thereby testing segments of the dsDNA in an iterative fashion until homology is found [144]. Following synapsis, the invading strand sets up a D-loop intermediate, whereby the 3′-end primes DNA synthesis using the duplex DNA as a template. It is presently unclear which polymerase(s) mediate D-loop extension in vivo, but Pol η (DNA polymerase η) can perform this function in vitro [145].
If the DSBs have occurred during the S- or G2-phases of the cell cycle, the homology search may capture the neighbouring intact sister chromatid as a repair template for potentially error-free repair [146,147]. Most somatic HR events in either yeast or mammalian cells entail copying only a short tract from the donor DNA molecule (STGC; ‘short tract’ gene conversion). A small proportion of HR events in mammalian cells entail LTGC (‘long tract’ gene conversion), in which several kilobases are copied from the donor [147,148]. Mutation of the Rad51 paralogues, XRCC3, Rad51C or XRCC2, skews HR in favour of LTGC [149–151]. However, it is not clear whether STGC and LTGC represent different outcomes of a common HR mechanism, or whether LTGC is the product of a distinct mechanism, such as BIR. Thus far, the longest gene conversions identified in mammalian cells are <10 kb [152]; much less than the hundreds of kilobases that can be copied during BIR in yeast [117].
Strand invasion into a homologous sequence forms a D-loop intermediate and the 3′-end of the invading strand is extended by a polymerase. If the D-loop captures the second end of the break, the HJs (Holliday junctions) formed could yield crossover or non-crossover products (Figure 2, pathway E). However, crossing over is rare during somatic HR [147,150,153]. The SDSA (synthesis-dependent strand annealing) model was advanced to explain this fact. One SDSA model proposes that, following extension by ‘bubble migration’ (i.e. a minimal migrating D-loop), the invading strand is displaced and pairs (i.e. anneals) with the processed second end of the break (Figure 2, pathway C). In contrast, the ‘double-strand break repair’ model posits an extended D-loop, which captures the second end of the break, leading to the formation of double HJs (Figure 2, pathway D). Although it seems likely that SDSA is the major HR mechanism in somatic mammalian cells, double HJs probably arise during other recombination processes, such as daughter strand gap repair. HJ resolution is therefore relevant to somatic HR and genomic instability.
Once a HJ has been formed, it is able to undergo branch migration along DNA, generating increasing or decreasing lengths of heteroduplex DNA depending on the direction of junction travel (reviewed in [154]). Specialized enzymes in prokaryotes promote branch migration, and human Rad54 shows a strong preference for binding to branched substrates that resemble one end of a D-loop and can promote branch migration in either the 3′ -to-5′ or 5′-to-3′ direction in an ATP-dependent manner [155]. Mammalian homologues of the Escherichia coli RecQ helicase, namely WRN (Werner’s syndrome protein), BLM and RECQ5β, can catalyse branch migration, but disrupt HJs and show anti-recombinogenic characteristics in vitro [156].
The resolution of a HJ is probably executed by several distinct enzyme complexes. The product of the Bloom’s syndrome gene, BLM, in complex with topoisomerase IIIa can dissolve double HJs to form non-crossover products [157]. Alternatively, the MUS81-EME1 complex may cleave HJs to produce crossovers with an exchange of flanking markers [158,159]. Recently, another HJ resolvase was identified in human cells, GEN1, which promotes junction resolution by a symmetrical cleavage mechanism that would be expected to give rise to crossovers and non-crossovers with equal efficiency [160]. There may also be other HJ resolvase activities yet to be identified. In this regard, four recent papers have demonstrated a role for SLX4 in HJ resolution in higher eukaryotes [124a–124d].
SPECIALIZED FUNCTIONS OF HR IN SOMATIC CELLS
During DNA replication, a lesion encountered on one of the parental strands may cause the DNA polymerase complex to stall, potentially collapsing the replication fork. Arrested forks may be processed to form a DSB or replication may be reinitiated downstream of the lesion, leaving a ssDNA lesion that cannot be filled in due to the presence of the blocking lesion. These so-called DSGs (daughter strand gaps) could be repaired via sister chromatid recombination in an error-free manner (reviewed in [161]). Studies in the fission yeast Schizosaccharomyces pombe revealed that a replication fork barrier is capable of promoting recombination and chromosomal rearrangements at that locus [162]. Treatment of cells with HU (hydroxyurea) induces replication fork arrest, generating tracts of ssDNA on the lagging strand, but generating few DSBs in normal cells [163]. Structurally, HU-induced ssDNA may resemble DSGs and the accumulation of BRCA1, BRCA2 and Rad51 at sites of replication arrest in HU-treated cells suggests a probable role for these proteins in mediating repair of DSGs at stalled replication forks [164,165].
Cells maintain telomeric DNA repeats at a critical length that allows the assembly of ‘T-loop’ structures that protect the chromosome ends. Telomeric capping sequesters the 3′ telomeric tail away from DNA damage sensors and processing activities within the cell (reviewed in [166]). Maintenance of telomere length normally requires telomerase, but this protein is non-essential in cells, indicating the existence of an alternative mechanism for telomere length maintenance. S. cerevisiae cells lacking telomerase gradually lose their telomeres and die, but rare survivors maintain telomeres through Rad52-dependent HR [167,168]. In these cases, telomere elongation can occur through BIR or gene conversion in which one telomere serves as a template for elongation of another [169,170].
In mammalian cells, the existence of an ALT (alternative lengthening of telomeres) pathway has been shown in cells lacking telomerase (reviewed in [171]). These cells exhibit sub-nuclear compartments containing telomeric DNA, telomere-binding proteins, recombination proteins such as Rad51 and Rad52, the MRN complex, RPA, and the WRN and BLM helicases [172]. In ALT cells, telomeric DNA is copied to other telomeres by means of HR and copy switching [173]. In support of this, Rad51D and Rad54 were reported to act at telomeres [174,175].
CHROMATIN RESPONSE IN HR
H2AX contributes to HR and SCR in a manner dependent upon the ability of H2AX to be phosphorylated on Ser139, and upon the ability of γH2AX to interact with MDC1 [88,95,114]. Consistent with its role as a critical γH2AX adaptor, MDC1 mediates H2AX-dependent HR, but the mechanisms responsible for this are not known. Possible mediators include RNF8 [100–102], MRN and BRCA1/BARD1 [96,98,176]; however, genetic analysis revealed that a MDC1 mutant lacking the domain required for recruitment of MRN, RNF8, BRCA1 or 53BP1 to chromatin nonetheless retains HR function [114]. This separation of function is underscored by the fact that the major HR function of BRCA1 is independent of H2AX and, indeed, independent of the E3 ubiquitin ligase activity of BRCA1 itself [88,177].
BRCA1 exists in a number of distinct complexes in mammalian cells. The BRCA1–BARD1–Abraxas–MERIT40– Rap80 complex is recruited to IR-induced foci in a manner dependent upon the UIM (ubiquitin-interacting motif) of Rap80 [103,178–180]. Lys63-linked polyubiquitin chains appear to be involved in DNA damage signalling, and recent studies have identified RNF8, a RING domain-containing E3 ubiquitin ligase as a key enzyme for this modification at DSBs [100–103]. RNF8 is also a direct binding partner of the MDC1 SQ-rich domain and mediator of both 53BP1 and BRCA1 recruitment to chromatin [100–103]. RNF8 probably ubiquitinates H2A via a second E3 ubiquitin ligase, RNF168, to reinforce BRCA1 recruitment via Rap80 and 53BP1 through an uncharacterized mechanism [104,105].
COMPETITION BETWEEN NHEJ AND HR
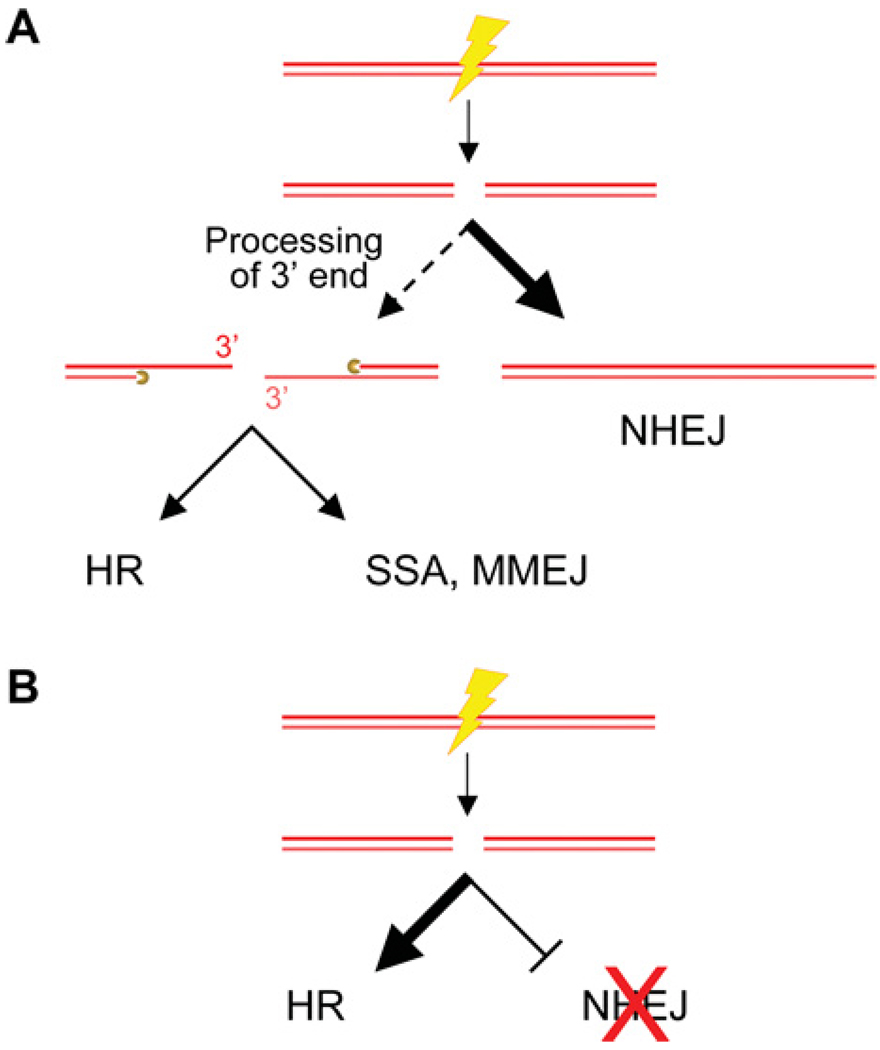
Cells lacking classical NHEJ genes reveal a DSB repair bias in favour of HR, suggesting that these two major pathways normally compete to repair DSBs [181]. During V(D)J recombination, the RAG proteins play a role in specifying the preference for repair by NHEJ in these cells [182]. However, other rules must apply for unscheduled DSBs. The balance between NHEJ and HR shifts during the cell cycle, presumably reflecting the availabilty of a sister chromatid synthesized during S-phase [146,147]. A study in chicken DT40 cells deficient in NHEJ or HR factors revealed that NHEJ mutants were highly sensitive to IR in the G1- and early S-phase of the cell cycle, whereas HR mutants were sensitive primarily in the S-/G2-phase [6]. Similar studies in mammalian cells demonstrated that NHEJ-deficient cells have reduced repair at all cell cycle stages, whereas HR-deficient cells have a minor defect in G1, but a greater impairment in S-/G2-/M-phase [183,184].
Recent evidence suggests that the shift from NHEJ to HR as the cell cycle progresses is actively regulated. Analysis of end resection at an HO-induced DSB at MAT in yeast revealed that G1-arrested cells failed to initiate efficient end resection, which prevented loading of RPA and Rad51, and blocked Mec1/ATR activation [185]. This effect correlated with low levels of activity of the major cyclin-dependent kinase, CDK1 (cyclin-dependent kinase 1)/Cdc28 and, critically, inhibition of CDK1 activity in G2-phase prevented end resection and checkpoint activation. Under these conditions, Mre11 persists at the DSB site, consistent with the idea that processing of the break has stalled. This suggests that CDK1 controls Mre11-associated nuclease function at a DSB, but not the recruitment of Mre11 to DNA ends. Cdk activity also regulates HR in fission yeast [186].
Sae2 controls the initiation of DNA end resection in both meiotic and mitotic yeast and is itself a DNA endonuclease [130,187]. Recently, Sae2-mediated control of DSB resection in yeast was shown to depend on its CDK phosphorylation status [188]. Mutation of Sae2 Ser267 to a non-phosphorylatable residue (S267A) causes an end-processing phenotype comparable with deletion of Sae2 [188]. In contrast, a S267E mutant that mimics constitutive phosphorylation complements these phenotypes and overcomes the need for CDK activity in DSB end resection. The Sae2-null and S267A mutants show delayed HR and enhanced NHEJ, whereas the S267E mutant showed slightly enhanced recombination and a decrease in NHEJ efficiency. Thus CDK1/Cdc28-mediated phosphorylation of Sae2 modulates the balance between NHEJ and HR during the cell cycle. These results support a model in which the commitment to DSB end resection is regulated to ensure that a cell engages the most appropriate DSB repair pathway to optimize genome stability (Figure 3A). The motif encompassing Ser267 of Sae2 is highly conserved amongst orthologues in higher eukaryotes, and mutation of the analogous residue in human CtIP also produces hypersensitivity to camptothecin [188]. These results suggest that similar CDK control of DNA end resection operates in other organisms. CtIP is one of several proteins that interact with the BRCA1 C-terminal tandem BRCT repeat, and this interaction is important for efficient end resection. This, in turn, suggests that the BRCA1-CtIP interaction influences the balance between HR and NHEJ [134].
Figure 3. Relationships between HR and NHEJ in mammalian cells.
(A) One of the early ‘choices’ in DSB repair is the extent to which the DNA ends are processed. In classical NHEJ, end resection may be minimal or absent. Should the ends be processed to yield a 3′ overhang, repair can occur through either HR, SSA or MMEJ. (B) Defects in NHEJ skew DSB repair in favour of HR.
CO-OPERATION BETWEEN NHEJ AND HR
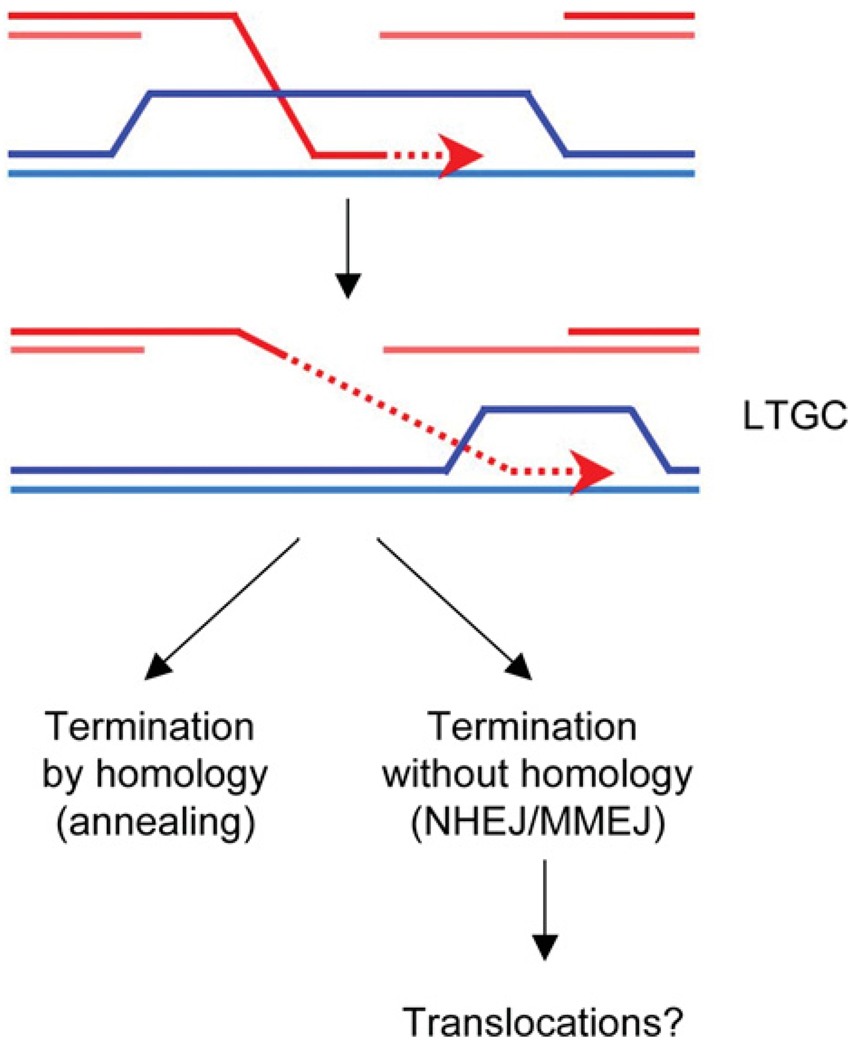
At the organismal level, HR and NHEJ clearly collaborate to suppress genomic instability. However, HR and NHEJ may also collaborate more directly to repair a subset of mammalian DSBs. This was revealed in work on a subpathway of HR termed LTGC. The majority of recombination events resolve after only a few hundred base pairs have been copied from the donor. However, a small proportion of HR events involve more extensive copying from the donor, generating a LTGC spanning several kilobases [147–149,150,189]. Some of these LTGC events terminate without homology, presumably by use of NHEJ [147] (Figure 4). In a study of interchromosomal recombination, direct sequencing of the joints demonstrated that repair had occurred by NHEJ, with microhomology observed at approximately half of the junctions [189]. The genes involved in this coupled mechanism remain to be identified. Work from our laboratory suggests that both XRCC4-dependent and XRCC4-independent NHEJ pathways are capable of terminating LTGC in mammalian cells (A.J. Hartlerode and R. Scully, unpublished work). Therefore, both classical NHEJ and MMEJ may participate in resolving LTGCs.
Figure 4. Model for termination of LTGC.
In many LTGC events, gene conversion is thought to be terminated by homologous pairing. However, in a proportion of events, gene conversion is terminated without the use of homology, by NHEJ/MMEJ. This type of termination might result in chromosomal translocations.
Coupling of HR and NHEJ has also been observed in a less direct manner during other forms of recombination. In a study of ectopic recombination in S. cerevisiae between two unlinked homologous loci, a novel class of gene conversion events was observed that included extensive lengths of non-homologous sequence [190]. Co-operation between HR and NHEJ has also been deduced in some gene targeting events [191–193]. In Drosophila melanogaster, examples of incomplete LTGC events have been identified where repair is completed by an end-joining pathway that is independent of DNA ligase IV [194].
CONCLUSIONS
Significant progress in understanding the regulation of DSB repair in mammalian cells has been made in recent years. However, it is clear that much remains to be understood about these repair pathways and the complex interactions between them. Discoveries made in yeast have greatly advanced the understanding of both HR and NHEJ; however, the relationship between DSB repair pathways in yeast and higher eukaryotes is not always clearcut. Many protein complexes involved in DSB repair appear to function in more than one pathway. This highlights the need for more sophisticated tools to simultaneously examine HR and NHEJ in mammalian cells. Elucidation of these important disease-associated DSB repair functions may reveal new therapeutic targets in cancer and other disease states.
ACKNOWLEDGEMENTS
We thank members of the Scully laboratory for discussions and comments on the review.
FUNDING
Our work is supported by the National Institutes of Health [grant numbers CA095175 and GM073894] and by awards from the Leukemia and Lymphoma Society and the Bloom’s Syndrome Foundation (to R.S.).
Abbreviations used
- 53BP1
p53-binding protein 1
- ALT
alternative lengthening of telomeres
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia mutated- and Rad3-related
- BARD1
BRCA1-associated RING domain 1
- BIR
break-induced replication
- BRCA
breast-cancer susceptibility gene
- CDK
cyclin-dependent kinase
- CSR
class switch recombination
- CtIP
CTBP (C-terminus-binding protein of adenovirus E1A)-interacting protein
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- DSB
double-strand break
- dsDNA
double-stranded DNA
- DSG
daughter strand gap
- HJ
Holliday junction
- HR
homologous recombination
- HU
hydroxyurea
- IR
ionizing radiation
- Ku
Ku70/Ku80
- LTGC
‘long tract’ gene conversion
- MDC1
mediator of DNA damage checkpoint 1
- MMEJ
microhomology-mediated end-joining
- MRN complex
Mre11–Rad50–NBS1 complex
- MRX complex
MRE11–RAD50–XRS2 complex
- NBS1
Nijmegen breakage syndrome 1
- NHEJ
non-homologous end-joining
- PALB2
partner and localizer of BRCA2
- PIKK
phosphoinositide 3-kinase-like family of protein kinase
- RING
really interesting new gene
- RNF
RING finger protein
- RPA
replication protein A
- S region
switch region
- SCR
sister chromatid recombination
- SDSA
synthesis-dependent strand annealing
- SSA
single-strand annealing
- ssDNA
single-stranded DNA
- STGC
‘short tract’ gene conversion
- X4-L4
complex containing XRCC4, DNA ligase IV and XLF
- XRCC
X-ray repair complementing defective repair in Chinese hamster cells
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd Edition. Washington: ASM Press; 2006. [Google Scholar]
- 2.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 5.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delacote F, Lopez BS. Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: the trans-S double-strand break repair model. Cell Cycle. 2008;7:33–38. doi: 10.4161/cc.7.1.5149. [DOI] [PubMed] [Google Scholar]
- 8.Cary RB, Peterson SR, Wang J, Bear DG, Bradbury EM, Chen DJ. DNA looping by Ku and the DNA-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 10.Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 12.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 14.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo S, Dynan WS. Geometry of a complex formed by double strand break repair proteins at a single DNA end: recruitment of DNA-PKcs induces inward translocation of Ku protein. Nucleic Acids Res. 1999;27:4679–4686. doi: 10.1093/nar/27.24.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeFazio LG, Stansel RM, Griffith JD, Chu G. Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J. 2002;21:3192–3200. doi: 10.1093/emboj/cdf299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, van Heems D, Ito E, Nakamura A, Sonoda E. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol. Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Dinkelmann M, Spehalski E, Stoneham T, Buis J, Wu Y, Sekiguchi JM, Ferguson DO. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat. Struct. Mol. Biol. 2009;16:808–813. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20b.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in clssical and altemative nonhomologous end joining. Nat. Struct. Mol. Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20c.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 21.Hopfner KP, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BA, Karcher A, Henderson B, Bodmer JL, McMurray CT. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 22.Lobachev K, Vitriol E, Stemple J, Resnick MA, Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 23.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 26.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Douglas P, Gupta S, Morrice N, Meek K, Lees-Miller SP. DNA-PK-dependent phosphorylation of Ku70/80 is not required for non-homologous end joining. DNA Repair. 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Yu Y, Wang W, Ding Q, Ye R, Chen D, Merkle D, Schriemer D, Meek K, Lees-Miller SP. DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair. 2003;2:1239–1252. doi: 10.1016/s1568-7864(03)00143-5. [DOI] [PubMed] [Google Scholar]
- 29.Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair. 2008;7:1680–1692. doi: 10.1016/j.dnarep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YG, Nnakwe C, Lane WS, Modesti M, Frank KM. Phosphorylation and regulation of DNA ligase IV stability by DNA-dependent protein kinase. J. Biol. Chem. 2004;279:37282–37290. doi: 10.1074/jbc.M401217200. [DOI] [PubMed] [Google Scholar]
- 32.Douglas P, Sapkota GP, Morrice N, Yu Y, Goodarzi AA, Merkle D, Meek K, Alessi DR, Lees-Miller SP. Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 2002;368:243–251. doi: 10.1042/BJ20020973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan DW, Chen BP, Prithivirajsingh S, Kurimasa A, Story MD, Qin J, Chen DJ. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 35.Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J. Biol. Chem. 2005;280:33839–33846. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- 36.Weterings E, Verkaik NS, Bruggenwirth HT, Hoeijmakers JH, van Gent DC. The role of DNA dependent protein kinase in synapsis of DNA ends. Nucleic Acids Res. 2003;31:7238–7246. doi: 10.1093/nar/gkg889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 38.Ma Y, Schwarz K, Lieber MR. The Artemis: DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair. 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Pluth JM, Cooper PK, Cowan MJ, Chen DJ, Yannone SM. Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair. 2005;4:556–570. doi: 10.1016/j.dnarep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 40.McElhinny SA, Ramsden DA. Sibling rivalry: competition between Pol X family members in V(D)J recombination and general double strand break repair. Immunol. Rev. 2004;200:156–164. doi: 10.1111/j.0105-2896.2004.00160.x. [DOI] [PubMed] [Google Scholar]
- 41.Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, Alt FW. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 42.Modesti M, Hesse JE, Gellert M. DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J. 1999;18:2008–2018. doi: 10.1093/emboj/18.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grawunder U, Zimmer D, Leiber MR. DNA ligase IV binds to XRCC4 via a motif located between rather than within its BRCT domains. Curr. Biol. 1998;8:873–876. doi: 10.1016/s0960-9822(07)00349-1. [DOI] [PubMed] [Google Scholar]
- 44.Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 46.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 49.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 51.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P, Lopez BS. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20902–20907. doi: 10.1073/pnas.0708541104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 54.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 55.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan-Hammarstrom Q, Jones AM, Lahdesmaki A, Zhou W, Gatti RA, Hammarstrom L, Gennery AR, Ehrenstein MR. Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J. Exp. Med. 2005;201:189–194. doi: 10.1084/jem.20040772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay JP. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 60.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv. Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 61.Gu Y, Jin S, Gao Y, Weaver DT, Alt FW. Ku70-deficient embryonic stem cells have increased ionizing radiosensitivity, defective DNA end-binding activity, and inability to support V(D)J recombination. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8076–8081. doi: 10.1073/pnas.94.15.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu C, Bogue MA, Lim DS, Hasty P, Roth DB. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 63.Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 64.Blackwell TK, Malynn BA, Pollock RR, Ferrier P, Covey LR, Fulop GM, Phillips RA, Yancopoulos GD, Alt FW. Isolation of scid pre-B cells that rearrange κ light chain genes: formation of normal signal and abnormal coding joins. EMBO J. 1989;8:735–742. doi: 10.1002/j.1460-2075.1989.tb03433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver DT, Alt FW. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 66.Lieber MR, Hesse JE, Lewis S, Bosma GC, Rosenberg N, Mizuuchi K, Bosma MJ, Gellert M. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell. 1988;55:7–16. doi: 10.1016/0092-8674(88)90004-9. [DOI] [PubMed] [Google Scholar]
- 67.Nicolas N, Moshous D, Cavazzana-Calvo M, Papadopoulo D, de Chasseval R, Le Deist F, Fischer A, de Villartay JP. A human severe combined immunodeficiency (SCID) condition with increased sensitivity to ionizing radiations and impaired V(D)J rearrangements defines a new DNA recombination/repair deficiency. J. Exp. Med. 1998;188:627–634. doi: 10.1084/jem.188.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rooney S, Sekiguchi J, Zhu C, Cheng HL, Manis J, Whitlow S, DeVido J, Foy D, Chaudhuri J, Lombard D, Alt FW. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 69.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat. Rev. Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 70.Iwasato T, Shimizu A, Honjo T, Yamagishi H. Circular DNA is excised by immunoglobulin class switch recombination. Cell. 1990;62:143–149. doi: 10.1016/0092-8674(90)90248-d. [DOI] [PubMed] [Google Scholar]
- 71.Catalan N, Selz F, Imai K, Revy P, Fischer A, Durandy A. The block in immunoglobulin class switch recombination caused by activation-induced cytidine deaminase deficiency occurs prior to the generation of DNA double strand breaks in switch mu region. J. Immunol. 2003;171:2504–2509. doi: 10.4049/jimmunol.171.5.2504. [DOI] [PubMed] [Google Scholar]
- 72.Dunnick W, Hertz GZ, Scappino L, Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manis JP, Dudley D, Kaylor L, Alt FW. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity. 2002;16:607–617. doi: 10.1016/s1074-7613(02)00306-0. [DOI] [PubMed] [Google Scholar]
- 74.Manis JP, Gu Y, Lansford R, Sonoda E, Ferrini R, Davidson L, Rajewsky K, Alt FW. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Casellas R, Nussenzweig A, Wuerffel R, Pelanda R, Reichlin A, Suh H, Qin XF, Besmer E, Kenter A, Rajewsky K, Nussenzweig MC. Ku80 is required for immunoglobulin isotype switching. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Riha K, Heacock ML, Shippen DE. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu. Rev. Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- 77.Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr. Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- 78.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ranganathan V, Heine WF, Ciccone DN, Rudolph KL, Wu X, Chang S, Hai H, Ahearn IM, Livingston DM, Resnick I. Rescue of a telomere length defect of Nijmegen breakage syndrome cells requires NBS and telomerase catalytic subunit. Curr. Biol. 2001;11:962–966. doi: 10.1016/s0960-9822(01)00267-6. [DOI] [PubMed] [Google Scholar]
- 80.d’Adda di Fagagna F, Hande MP, Tong WM, Roth D, Lansdorp PM, Wang ZQ, Jackson SP. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 81.Espejel S, Martin M, Klatt P, Martin-Caballero J, Flores JM, Blasco MA. Shorter telomeres, accelerated ageing and increased lymphoma in DNA-PKcs-deficient mice. EMBO Rep. 2004;5:503–509. doi: 10.1038/sj.embor.7400127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hande P, Slijepcevic P, Silver A, Bouffler S, van Buul P, Bryant P, Lansdorp P. Elongated telomeres in scid mice. Genomics. 1999;56:221–223. doi: 10.1006/geno.1998.5668. [DOI] [PubMed] [Google Scholar]
- 83.Myung K, Ghosh G, Fattah FJ, Li G, Kim H, Dutia A, Pak E, Smith S, Hendrickson EA. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol. Cell. Biol. 2004;24:5050–5059. doi: 10.1128/MCB.24.11.5050-5059.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 85.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 87.Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR. AID is required to initiate Nbs1/γ -H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, Scully R. Control of sister chromatid recombination by histone H2AX. Mol. Cell. 2004;16:1017–1025. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 91.Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat. Cell Biol. 2002;4:993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- 92.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 93.Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH. Formation of dynamic γ -H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol. Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 96.Goldberg M, Stucki M, Falck J, D’Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 97.Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 99.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 100.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 103.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 105.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 106.Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F. 53BP1 is required for class switch recombination. J. Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morales JC, Xia Z, Lu T, Aldrich MB, Wang B, Rosales C, Kellems RE, Hittelman WN, Elledge SJ, Carpenter PB. Role for the BRCA1 C-terminal repeats (BRCT) protein 53BP1 in maintaining genomic stability. J. Biol. Chem. 2003;278:14971–14977. doi: 10.1074/jbc.M212484200. [DOI] [PubMed] [Google Scholar]
- 112.Ward IM, Minn K, van Deursen J, Chen J. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 2003;23:2556–2563. doi: 10.1128/MCB.23.7.2556-2563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 114.Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- 117.Llorente B, Smith CE, Symington LS. Break-induced replication: what is it and what is it for? Cell Cycle. 2009;7:859–864. doi: 10.4161/cc.7.7.5613. [DOI] [PubMed] [Google Scholar]
- 118.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 119.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–443. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 120.Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- 121.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 122.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 123.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 124.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 124a.Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124b.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yaten JR, 3rd, Russell P. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124c.Muñoz IM, Hain K, Déclais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol. Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 124d.Andersen SL, Bergstralh DT, Kohl KP, La Rocque JR, Moore CB, Sekelsky J. Drosophila MUS312 and the vertebrate ortholog BTBD12 interact with DNA structure-specific endonucleases in DNA repair and recombination. Mol. Cell. 2009;35:128–135. doi: 10.1016/j.molcel.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 127.Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 129.Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 130.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/ Xrs2 complex. Mol. Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking BRCA2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 138.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 139.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 140.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 141.San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J. Biol. Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McPherson JP, Hande MP, Poonepalli A, Lemmers B, Zablocki E, Migon E, Shehabeldin A, Porras A, Karaskova J, Vukovic B. A role for Brca1 in chromosome end maintenance. Hum. Mol. Genet. 2006;15:831–838. doi: 10.1093/hmg/ddl002. [DOI] [PubMed] [Google Scholar]
- 143.Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Involvement of BRCA2 in DNA repair. Mol. Cell. 1998;1:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 144.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 145.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 146.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Puget N, Knowlton M, Scully R. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair. 2005;4:149–161. doi: 10.1016/j.dnarep.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nagaraju G, Hartlerode A, Kwok A, Chandramouly G, Scully R. XRCC2 and XRCC3 regulate the balance between short- and long-tract gene conversion between sister chromatids. Mol. Cell. Biol. 2009;29:4283–4294. doi: 10.1128/MCB.01406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nagaraju G, Odate S, Xie A, Scully R. Differential regulation of short-and long-tract gene conversion between sister chromatids by Rad51C. Mol. Cell. Biol. 2006;26:8075–8086. doi: 10.1128/MCB.01235-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Brenneman MA, Wagener BM, Miller CA, Allen C, Nickoloff JA. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination. Mol. Cell. 2002;10:387–395. doi: 10.1016/s1097-2765(02)00595-6. [DOI] [PubMed] [Google Scholar]
- 152.Stark JM, Jasin M. Extensive loss of heterozygosity is suppressed during homologous repair of chromosomal breaks. Mol. Cell. Biol. 2003;23:733–743. doi: 10.1128/MCB.23.2.733-743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Richardson C, Moynahan ME, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu Y, West SC. Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell Biol. 2004;5:937–944. doi: 10.1038/nrm1502. [DOI] [PubMed] [Google Scholar]
- 155.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 156.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 2008;33:609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 158.Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 159.Constantinou A, Chen XB, McGowan CH, West SC. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 161.Nagaraju G, Scully R. Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair. 2007;6:1018–1031. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell. 2005;121:689–702. doi: 10.1016/j.cell.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 163.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 164.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Scully R, Puget N, Vlasakova K. DNA polymerase stalling, sister chromatid recombination and the BRCA genes. Oncogene. 2000;19:6176–6183. doi: 10.1038/sj.onc.1203971. [DOI] [PubMed] [Google Scholar]
- 166.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 167.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 168.Le S, Moore JK, Haber JE, Greider CW. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pluta AF, Zakian VA. Recombination occurs during telomere formation in yeast. Nature. 1989;337:429–433. doi: 10.1038/337429a0. [DOI] [PubMed] [Google Scholar]
- 170.Wang SS, Zakian VA. Telomere-telomere recombination provides an express pathway for telomere acquisition. Nature. 1990;345:456–458. doi: 10.1038/345456a0. [DOI] [PubMed] [Google Scholar]
- 171.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 172.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 173.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat. Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 174.Jaco I, Munoz P, Goytisolo F, Wesoly J, Bailey S, Taccioli G, Blasco MA. Role of mammalian Rad54 in telomere length maintenance. Mol. Cell. Biol. 2003;23:5572–5580. doi: 10.1128/MCB.23.16.5572-5580.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tarsounas M, Munoz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, West SC. Telomere maintenance requires the RAD51D recombination/repair protein. Cell. 2004;117:337–347. doi: 10.1016/s0092-8674(04)00337-x. [DOI] [PubMed] [Google Scholar]
- 176.Lou Z, Chini CC, Minter-Dykhouse K, Chen J. Mediator of DNA damage checkpoint protein 1 regulates BRCA1 localization and phosphorylation in DNA damage checkpoint control. J. Biol. Chem. 2003;278:13599–13602. doi: 10.1074/jbc.C300060200. [DOI] [PubMed] [Google Scholar]
- 177.Reid LJ, Shakya R, Modi AP, Lokshin M, Cheng JT, Jasin M, Baer R, Ludwig T. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20876–20881. doi: 10.1073/pnas.0811203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 179.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002;30:3454–3463. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 183.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hinz JM, Yamada NA, Salazar EP, Tebbs RS, Thompson LH. Influence of double-strand-break repair pathways on radiosensitivity throughout the cell cycle in CHO cells. DNA Repair. 2005;4:782–792. doi: 10.1016/j.dnarep.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 185.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Caspari T, Murray JM, Carr AM. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 2002;16:1195–1208. doi: 10.1101/gad.221402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiaeSae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 188.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell. Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bailis AM, Arthur L, Rothstein R. Genome rearrangement in top3 mutants of Saccharomyces cerevisiae requires a functional RAD1 excision repair gene. Mol. Cell. Biol. 1992;12:4988–4993. doi: 10.1128/mcb.12.11.4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Belmaaza A, Chartrand P. One-sided invasion events in homologous recombination at double-strand breaks. Mutation Res. 1994;314:199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 192.Puchta H. Double-strand break-induced recombination between ectopic homologous sequences in somatic plant cells. Genetics. 1999;152:1173–1181. doi: 10.1093/genetics/152.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.McVey M, Radut D, Sekelsky JJ. End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent. Genetics. 2004;168:2067–2076. doi: 10.1534/genetics.104.033902. [DOI] [PMC free article] [PubMed] [Google Scholar]