SUMMARY
Cannibalism is a mechanism to delay sporulation in Bacillus subtilis. Cannibal cells express the skf and sdp toxin systems to lyse a fraction of their sensitive siblings. The lysed cells release nutrients that serve to feed the community, effectively delaying spore formation. Here we provide evidence that the subpopulation of cells that differentiates into cannibals is the same subpopulation that produces the extracellular matrix that holds cells together in biofilms. Cannibalism and matrix formation are both triggered in response to the signaling molecule surfactin. Nutrients released by the cannibalized cells are preferentially used by matrix-producing cells, as they are the only cells expressing resistance to the Skf and Sdp toxins. As a result this subpopulation increases in number and matrix production is enhanced when cannibalism toxins are produced. The cannibal/matrix-producing subpopulation is also generated in response to antimicrobials produced by other microorganisms and may thus constitute a defense mechanism to protect B. subtilis from the action of antibiotics in natural settings.
Keywords: Bacillus, Biofilm, Cell Death, Differentiation, Matrix
INTRODUCTION
The ability to differentiate into many distinct cell types is a hallmark of the soil-dwelling, gram-positive bacterium Bacillus subtilis. For example, in nutrient limiting conditions a subpopulation of cells differentiates into spores (Errington, 2003, Piggot & Hilbert, 2004). Spore development is energy intensive and, once committed, cells may not exit this state for prolonged periods. Thus, this bacterium has evolved mechanisms to delay entry into sporulation as long as possible. It accomplishes this by having the capacity to direct a subpopulation of cells down a differentiation pathway that gives rise to so-called cannibals (Claverys & Havarstein, 2007, Ellermeier et al., 2006, Gonzalez-Pastor et al., 2003). Cannibal cells are resistant to two toxins, Skf and Sdp, that they secrete to kill a fraction of their siblings. As a result, cannibals overcome nutritional limitation and delay the onset of sporulation.
In addition to spores and cannibals, B. subtilis can undergo several other developmental processes. For instance, within multicellular aggregates known as biofilms a subpopulation of cells differentiates to produce an extracellular matrix that encases the community (Branda et al., 2006, Chai et al., 2008, Chu et al., 2006, Vlamakis et al., 2008). This specialized subpopulation is termed matrix-producing because it highly expresses the epsA-O (henceforth eps) and yqxM-sipW-tasA (henceforth yqxM) operons that encode the machinery to produce an exopolysaccharide (Eps) and the protein TasA, respectively. Eps and TasA are the two main components of the extracellular matrix.
The three developmental pathways described above for B. subtilis, sporulation, cannibalism, and matrix production, are strongly interconnected. They are activated by the same master regulatory protein, Spo0A (Branda et al., 2004, Ellermeier et al., 2006, Fujita et al., 2005, Gonzalez-Pastor et al., 2003, Kearns et al., 2005). However, Spo0A does not regulate these three pathways identically. For instance, the subpopulation of sporulating cells, which derives from the subpopulation of matrix-producing cells, shows different spatio-temporal distribution within the biofilm when compared to the matrix producers (Vlamakis et al., 2008).
The differential expression of Spo0A-regulated genes is thought to be due to the fact that different promoters respond to different concentrations of phosphorylated Spo0A (Spo0A~P) in the cell (Fujita et al., 2005, Veening et al., 2008). Spo0A can be phosphorylated, either directly or indirectly, by the action of five different kinases (KinA-E) (Jiang et al., 2000, LeDeaux et al., 1995). Presumably depending on the ability of those kinases to phosphorylate Spo0A, different levels of phosphorylation can be reached. Lower levels of Spo0A~P suffice for the expression of genes involved in matrix production and cannibalism, whereas higher levels of Spo0A~P are necessary to trigger sporulation (Fujita et al., 2005). Depending on the ability of each kinase to sense different signals, the three differentiation pathways may be activated in distinct subpopulations of cells. Despite many efforts, the nature of most of the signals sensed by the kinases remains a mystery.
We recently identified the lipopeptide surfactin as a “quorum-sensing” molecule produced by B. subtilis that appears to activate the membrane histidine kinase KinC with the consequent phosphorylation of Spo0A (Lopez et al., 2009). We presume this results in low levels of Spo0A~P because KinC activation leads to transcription of sinI, a gene whose promoter requires low levels of Spo0A~P for activation (Fujita et al., 2005). In addition, we have demonstrated that sinI expression displays bimodality, i.e. only a subpopulation of the cells expresses sinI after KinC activation by surfactin (Chai et al., 2008, Lopez et al., 2009). In this subpopulation, the antirepressor SinI antagonizes the repressor SinR, allowing expression of the eps and yqxM operons leading to matrix production (Kearns et al., 2005).
Cannibalism and matrix production are both are activated by low levels of Spo0A~P (Fujita et al., 2005). Low levels of Spo0A~P also directly induce the expression of the skfA-H operon and indirectly, by repressing the repressor AbrB, the expression of the sdpABC operon. These operons encode the two cannibalism toxins, Skf and Sdp (Fujita et al., 2005, Gonzalez-Pastor et al., 2003). Given that both processes are coordinated by low levels of Spo0A~P, we postulated that matrix production and cannibalism could be triggered by the same signaling molecule, surfactin.
In this paper, we show that surfactin triggers both matrix and cannibal toxin production in the same subpopulation of cells. This subpopulation secretes cannibalism toxins into the milieu, concomitant with expression of the immunity machinery used to resist the action of these toxins. At the same time, these cells produce the extracellular matrix that encases them in the biofilm. Surrounded by the toxins, only matrix producers are favored to grow, as they are able to use the nutrients released by their killed siblings. This phenomenon increases the number of matrix-producing cells within the population. In turn, with the increase in the relative number of matrix-producing cells, these communities are able to produce more extracellular matrix. We propose that this behavior is a mechanism to eliminate cell types that are no longer required for the development of the community and to promote the subpopulation of matrix producers.
RESULTS
Surfactin induces matrix production and cannibalism in the same subpopulation
To test the hypothesis that matrix and cannibal toxin production occur in the same subpopulation we fused specific promoters to genes encoding different fluorescent proteins and monitored their expression in single cells using flow cytometry. Extending earlier work by E. Hobbs (Hobbs, 2006), we first tested whether the genes for Skf synthesis were induced by surfactin in a KinC-dependent manner, as is the case for the matrix operon yqxM (Lopez et al., 2009).
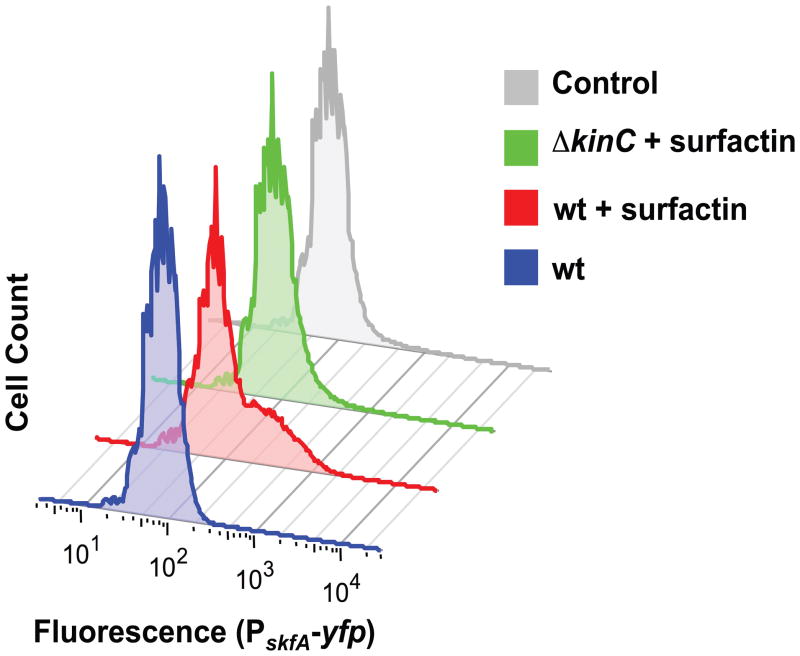
We followed Skf gene expression using the reporter PskfA-yfp in the wild-type strain grown in LB medium, where surfactin production is not detectable (Lopez et al., 2009). Under these conditions, we did not observe any reporter activation (Fig. 1 blue trace). The addition of purified surfactin to the medium resulted in a subpopulation of cells highly expressing the reporter (Fig. 1 red trace). The skfA-expressing subpopulation was not observed in cultures of the ΔkinC mutant when surfactin was added (Fig. 1 green trace). These results indicate that the expression of cannibalism genes is indeed triggered by the same signaling molecule that triggers matrix production.
Fig. 1.
Activation of cannibalism by the quorum-sensing molecule surfactin. Flow cytometry monitoring the expression of the reporter PskfA-yfp in LB cultures treated with surfactin after 8 h of incubation. The addition of surfactin induced higher expression of the reporter in a subpopuation of cells, as evidenced by the shoulder observed in the expression profile of treated cultures. This subpopulation was not observed when surfactin was added in the absence of the membrane kinase KinC. Fluorescence is shown in arbitrary units.
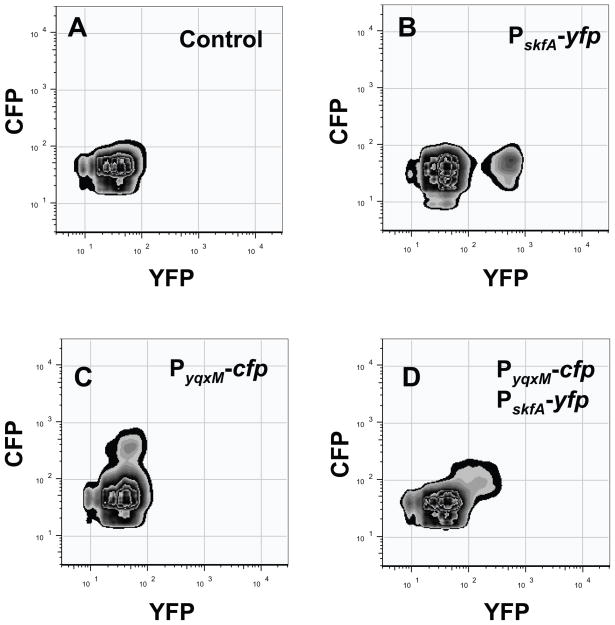
It therefore seemed feasible that the fraction of cells sensing surfactin and thus, differentiating into matrix producers, could also differentiate into cannibals. We monitored the subpopulation of matrix producers and cannibals using the reporters PyqxM-cfp and PskfA-yfp in single and double-labeled strains to simultaneously measure YFP- and CFP-producing cells in the same strain using flow cytometry. We plotted the results in graphs where CFP fluorescence is on the y-axis and YFP fluorescence is on the x-axis. The number of cells expressing different levels of fluorescence was then plotted on a z-axis perpendicular to the plane of the x- and y-axes, as shown by the isolines. Fig. 2A presents the control of background fluorescence for both CPF and YFP in a strain harboring no fluorescent protein genes. We first distinguished each subpopulation of cells (matrix producers and cannibals) in each fluorescence channel using single-labeled strains as controls (PskfA-yfp and PyqxM-cfp in Fig. 2B and Fig. 2C respectively). The double-labeled strain Pskf-yfp, PyqxM-cfp in Fig. 2D showed a single subpopulation of fluorescent cells, and these cells were expressing both YFP and CFP. This finding indicates both cell differentiation pathways are coordinately activated in the same subpopulation.
Fig. 2.
Subpopulations of cannibals and matrix producers are the same cells. Flow cytometry monitoring the subpopulations of cannibals (PskfA-yfp) and matrix producers (PyqxM-cfp). Fluorescence intensity for the YFP channel is presented on the X-axis and for the CFP channel is presented on the Y-axis. Fluorescence is measured in arbitrary units. (A) Wild-type strain expressing no fluorescence protein was used as the negative control. (B) Strain harboring only the PskfA-yfp (cannibalism) reporter. (C) Strain harboring only the PyqxM-cfp (matrix) reporter. (D) Double-labeled strain PskfA-yfp, PyqxM-cfp.
Mutations that increase cannibalism
Given that the same cells express both cannibalism and matrix production genes, we were compelled to study the influence of cannibalism toxins on biofilm morphology. To this end, we deleted genes predicted to have a role either in the production of, or sensitivity to, cannibalism toxins. Then, we analyzed biofilm development in those mutant strains.
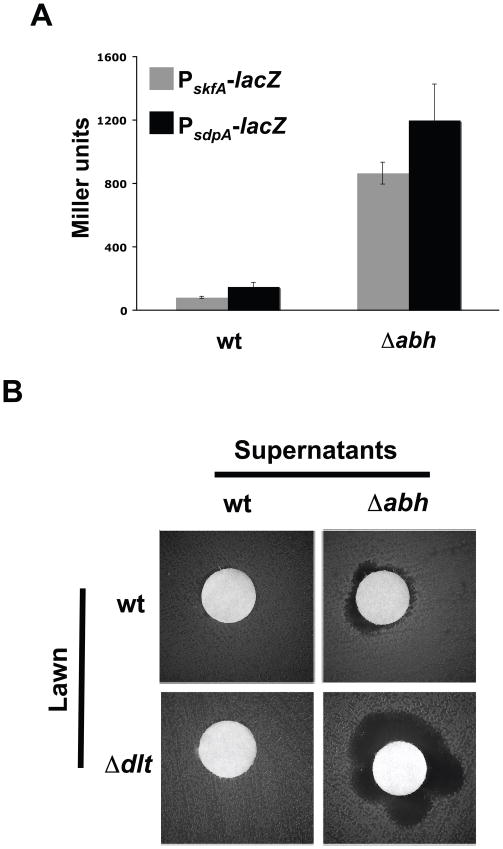
We first attempted to obtain mutants with an increase in the expression of the skf and sdp operons. Through literature searches, we identified two regulators proposed to negatively regulate the expression of several antimicrobials, including cannibalism toxins. These regulators were identified as AbrB (Antibiotic regulator of Bacillus subtilis) and Abh (AbrB-homolog) (Bobay et al., 2006, Trowsdale et al., 1979, Trowsdale et al., 1978, Yao & Strauch, 2005, Zuber & Losick, 1987). Mutants lacking either of these two regulators have increased expression of antimicrobial genes and more specifically, the cannibalism toxins (Strauch et al., 2007). In the ΔabrB mutant, biofilm development was altered independent of its effect associated with cannibalism expression (Fig. S1). This was to expected as we had previously shown that AbrB directly regulates the expression of matrix genes (Chu et al., 2008). Thus, we did not pursue the ΔabrB mutant further. Unlike AbrB, the Abh regulator is believed to regulate the expression of genes involved in matrix production indirectly (Strauch et al., 2007), a point to which we return below. To determine if the Δabh mutant has increased expression of the cannibalism operons in the strain used in our study, we made lacZ transcriptional reporter fusions to the two cannibalism operons skf and sdp and measured the level of β-galactosidase activity in the Δabh mutant compared to the wild type. Deletion of abh caused a remarkable increase in expression of both skf and sdp operons (Fig. 3A). Thus, abh indeed functions to negatively regulate cannibalism and the Δabh mutant overexpresses both the skf and sdp operons.
Fig. 3.
The Δabh Δdlt mutant over-expresses cannibalism genes. (A)β-galactosidase assay of the transcriptional gene expression of the cannibalism operons (skf and sdp) in the absence of their repressor Abh. Assays were performed on extracts of cells grown in MSgg medium for 72 h. (B) Antibiogram of the supernatant produced by wild type or the Δabh mutant in a disc over a lawn of wild type or the Δdlt mutant strain. Lawns and extracts were obtained from cells grown in MSgg medium. Assay was performed on MSgg medium for 24 h prior to imaging.
In addition to identifying mutants that overproduce the cannibalism toxins, we also searched for mutants that might be more sensitive to the action of these toxins. We once again selected targets based on previous publications. We focused our search on structures located in the cell envelope of B. subtilis, known to provide resistance to the action of antimicrobial peptides. The dltABCDE operon (henceforth dlt) encodes the protein machinery responsible for the D-alanine esterification of cell wall teichoic acids (Perego et al., 1995). The products of the dlt operon introduce positive charges in the cell envelope, making it a repulsive barrier against some peptide toxins (Cao & Helmann, 2004, Hyyrylainen et al., 2007, Perego et al., 1995). We reasoned that a Δdlt mutant lacking the protective barrier in the cell envelope would be more sensitive to the action of the cannibalism toxins. To test the sensitivity of a Δdlt mutant, a classical antibiogram experiment was performed using a lawn of the Δdlt mutant overlayed with a disc containing filter-sterilized supernatant of the Δabh mutant, which overproduces antimicrobials, as explained above. In this assay, the Δdlt mutant had a larger zone of clearance than the wild type in response to the Δabh supernatant (Fig. 3B). These results indicate that the Δdlt mutant is more sensitive to the action of antimicrobials than the wild type, although the wild-type supernatant did not visibly affect viability of the Δdlt mutant (Fig. 3B). However, the Δabh mutant does indeed produce more antimicrobials as can be observed in the ability of its supernatant to generate a killing zone when spotted over the wild-type lawn (Fig. 3B).
Our next step was to generate a strain lacking both the dlt and abh genes. As shown in the following section, the ΔdltΔabh mutant does not appear to have a growth defect compared to wild type, yet it displays a stronger response to cannibalism. It highly produces the cannibalism toxins and is at the same time more sensitive to their action. We used this mutant to study the influence of hypercannibalism in biofilm development.
Hypercannibalism influences biofilm development
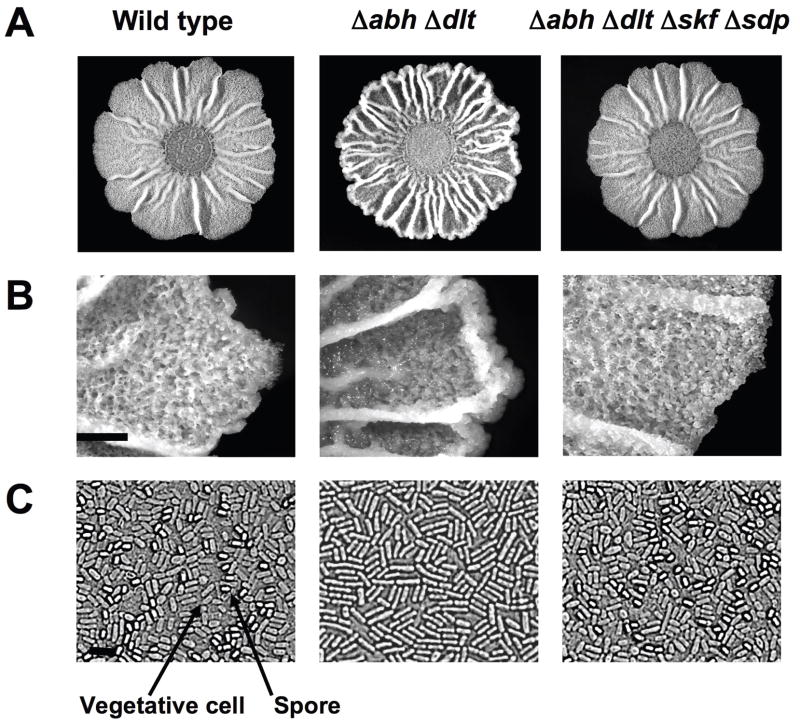
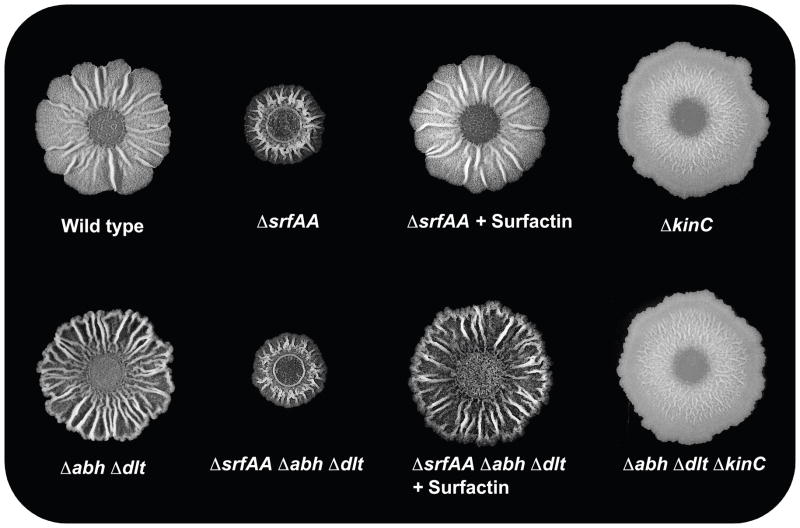
To study the effect of the Skf and Sdp toxins on biofilm development in greater detail, colony morphology of the hypercannibal mutant (ΔdltΔabh) was investigated. To do this, a spot of liquid culture was placed on the solid defined medium MSgg (Branda et al., 2001). After three days of incubation, biofilm colonies exhibit several morphological features indicative of the differentiation of distinct cell subpopulations (Fig. 4A). For example, the production of the extracellular matrix in wild-type cells results in the formation of wrinkles on the surface of the colony, which is a feature that can be correlated with the presence of matrix-producing cells (Branda et al., 2004). Similarly, the raising of aerial structures on the surface of the biofilm is indicative of the presence of a subpopulation of sporulating-cells, since the spores localized in the apical area of these structures (Branda et al., 2001) (Fig. 4B).
Fig. 4.
The hypercannibal ΔdltΔabh mutant overproduced extracellular matrix and sporulating-cells are absent. (A) Colony pictures of the strains grown on MSgg medium for 72 h. Scale bar is 1 cm. (B) Magnified images detailing the occurrence of aerial structures where spores localize on the surface of the biofilm. Scale bar is 1 mm. (C) Microscopy pictures used to determine the presence of spores in each biofilm. Representative spores and vegetative cells are denoted using arrows. Scale bar is 3 μm.
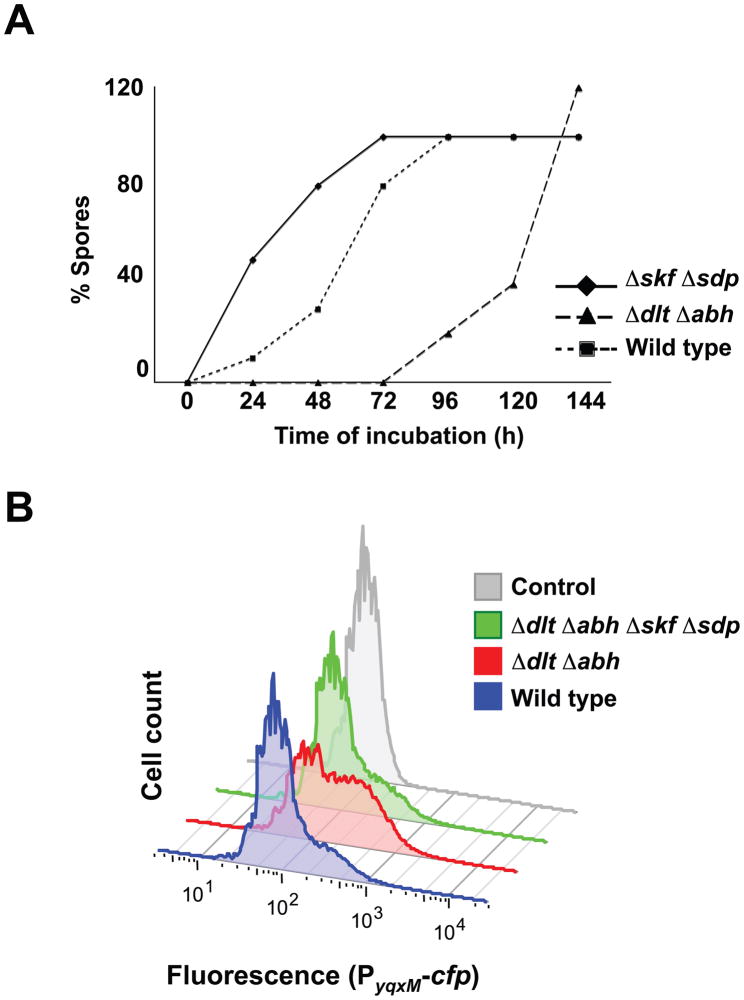
We compared the morphology of the biofilm from the hypercannibal mutant (ΔdltΔabh) to wild-type cells grown on MSgg medium. Strikingly, the hypercannibal mutant displayed increased wrinkling compared to wild type, suggesting an increase in the production of extracellular matrix in these colonies (Fig. 4A). In addition, analysis at higher magnification revealed that the hypercannibal mutant lacked the aerial structures (noticeable in between the large wrinkles) where spores preferentially are localized (Fig. 4B). Microscopic analysis showed an absence of the subpopulation of sporulating cells in the hypercannibal mutant (Fig. 4C). However, quantification of the number of viable spores over time showed that sporulation was not abolished in the hypercannibal mutant but rather delayed. After six days of biofilm development, the number of spores observed in the hypercannibal mutant was comparable to the wild type (Fig. 5A). These results further validate the proposal that cannibalism serves to delay sporulation. The original observation of Gonzalez-Pastor et al. was that mutants unable to make the toxins sporulated early (Gonzalez-Pastor et al., 2003). This result is recapitulated in Fig. 5A (Diamonds ΔskfΔsdp). Now we show the complement, when toxins are overproduced sporulation is further delayed (Fig. 5A, triangles, ΔdltΔabh).
Fig. 5.
(A) Sporulation is delayed in the hypercannibal ΔdltΔabh mutant. Quantification of vegetative CFU after heat treatment over time in different mutants. Samples were grown as colonies on MSgg and incubated at 30°C for the indicated time prior to analysis. Percent of spores is compared to the number of wild-type spores at day six of the assay. (B) Hypercannibalism increased the subpopulation of matrix-producing cells. Flow cytometry monitoring matrix producers using the reporter PyqxM-cfp. The ΔdltΔabh mutant showed a two-fold increase in cells differentiated as matrix producers compared to the wild type.
Both developmental alterations, hyperwrinkling and sporulation delay, might be explained by the action of the cannibalism toxins on the cell population. Since matrix producers secrete the cannibalism toxins and express the resistance machinery to these, an increase in toxin production should also increase the fraction of matrix producers in the population. This is because the non-matrix-producing cells are sensitive to the toxins and thus killed, ultimately serving as food for the toxin-resistant matrix producers. Thus, a greater proportion of matrix producers constitute the cell community, leading to higher production of extracellular matrix. Additionally, since cells sensitive to the toxins lyse, the process of sporulation is delayed, presumably due to the nutrients released. To test this hypothesis, we used flow cytometry to quantify the subpopulation of matrix producers in the hypercannibal mutant in comparison to the wild type. Matrix-producing cells were monitored in both the hypercannibal mutant (Δdlt Δabh) and wild type by measuring expression of the transcriptional reporter PyqxM-yfp. Results indicated that the subpopulation of matrix producers was increased in the hypercannibal mutant in comparison to the wild type (Fig. 5B). Interestingly, we did not observe any increase in the intensity of the signal but rather the number of cells expressing the reporter was greater.
To distinguish whether the increase in matrix producers in the hypercannibal mutant is exclusively related to cannibalism or if other effects associated with the mutations were involved, we deleted both skf and sdp operons in the hypercannibal background. The quadruple mutant (ΔdltΔabhΔskfΔsdp) showed a complete abrogation of the hypercannibalism phenotype. The level of colony wrinkling was comparable to the wild type indicating that matrix was not over-expressed, and the aerial structures were restored and harbored spores (Fig. 4). Measuring expression of the transcriptional reporter PyqxM-yfp in the quadruple mutant demonstrated that the subpopulation of matrix producers was restored to levels comparable to the wild type (Fig. 5B). These experiments indicated that the cannibalism genes are the principal factors inducing developmental changes in the hypercannibal mutant. We also conclude that the previously reported effect of Δabh on transcription from the yqxM promoter (Strauch et al., 2007) is an indirect consequence of its effect on skf and sdp expression.
Hypercannibalism depends on surfactin signaling
The quorum-sensing molecule surfactin activates both matrix production and cannibalism in the same subpopulation of cells via activation of the membrane kinase KinC (See Fig. 1 and 2) (Lopez et al., 2009). It therefore follows that the morphological defect associated with hypercannibalism depends on the ability of cells to sense surfactin as a signal molecule. We used the hypercannibal mutant as a background strain and inhibited the production of surfactin by deleting the srf operon (Δsrf). The colony morphologies of the Δsrf and ΔdltΔabhΔsrf mutants were indistinguishable, suggesting that the absence of surfactin also inhibits the expression of the cannibalism toxins and decreases matrix production (Fig 6). Furthermore, the addition of purified surfactin to the Δsrf and ΔdltΔabhΔsrf mutants restored the previous morphology described for the wild type and the hypercannibal mutant (Fig. 6). Consistent with a requirement for surfactin signaling, elimination of the surfactin-sensing kinase KinC in ΔkinC and ΔdltΔabhΔkinC mutants also led to a loss of wrinkled colony morphology, which is consistent with a decrease in matrix production. Again, the ΔkinC and the ΔdltΔabhΔkinC mutants were phenotypically indistinguishable, suggesting that the cannibalism genes are not expressed when the surfactin-signaling pathway is silenced.
Fig. 6.
The signaling molecule surfactin and the histidine kinase KinC are essential for the hypercannibalism phenotype. Effect of the Δsrf and the ΔkinC mutations on wild-type and hypercannibalism ΔdltΔabh mutant biofilms. Colonies were grown on MSgg at 30°C and photographed after 72 h.
Effect of other antimicrobial peptides
The subpopulation of matrix producers is resistant to the action of the cannibalism toxins due to concomitant expression of the immunity machinery required to detoxify the toxins (Gonzalez-Pastor et al., 2003). Interestingly, the protein machineries described in B. subtilis for the detoxification of antimicrobials work promiscuously for other toxins produced by different bacteria (Butcher & Helmann, 2006, Lewis, 1999). We speculate this promiscuity could be used to detoxify a large number of toxins secreted by competitors. To investigate this, we asked if another antimicrobial peptide could mimic the cannibalism effect in B. subtilis.
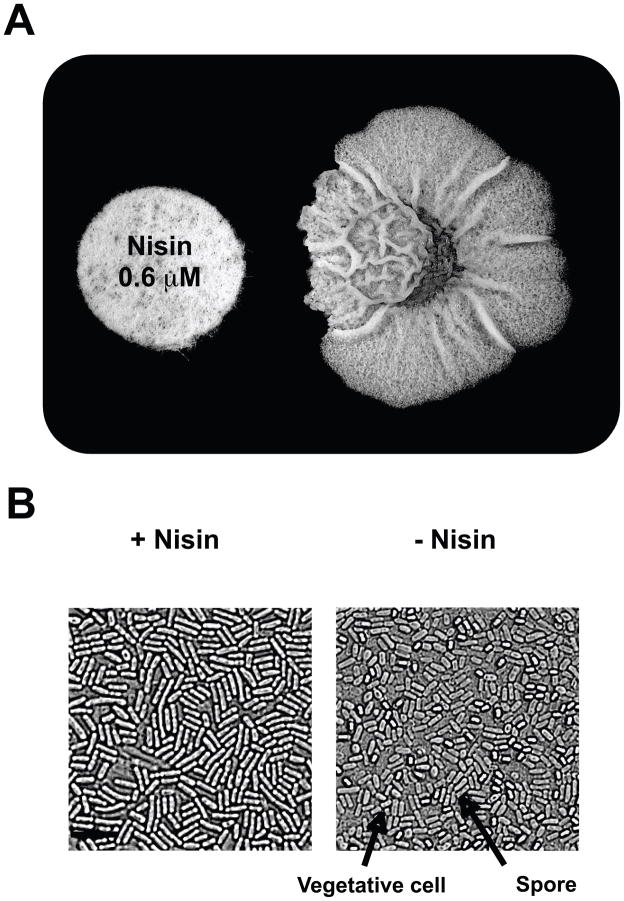
Because the molecular structures of the cannibalism toxins have not yet been elucidated, we looked for a natural product that might have similar interactions with B. subtilis as does Skf. We focused on nisin, produced by Lactococcus lactis (Mattick & Hirsch, 1944). Nisin is a peptide that exhibits antimicrobial activity towards other gram-positive bacteria, including B. subtilis (Buchman et al., 1988). Like Skf, nisin is also repelled by the barrier that the dlt operon provides to the cell wall of B. subtilis (Cao & Helmann, 2004). We assayed the effect of nisin on the biofilm of B. subtilis by simulating the secretion of nisin from a neighboring colony using an antibiogram disc impregnated with 0.6 μM of purified antibiotic. This concentration did not affect growth. After three days of incubation near the artificial hostile neighbor, the biofilm of wild-type B. subtilis displayed a remarkable increase in wrinkles in the area closest to the disc, suggesting an increase in the production of extracellular matrix in this region of the colony. Moreover, the region affected by nisin was also characterized by the absence of spores (Fig. 7A and B). The increase in matrix production and decrease in sporulation observed in the nisin-treated cells mimics the hypercannibal effect observed in the Δdlt Δabh mutant (see Fig 4). We propose that other antimicrobial peptides, including nisin, similar to the cannibalism toxins, caused the same developmental effect cannibalism exerts on the biofilms of B. subtilis.
Fig. 7.
Other peptide antimicrobials mimic the effect of cannibalism. (A) Disc impregnated with nisin (0.6 μM) set near the growth area of B. subtilis. Image was taken after 72 h of incubation on a MSgg plate at 30°C. (B) Microscopy images detailing the occurrence of spores in the region affected and non-affected with nisin. Scale bar is 3μm. Representative spores and vegetative cells are denoted with arrows.
DISCUSSION
We have presented evidence that cannibalism is involved in the development of biofilm communities in B. subtilis. The extracellular matrix of the biofilm and the Skf and Sdp cannibalism toxins are produced by the same subpopulation of cells. This subpopulation also produces the resistance machinery for the toxins. When Skf and Sdp are secreted and massive cell killing ensues the matrix producers are able to thrive using the nutrients released by lysed siblings that are sensitive to the toxins. In turn, release of nutrients promotes growth of the subpopulation of matrix producers, which increases the production of extracellular matrix and causes a temporary inhibition of sporulation.
Cannibalism functions as a mechanism of programmed cell death (PCD) to modulate diverse developmental processes in bacteria (Engelberg-Kulka et al., 2006). For example, PCD participates in the formation of fruiting bodies in Myxococcus xanthus, which a portion of cells are killed prior to differentiation of myxospores (Nariya & Inouye, 2008). In contrast, cannibalism in B. subtilis delays sporulation (Gonzalez-Pastor et al., 2003) and inhibits the formation of the aerial structures where spores localize. In Streptococcus pneumoniae, PCD is driven by a subpopulation of competent cells that lyse the non-competent siblings present in the same niche (Claverys et al., 2007, Guiral et al., 2005). In contrast to what is observed in M. xanthus and B. subtilis, the fratricide process in S. pneumoniae is triggered by excess of nutrients rather than starvation. This argues for other additional functions, aside from nutrient release, associated with PCD in bacteria.
Although the secretion of toxins promotes different developmental processes in these distinct model organisms, they might have a common feature that defines the functionality of PCD in bacteria. The antimicrobial action of the toxins promotes the survival of a sustainable subpopulation while eliminating other subpopulations no longer required to deal with external aggressions, such as nutrient limitation, pH changes, presence of antibiotics or the immune response (Claverys & Havarstein, 2007, Lewis, 2000). Based on this idea, we propose an additional role for cannibalism in B. subtilis. This would be to promote the growth of the subpopulation of matrix producers and boost the production of extracellular matrix. Other cell types will be killed by the action of the Skf and Sdp toxins. However, the remarkable resistance of spores to external damages protects them from the action of these cannibalism toxins (Setlow, 2006, Wang et al., 2006). Yet, sporulation is itself delayed by the release of nutrients from the killed cells.
Coordinated expression of cannibalism and matrix production in a subpopulation of cells is triggered by the quorum-sensing molecule surfactin. This could result in a fitness advantage in natural habitats, where producing an extracellular matrix would provide protection at the same time as an effective arsenal of toxins to kill neighboring bacteria who may be competing for the same resources. Consistent with this, it has been reported that B. subtilis preferentially uses the Skf and Sdp toxins to kill other bacteria rather than itself in mixed cultures (Lin et al., 2001, Nandy et al., 2007).
The chances that other soil bacteria will trigger cannibalism in B. subtilis increases with the ability of other soil microorganisms to produce natural products that mimic the action of the signaling molecule surfactin. The membrane kinase KinC senses leakage of potassium caused by the action of surfactin in the cell membrane, rather than the structure of the molecule itself. This mechanism of activation allows cells to promiscuously sense the presence of other small-molecules with physiological effects similar to those of surfactin. These natural products include the structurally unrelated compounds nystatin, amphotericin, valinomycin and gramicidin which can all induce the quorum-sensing pathway and cause differentiation of the subpopulation of matrix producers (Lopez et al., 2009). Additionally, other antimicrobial peptides secreted by soil bacteria could mimic the effect of the cannibalism toxins, due to the ability of the resistance machinery to work non-specifically for several similar molecules (Butcher & Helmann, 2006). This ability to sense small-molecules produced by a range of diverse soil microorganisms suggests a broad mechanism that B. subtilis uses to respond to surrounding bacterial communities by altering its development.
EXPERIMENTAL PROCEDURES
Strains, media and culture conditions
B. subtilis strain NCIB3610 was used as the wild type. For biofilm formation experiments 3μl of a cell suspension were spotted onto defined MSgg medium supplemented with 1.5% Bacto agar and incubated at 30 C for three days (Branda et al., 2001). For Fig 1, 20 μM purified surfactin (Sigma) was added to LB broth prior to inoculation and cells were incubated for 8 h prior to harvesting for flow cytometry. For Fig 6, surfactin was added by spotting 3 μl of 10 mM purified surfactin onto the surface of the MSgg agar prior to inoculation.
Strains, reporter construction and measurements
Strains used and generated in this work are listed in table S1. Deletion mutants Δabh::km, Δdlt::tet, Δdlt::km, Δskf::cm, Δsdp::spc, were generated using long flanking homology PCR (Wach, 1996) (using the primers listed in table S2). Transcriptional fusions to lacZ were cloned in pDG1663 vector using primers listed in table 2 (Guerout-Fleury et al., 1996) and inserted into the amyE locus by natural competence. All constructions were transferred to NCIB3610 and mutant strains when required by SPP1 phage transduction (Yasbin & Young, 1974). β-galactosidase assays and spore counting measurements were performed according to the literature (Kearns et al., 2005, Vlamakis et al., 2008).
Image capture and analysis
Colonies were photographed using a Zeiss Stemi SV6 Stereoscope connected to a color AxioCam®. Microscopy images were taken on a Nikon Eclipse TE2000-U microscope equipped with an X-cite 120 illumination system, using a Hamamatsu digital camera model ORCA-ER. Fluorescence signal was detected with a Ex436/500 filter. Image processing was done using MetaMorph® Software and Photoshop®.
Flow cytometry
Biofilms were harvested, resuspended in 1 ml of PBS buffer and dispersed with three pulses of mild sonication. Cells were fixed in 4% paraformaldehyde for 7 min, washed with PBS and resuspended in GTE buffer (50mM Glucose, 10 mM EDTA pH 8, 20 mM Tris-HCl pH 8). Mild sonication of the samples was required to obtain single cells (Branda et al., 2006).
For flow cytometric analysis, cells were diluted 1:100 in PBS and measured on a BD LSR II flow cytometer (BD Biosciences) using a solid state laser. For YFP fluorescence, we used a laser excitation of 488 nm coupled with 530/30 and 505LP sequential filters. For CFP fluorescence, we used laser excitation at 405 nm coupled with 408/40 and 460LP sequential filters. The photomultiplier voltage was set between 400 and 500 V. Every sample was analyzed measuring 50,000 events using FACS Diva (BD Biosciences) software to capture the data. Further analysis was performed using FlowJo 8.7.2 software (http://www.flowjo.com).
Supplementary Material
Acknowledgments
We thank the core facilities of the Harvard FAS Center for Systems Biology and P. Rogers and B. Tilton for guidance using flow cytometry. We thank A. Earl, Y. Chai, F. Chu, A. McLoon, D. Romero, and C. Aguilar for helpful discussions. This work was funded by the BASF Advance Research Initiative at Harvard (BARI) to R.K., R.L. D.L, and H.V. Funding also from NIH grant GM58213 to R.K and GM18568 to R.L. D.L. was the recipient of a postdoctoral fellowship from the Fundación Séneca, Comunidad Autónoma de la Región de Murcia (Spain).
References
- Bobay BG, Mueller GA, Thompson RJ, Murzin AG, Venters RA, Strauch MA, Cavanagh J. NMR structure of AbhN and comparison with AbrBN: FIRST insights into the DNA binding promiscuity and specificity of AbrB-like transition state regulator proteins. J Biol Chem. 2006;281:21399–21409. doi: 10.1074/jbc.M601963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman GW, Banerjee S, Hansen JN. Structure, expression, and evolution of a gene encoding the precursor of nisin, a small protein antibiotic. J Biol Chem. 1988;263:16260–16266. [PubMed] [Google Scholar]
- Butcher BG, Helmann JD. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- Cao M, Helmann JD. The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J Bacteriol. 2004;186:1136–1146. doi: 10.1128/JB.186.4.1136-1146.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- Chu F, Kearns DB, McLoon A, Chai Y, Kolter R, Losick R. A novel regulatory protein governing biofilm formation in. Bacillus subtilis Mol Microbiol. 2008;68:1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d’etre. Nat Rev Microbiol. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- Claverys JP, Martin B, Havarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;64:1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2006;2:e135. doi: 10.1371/journal.pgen.0020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Fujita M, Gonzalez-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier NP. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs EC. Dept of Molecular and Cellular Biology. Cambirdge: Harvard University; 2006. Control of cannibalism in Bacillus subtilis. [Google Scholar]
- Hyyrylainen HL, Pietiainen M, Lunden T, Ekman A, Gardemeister M, Murtomaki-Repo S, Antelmann H, Hecker M, Valmu L, Sarvas M, Kontinen VP. The density of negative charge in the cell wall influences two-component signal transduction in Bacillus subtilis. Microbiology. 2007;153:2126–2136. doi: 10.1099/mic.0.2007/008680-0. [DOI] [PubMed] [Google Scholar]
- Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- LeDeaux JR, Yu N, Grossman AD. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:861–863. doi: 10.1128/jb.177.3.861-863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Multidrug resistance: Versatile drug sensors of bacterial cells. Curr Biol. 1999;9:R403–407. doi: 10.1016/s0960-9822(99)80254-1. [DOI] [PubMed] [Google Scholar]
- Lewis K. Programmed death in bacteria. Microbiol Mol Biol Rev. 2000;64:503–514. doi: 10.1128/mmbr.64.3.503-514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Qu LJ, Gu H, Chen Z. A 3.1-kb genomic fragment of Bacillus subtilis encodes the protein inhibiting growth of Xanthomonas oryzae pv. oryzae. J Appl Microbiol. 2001;91:1044–1050. doi: 10.1046/j.1365-2672.2001.01475.x. [DOI] [PubMed] [Google Scholar]
- Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A. 2009;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandy SK, Bapat PM, Venkatesh KV. Sporulating bacteria prefers predation to cannibalism in mixed cultures. FEBS Lett. 2007;581:151–156. doi: 10.1016/j.febslet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem. 1995;270:15598–15606. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]
- Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr Opin Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Strauch MA, Bobay BG, Cavanagh J, Yao F, Wilson A, Le Breton Y. Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J Bacteriol. 2007;189:7720–7732. doi: 10.1128/JB.01081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J, Chen SM, Hoch JA. Genetic analysis of a class of polymyxin resistant partial revertants of stage O sporulation mutants of Bacillus subtilis: map of the chromosome region near the origin of replication. Mol Gen Genet. 1979;173:61–70. doi: 10.1007/BF00267691. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Sheflett M, Hoch JA. New cluster of ribosomal genes in Bacillus subtilis with regulatory role in sporulation. Nature. 1978;272:179–181. doi: 10.1038/272179a0. [DOI] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bethedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wang ST, Setlow B, Conlon EM, Lyon JL, Imamura D, Sato T, Setlow P, Losick R, Eichenberger P. The forespore line of gene expression in Bacillus subtilis. J Mol Biol. 2006;358:16–37. doi: 10.1016/j.jmb.2006.01.059. [DOI] [PubMed] [Google Scholar]
- Yao F, Strauch MA. Independent and interchangeable multimerization domains of the AbrB, Abh, and SpoVT global regulatory proteins. J Bacteriol. 2005;187:6354–6362. doi: 10.1128/JB.187.18.6354-6362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P, Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.