Abstract
In the title compound, C14H10Cl2N2O2, the dihedral angle between the two aromatic rings is 17.39 (4)°. An intramolecular O—H⋯O hydrogen bond forms a six-membered R(6)1 1 ring motif. In the crystal structure, intermolecular N—H⋯O and O—H⋯O hydrogen-bonding interactions occur.
Related literature
For the biological activity of Schiff bases, see: El-Masry et al. (2000 ▶); Samadhiya & Halve (2001 ▶). For the synthesis of Schiff bases, see: Siddiqui et al. (2006 ▶); Iqbal et al. (2007 ▶). For applications of Schiff bases, see: Mookherjee et al. (1989 ▶); Kumar et al. (2009 ▶). For graph-set notation, see: Bernstein et al. (1995 ▶).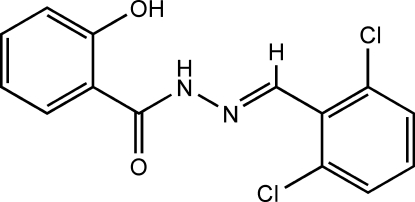
Experimental
Crystal data
C14H10Cl2N2O2
M r = 309.14
Monoclinic,

a = 7.5029 (6) Å
b = 23.8363 (13) Å
c = 8.0286 (7) Å
β = 109.860 (6)°
V = 1350.45 (18) Å3
Z = 4
Mo Kα radiation
μ = 0.48 mm−1
T = 180 K
0.34 × 0.26 × 0.18 mm
Data collection
Stoe IPDS-2T diffractometer
Absorption correction: multi-scan (SADABS; Bruker, 2007 ▶) T min = 0.856, T max = 0.921
21101 measured reflections
2958 independent reflections
2455 reflections with I > 2σ(I)
R int = 0.041
Refinement
R[F 2 > 2σ(F 2)] = 0.029
wR(F 2) = 0.074
S = 1.03
2958 reflections
191 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.24 e Å−3
Δρmin = −0.39 e Å−3
Data collection: X-AREA (Stoe & Cie, 1999 ▶); cell refinement: X-AREA; data reduction: X-AREA; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶) and PLATON (Spek, 2009 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON.
Supplementary Material
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810035385/im2223sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810035385/im2223Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯N2i | 0.85 (2) | 2.38 (2) | 3.165 (2) | 153 (2) |
| N1—H1⋯O1i | 0.85 (2) | 2.45 (2) | 3.158 (2) | 140 (1) |
| O2—H2A⋯O1 | 0.90 (2) | 1.78 (2) | 2.608 (2) | 153 (2) |
Symmetry code: (i)  .
.
Acknowledgments
HLS is grateful to the Institute of Chemistry, University of the Punjab, for financial support.
supplementary crystallographic information
Comment
Schiff base reaction products of alkyl anthranilates and their derivatives have been employed in augmenting the aroma or taste of consumable materials including perfume compositions, colognes, perfumed articles, foodstuffs, chewing gums and beverages (Mookherjee et al., 1989). Related compounds have also shown to exhibit biological activities such as antibacterial, antimicrobial (El-Masry et al., 2000), and were investigated as herbicides (Samadhiya et al., 2001). Further, Schiff bases have also been employed as ligands for complexation of metal ions (Kumar et al., 2009). With this perspective of widespread applications of Schiff bases we embarked on the synthesis, characterization and biological evaluation of this class of compounds (Siddiqui et al., 2006; Iqbal et al., 2007). Herein, we report the synthesis and crystal structure of the title compound.
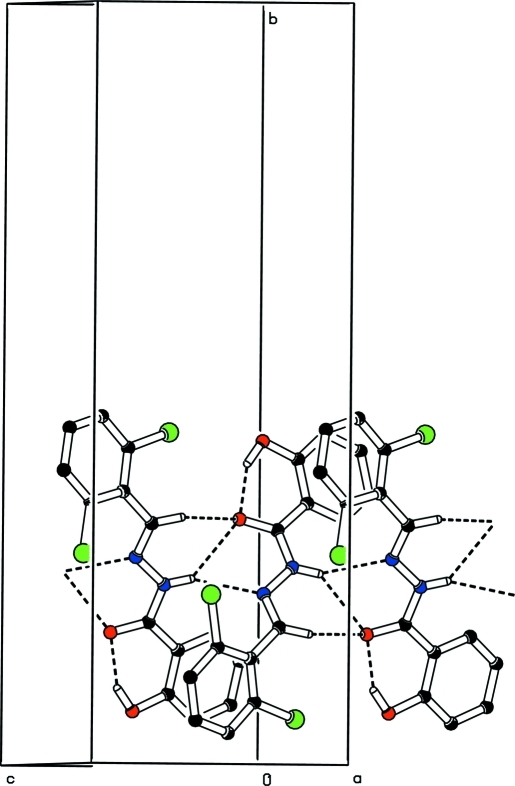
The title compound is presented in Fig.1. The two aromatic ring systems in the hydrazide are inclined at an angle of 17.39 (0.04) ° with respect to each other. The structure possesses classical inter and intra molecular hydrogen bonding. The intramolecular O–H···O type hydrogen bonding forms six membered ring motif R(6)11 (Bernstein et al., 1995) which inclines at an angle of 9.73 (0.14) ° with respect to aromatic C1–C6. The intermolecular C–H···O and N—H···N type of hydrogen bonding forms nine membered ring motif R(9)22 (Bernstein et al., 1995) where N–H···O type of hydrogen bonding interveins to form a six and a five membered ring system R(6)21 and R(5)12(Bernstein et al., 1995), respectively (Fig. 2, table 1).
Experimental
A mixture of 2-hydroxy-benzoic acid hydrazide (1.5 g, 10.0 mmol) and 2,6-dichlorobenzaldehyde (1.7 g, 10.0 mmol) in absolute ethanol (20 ml) was heated to reflux (2 hrs.), cooled to room temperature and filtered. The off-white precipitates were washed with the same solvent and dried at room temperature to yield 2.8 g of off-white, needle-like crystals of the title compound (9.1 mmol, 90.6%). Suitable crystals were grown from a solution of CH3OH by slow evaporation at room temperature.
Refinement
All aromatic H-atoms were positioned geometrically with C—H = 0.95 Å and refined using riding model with Uiso(H) = 1.2 Ueq(C), while the imine hydrogen was located in difference map and was refined with C—H = 0.95 (2) Å and Uiso(H) = 1.2 Ueq(C8). N–H and O–H H atoms also were located in difference map and were refined with N—H = 0.86 (2) Å and O—H = 0.89 (2) Å and Uiso(H) = 1.2 Ueq(N) and Uiso(H) = 1.5 Ueq(O), respectively.
Figures
Fig. 1.
Molecular structure of the title compound with thermal ellipsoids drawn at the 50% probability level.
Fig. 2.
Packing diagram of the crystal structure showing hydrogen bonding as dashed lines.
Crystal data
| C14H10Cl2N2O2 | F(000) = 632 |
| Mr = 309.14 | Dx = 1.520 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 19179 reflections |
| a = 7.5029 (6) Å | θ = 1.7–29.5° |
| b = 23.8363 (13) Å | µ = 0.48 mm−1 |
| c = 8.0286 (7) Å | T = 180 K |
| β = 109.860 (6)° | Needles, white |
| V = 1350.45 (18) Å3 | 0.34 × 0.26 × 0.18 mm |
| Z = 4 |
Data collection
| Stoe IPDS-2T diffractometer | 2958 independent reflections |
| Radiation source: fine-focus sealed tube | 2455 reflections with I > 2σ(I) |
| graphite | Rint = 0.041 |
| ω scans | θmax = 27.0°, θmin = 1.7° |
| Absorption correction: multi-scan (SADABS; Bruker, 2007) | h = −9→9 |
| Tmin = 0.856, Tmax = 0.921 | k = −30→30 |
| 21101 measured reflections | l = −10→10 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.029 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.074 | w = 1/[σ2(Fo2) + (0.0408P)2 + 0.2354P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.03 | (Δ/σ)max = 0.001 |
| 2958 reflections | Δρmax = 0.24 e Å−3 |
| 191 parameters | Δρmin = −0.39 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0085 (19) |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.66364 (6) | 0.562338 (15) | 0.52580 (5) | 0.03982 (12) | |
| Cl2 | 0.28455 (5) | 0.723630 (17) | 0.06158 (5) | 0.03924 (12) | |
| O1 | 0.77275 (17) | 0.82495 (4) | 0.34242 (14) | 0.0369 (3) | |
| O2 | 0.84369 (18) | 0.92793 (5) | 0.45517 (16) | 0.0431 (3) | |
| H2A | 0.809 (3) | 0.8981 (10) | 0.383 (3) | 0.065* | |
| N1 | 0.72285 (16) | 0.76179 (5) | 0.53258 (15) | 0.0239 (2) | |
| H1 | 0.720 (2) | 0.7541 (7) | 0.636 (2) | 0.029* | |
| N2 | 0.63961 (16) | 0.72631 (5) | 0.39193 (15) | 0.0233 (2) | |
| C1 | 0.9163 (2) | 0.90454 (6) | 0.6187 (2) | 0.0308 (3) | |
| C2 | 1.0154 (2) | 0.93943 (7) | 0.7591 (2) | 0.0383 (4) | |
| H2 | 1.0262 | 0.9784 | 0.7396 | 0.046* | |
| C3 | 1.0976 (2) | 0.91717 (7) | 0.9261 (2) | 0.0409 (4) | |
| H3 | 1.1648 | 0.9411 | 1.0215 | 0.049* | |
| C4 | 1.0837 (2) | 0.86024 (7) | 0.9572 (2) | 0.0374 (4) | |
| H4 | 1.1431 | 0.8452 | 1.0723 | 0.045* | |
| C5 | 0.9828 (2) | 0.82576 (6) | 0.81912 (18) | 0.0281 (3) | |
| H5 | 0.9726 | 0.7869 | 0.8404 | 0.034* | |
| C6 | 0.89542 (19) | 0.84719 (6) | 0.64844 (18) | 0.0246 (3) | |
| C7 | 0.79269 (19) | 0.81097 (6) | 0.49710 (18) | 0.0246 (3) | |
| C8 | 0.56322 (19) | 0.68201 (6) | 0.42645 (18) | 0.0242 (3) | |
| H8 | 0.562 (2) | 0.6739 (7) | 0.542 (2) | 0.029* | |
| C9 | 0.47973 (19) | 0.64095 (6) | 0.28383 (17) | 0.0252 (3) | |
| C10 | 0.5196 (2) | 0.58369 (6) | 0.31608 (19) | 0.0286 (3) | |
| C11 | 0.4494 (2) | 0.54288 (7) | 0.1881 (2) | 0.0379 (4) | |
| H11 | 0.4807 | 0.5045 | 0.2145 | 0.045* | |
| C12 | 0.3331 (3) | 0.55883 (8) | 0.0214 (2) | 0.0439 (4) | |
| H12 | 0.2849 | 0.5313 | −0.0681 | 0.053* | |
| C13 | 0.2862 (2) | 0.61464 (8) | −0.0162 (2) | 0.0406 (4) | |
| H13 | 0.2048 | 0.6254 | −0.1307 | 0.049* | |
| C14 | 0.3588 (2) | 0.65488 (6) | 0.11426 (19) | 0.0297 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0478 (2) | 0.02692 (19) | 0.0405 (2) | 0.00367 (16) | 0.00954 (17) | 0.00623 (15) |
| Cl2 | 0.03272 (19) | 0.0378 (2) | 0.0399 (2) | −0.00243 (15) | 0.00277 (15) | 0.01388 (16) |
| O1 | 0.0530 (7) | 0.0320 (6) | 0.0225 (5) | −0.0099 (5) | 0.0088 (5) | 0.0033 (4) |
| O2 | 0.0526 (7) | 0.0255 (5) | 0.0402 (7) | −0.0026 (5) | 0.0013 (5) | 0.0067 (5) |
| N1 | 0.0302 (6) | 0.0239 (6) | 0.0174 (5) | −0.0030 (4) | 0.0079 (5) | −0.0014 (4) |
| N2 | 0.0253 (5) | 0.0229 (5) | 0.0214 (5) | 0.0000 (4) | 0.0077 (4) | −0.0026 (4) |
| C1 | 0.0309 (7) | 0.0255 (7) | 0.0350 (8) | 0.0002 (5) | 0.0097 (6) | 0.0000 (6) |
| C2 | 0.0373 (8) | 0.0269 (7) | 0.0494 (9) | −0.0047 (6) | 0.0130 (7) | −0.0102 (7) |
| C3 | 0.0394 (8) | 0.0431 (9) | 0.0382 (9) | −0.0097 (7) | 0.0104 (7) | −0.0166 (7) |
| C4 | 0.0375 (8) | 0.0470 (9) | 0.0261 (7) | −0.0070 (7) | 0.0088 (6) | −0.0059 (6) |
| C5 | 0.0273 (7) | 0.0315 (7) | 0.0257 (7) | −0.0033 (5) | 0.0092 (6) | −0.0009 (5) |
| C6 | 0.0237 (6) | 0.0254 (7) | 0.0249 (7) | −0.0003 (5) | 0.0084 (5) | −0.0026 (5) |
| C7 | 0.0269 (7) | 0.0229 (6) | 0.0232 (7) | 0.0022 (5) | 0.0076 (5) | 0.0011 (5) |
| C8 | 0.0274 (7) | 0.0229 (6) | 0.0225 (7) | 0.0012 (5) | 0.0087 (5) | 0.0011 (5) |
| C9 | 0.0272 (6) | 0.0257 (7) | 0.0250 (7) | −0.0032 (5) | 0.0118 (5) | −0.0010 (5) |
| C10 | 0.0310 (7) | 0.0272 (7) | 0.0305 (7) | −0.0025 (6) | 0.0142 (6) | −0.0012 (5) |
| C11 | 0.0469 (9) | 0.0279 (7) | 0.0450 (9) | −0.0067 (6) | 0.0238 (8) | −0.0096 (7) |
| C12 | 0.0539 (10) | 0.0437 (10) | 0.0386 (9) | −0.0164 (8) | 0.0214 (8) | −0.0176 (7) |
| C13 | 0.0431 (9) | 0.0520 (10) | 0.0255 (7) | −0.0143 (8) | 0.0102 (7) | −0.0048 (7) |
| C14 | 0.0299 (7) | 0.0324 (7) | 0.0279 (7) | −0.0058 (6) | 0.0114 (6) | 0.0022 (6) |
Geometric parameters (Å, °)
| Cl1—C10 | 1.7395 (15) | C4—H4 | 0.9500 |
| Cl2—C14 | 1.7364 (16) | C5—C6 | 1.399 (2) |
| O1—C7 | 1.2448 (17) | C5—H5 | 0.9500 |
| O2—C1 | 1.3582 (19) | C6—C7 | 1.4763 (18) |
| O2—H2A | 0.90 (2) | C8—C9 | 1.4745 (19) |
| N1—C7 | 1.3533 (18) | C8—H8 | 0.954 (17) |
| N1—N2 | 1.3788 (15) | C9—C14 | 1.396 (2) |
| N1—H1 | 0.854 (18) | C9—C10 | 1.402 (2) |
| N2—C8 | 1.2761 (18) | C10—C11 | 1.383 (2) |
| C1—C2 | 1.395 (2) | C11—C12 | 1.379 (3) |
| C1—C6 | 1.406 (2) | C11—H11 | 0.9500 |
| C2—C3 | 1.378 (2) | C12—C13 | 1.383 (3) |
| C2—H2 | 0.9500 | C12—H12 | 0.9500 |
| C3—C4 | 1.390 (3) | C13—C14 | 1.388 (2) |
| C3—H3 | 0.9500 | C13—H13 | 0.9500 |
| C4—C5 | 1.381 (2) | ||
| C1—O2—H2A | 103.3 (15) | O1—C7—C6 | 121.12 (12) |
| C7—N1—N2 | 117.32 (11) | N1—C7—C6 | 117.68 (12) |
| C7—N1—H1 | 121.9 (11) | N2—C8—C9 | 118.97 (12) |
| N2—N1—H1 | 120.5 (11) | N2—C8—H8 | 122.3 (10) |
| C8—N2—N1 | 116.20 (11) | C9—C8—H8 | 118.7 (10) |
| O2—C1—C2 | 117.72 (14) | C14—C9—C10 | 116.02 (13) |
| O2—C1—C6 | 122.16 (13) | C14—C9—C8 | 124.31 (13) |
| C2—C1—C6 | 120.12 (14) | C10—C9—C8 | 119.67 (12) |
| C3—C2—C1 | 119.77 (15) | C11—C10—C9 | 122.96 (15) |
| C3—C2—H2 | 120.1 | C11—C10—Cl1 | 117.91 (12) |
| C1—C2—H2 | 120.1 | C9—C10—Cl1 | 119.14 (11) |
| C2—C3—C4 | 120.96 (15) | C12—C11—C10 | 118.86 (15) |
| C2—C3—H3 | 119.5 | C12—C11—H11 | 120.6 |
| C4—C3—H3 | 119.5 | C10—C11—H11 | 120.6 |
| C5—C4—C3 | 119.43 (15) | C11—C12—C13 | 120.47 (15) |
| C5—C4—H4 | 120.3 | C11—C12—H12 | 119.8 |
| C3—C4—H4 | 120.3 | C13—C12—H12 | 119.8 |
| C4—C5—C6 | 121.02 (14) | C12—C13—C14 | 119.64 (16) |
| C4—C5—H5 | 119.5 | C12—C13—H13 | 120.2 |
| C6—C5—H5 | 119.5 | C14—C13—H13 | 120.2 |
| C5—C6—C1 | 118.64 (13) | C13—C14—C9 | 122.02 (15) |
| C5—C6—C7 | 122.17 (12) | C13—C14—Cl2 | 117.22 (12) |
| C1—C6—C7 | 119.07 (12) | C9—C14—Cl2 | 120.70 (11) |
| O1—C7—N1 | 121.20 (12) | ||
| C7—N1—N2—C8 | −175.33 (12) | N1—N2—C8—C9 | −177.11 (11) |
| O2—C1—C2—C3 | −177.58 (15) | N2—C8—C9—C14 | −47.26 (19) |
| C6—C1—C2—C3 | 1.9 (2) | N2—C8—C9—C10 | 133.40 (14) |
| C1—C2—C3—C4 | 0.1 (2) | C14—C9—C10—C11 | 1.8 (2) |
| C2—C3—C4—C5 | −1.3 (2) | C8—C9—C10—C11 | −178.78 (13) |
| C3—C4—C5—C6 | 0.4 (2) | C14—C9—C10—Cl1 | −178.18 (10) |
| C4—C5—C6—C1 | 1.6 (2) | C8—C9—C10—Cl1 | 1.21 (18) |
| C4—C5—C6—C7 | 177.71 (13) | C9—C10—C11—C12 | −0.7 (2) |
| O2—C1—C6—C5 | 176.71 (13) | Cl1—C10—C11—C12 | 179.27 (12) |
| C2—C1—C6—C5 | −2.8 (2) | C10—C11—C12—C13 | −0.6 (2) |
| O2—C1—C6—C7 | 0.5 (2) | C11—C12—C13—C14 | 0.8 (2) |
| C2—C1—C6—C7 | −178.98 (13) | C12—C13—C14—C9 | 0.4 (2) |
| N2—N1—C7—O1 | 4.02 (19) | C12—C13—C14—Cl2 | −176.77 (13) |
| N2—N1—C7—C6 | −175.26 (11) | C10—C9—C14—C13 | −1.6 (2) |
| C5—C6—C7—O1 | −155.05 (14) | C8—C9—C14—C13 | 179.01 (14) |
| C1—C6—C7—O1 | 21.0 (2) | C10—C9—C14—Cl2 | 175.41 (10) |
| C5—C6—C7—N1 | 24.23 (19) | C8—C9—C14—Cl2 | −3.95 (19) |
| C1—C6—C7—N1 | −159.71 (13) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···N2i | 0.85 (2) | 2.38 (2) | 3.165 (2) | 153 (2) |
| N1—H1···O1i | 0.85 (2) | 2.45 (2) | 3.158 (2) | 140 (1) |
| O2—H2A···O1 | 0.90 (2) | 1.78 (2) | 2.608 (2) | 153 (2) |
Symmetry codes: (i) x, −y+3/2, z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IM2223).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Bruker (2007). SADABS Bruker AXS Inc., Madison, Wisconsin, USA.
- El-Masry, A. H., Fahmy, H. H. & Abdelwahed, S. H. A. (2000). Molecules, 5, 1429–1438.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Iqbal, A., Siddiqui, H. L., Ashraf, C. M., Ahmad, M. & Weaver, G. W. (2007). Molecules, 12, 245–254. [DOI] [PMC free article] [PubMed]
- Kumar, S., Dhar, D. N. & Saxena, P. N. (2009). J. Sci. Ind. Res.68, 181–187.
- Mookherjee, B. D., Trenkle, R. W., Calderone, N., Schreck, L. & Sands, K. P. (1989). US Patent 4 839 083 (58 pp.).
- Samadhiya, S. & Halve, A. (2001). Orient. J. Chem.17, 119–122.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Siddiqui, H. L., Iqbal, A., Ahmad, S. & Weaver, G. W. (2006). Molecules, 11, 206–211. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (1999). X-AREA Stoe & Cie GmbH, Darmstadt, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks I, global. DOI: 10.1107/S1600536810035385/im2223sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810035385/im2223Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report