Abstract
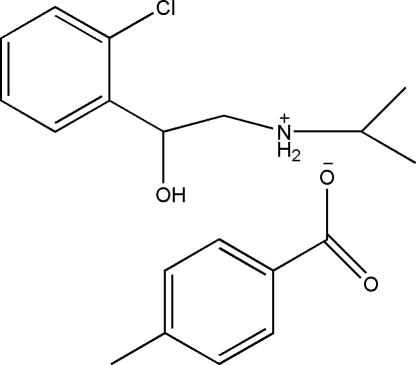
The title compound, C11H17ClNO+·C8H7O2 −, was obtained by the reaction of chlorprenaline {or 1-(2-chlorophenyl)-2-[(1-methylethyl)amino]ethanol} and p-toluic acid. The chlorprenaline is twisted moderately with a C—C—C—C torsion angle of 109.6 (2)°. The two molecules are linked by classical O—H⋯O and N—H⋯O hydrogen bonds. Further N—H⋯O hydrogen bonds link two of these units into dimers.
Related literature
For related structures, see: Feng et al. (2010 ▶); Takwale & Pant (1971 ▶); Tang et al. (2009a
▶,b
▶).
Experimental
Crystal data
C11H17ClNO+·C8H7O2 −
M r = 349.84
Monoclinic,
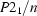
a = 8.5966 (4) Å
b = 8.1288 (3) Å
c = 26.8949 (12) Å
β = 91.600 (1)°
V = 1878.68 (14) Å3
Z = 4
Mo Kα radiation
μ = 0.22 mm−1
T = 296 K
0.37 × 0.30 × 0.22 mm
Data collection
Rigaku R-AXIS RAPID/ZJUG CCD diffractometer
Absorption correction: multi-scan (ABSCOR; Higashi, 1995 ▶) T min = 0.913, T max = 0.953
27924 measured reflections
4271 independent reflections
2763 reflections with I > 2σ(I)
R int = 0.036
Refinement
R[F 2 > 2σ(F 2)] = 0.042
wR(F 2) = 0.125
S = 1.00
4271 reflections
222 parameters
H-atom parameters constrained
Δρmax = 0.21 e Å−3
Δρmin = −0.31 e Å−3
Data collection: PROCESS-AUTO (Rigaku/MSC, 2006 ▶); cell refinement: PROCESS-AUTO; data reduction: CrystalStructure (Rigaku/MSC, 2007 ▶); program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810033878/rk2225sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810033878/rk2225Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H101⋯O3 | 0.82 | 1.88 | 2.6986 (18) | 173 |
| N1—H103⋯O2 | 0.90 | 1.91 | 2.7835 (18) | 164 |
| N1—H102⋯O3i | 0.90 | 1.89 | 2.7824 (19) | 174 |
Symmetry code: (i) 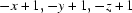 .
.
Acknowledgments
This project was supported by the Zhejiang Provincial Natural Science Foundation of China (grant No. Y206174).
supplementary crystallographic information
Comment
A recent study reports the structure of bis{N-[2-(2-chlorophenyl)-2-hydroxyethyl]propan-2-aminium} oxalate (Tang et al., 2009b), which was synthesized by oxalic acid and chlorprenaline (Tang et al., 2009a). Here using p-toluic acid instead of oxalic acid and following a similar synthetic procedure yields the title compound, I.
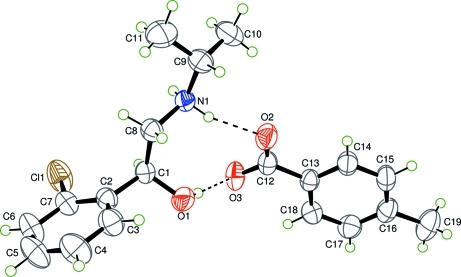
In I, the chlorprenaline molecule and the p-toluic molecule are linked to each other by the classical N1—H103···O2 hydogen bond [2.7835 (18)Å] and the O1—H101···O3 hydogen bond [2.6986 (18)Å] (Fig. 1 & Table 1). The chlorprenaline in I are twisted moderately as compared with those of other compounds. The C7—C2—C1—C8 torsion angle of 109.6 (2)° is larger than the value of the similar torsion angle of 91.9 (2)° (Tang et al., 2009a). The C12–O2 distance of 1.248 (2)Å is much shorter than the similar distance of 1.292 (8)Å (Takwale & Pant, 1971). The C9–N1 distance of 1.507 (2)Å is longer than the value of the similar bond distance of 1.473 (4)Å (Tang et al., 2009b), as similar as the value of the similar bond distance of 1.503 (2)Å (Feng et al., 2010).
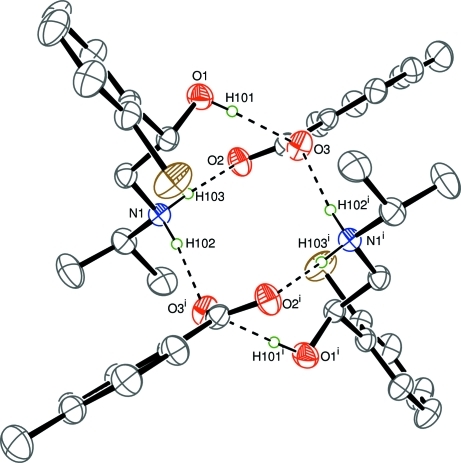
Classical O—H···O and N—H···O hydrogen bonds are found in the cystal structure (Fig. 2) are essential forces in crystal formation.
Experimental
Racemic chlorprenaline was prepared by chlorprenaline hydrochloride purchased from ShangHai Shengxin Medicine & Chemical Co., Ltd. ShangHai, China. Chlorprenaline hydrochloride and NaOH in a molar ratio of 1:1 were mixed and dissolved in a methanol–water solution (1:1 v/v). The precipitate formed was filtered off, washed with water and dried. It was used without further purification. Racemic chlorprenaline (0.5 g, 0.0023 mol) was dissolved in methanol (7 ml) and then p-toluylic acid (0.31 g, 0.0023 mol) was added.The mixture was dissovled by heating to 343 K where a clear solution resulted. The resulting solution was concentrated at ambient temperature. Colourless crystals of title compound separated from the solution in about 70% yield after one day.
Refinement
All of the H atoms were placed in calculated positions and allowed to ride on their parent atoms at distances of 0.93Å (aromatic), 0.98Å (methine), 0.97Å (methylene), 0.96Å (methyl) 0.82Å (hydroxyl) and 0.90Å (amine), with Uiso(H) = 1.2–1.5 Ueq(parent atom).
Figures
Fig. 1.
The asymmetric unit of I with atom numbering scheme. Displacement ellipsoids are presented at 50% probability level. H atoms are presented as a small spheres of arbitrary radius.
Fig. 2.
The two units of I with atom labels. The dashed lines indicate hydrogen bonds.
Crystal data
| C11H17ClNO+·C8H7O2− | F(000) = 744 |
| Mr = 349.84 | Dx = 1.237 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 17098 reflections |
| a = 8.5966 (4) Å | θ = 3.0–27.4° |
| b = 8.1288 (3) Å | µ = 0.22 mm−1 |
| c = 26.8949 (12) Å | T = 296 K |
| β = 91.600 (1)° | Chunk, colourless |
| V = 1878.68 (14) Å3 | 0.37 × 0.30 × 0.22 mm |
| Z = 4 |
Data collection
| Rigaku R-AXIS RAPID/ZJUG CCD diffractometer | 4271 independent reflections |
| Radiation source: rotate anode | 2763 reflections with I > 2σ(I) |
| graphite | Rint = 0.036 |
| Detector resolution: 10.00 pixels mm-1 | θmax = 27.4°, θmin = 3.0° |
| φ and ω scans | h = −11→11 |
| Absorption correction: multi-scan (ABSCOR; Higashi, 1995) | k = −10→9 |
| Tmin = 0.913, Tmax = 0.953 | l = −34→34 |
| 27924 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.042 | H-atom parameters constrained |
| wR(F2) = 0.125 | w = 1/[σ2(Fo2) + (0.0502P)2 + 0.6753P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max < 0.001 |
| 4271 reflections | Δρmax = 0.21 e Å−3 |
| 222 parameters | Δρmin = −0.31 e Å−3 |
| 0 restraints | Extinction correction: SHELXL97 (Sheldrick, 2008), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.0084 (10) |
Special details
| Geometry. All s.u.'s (except the s.u. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell s.u.'s are taken into account individually in the estimation of s.u.'s in distances, angles and torsion angles; correlations between s.u.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell s.u.'s is used for estimating s.u.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.29951 (9) | 0.54161 (7) | 0.65218 (2) | 0.0833 (2) | |
| O1 | 0.27619 (16) | 0.21200 (19) | 0.53119 (5) | 0.0615 (4) | |
| H101 | 0.2803 | 0.2754 | 0.5076 | 0.092* | |
| O2 | 0.49211 (17) | 0.21720 (18) | 0.43539 (5) | 0.0619 (4) | |
| N1 | 0.62036 (16) | 0.27020 (17) | 0.53021 (5) | 0.0420 (3) | |
| H102 | 0.6383 | 0.3783 | 0.5348 | 0.050* | |
| H103 | 0.5619 | 0.2592 | 0.5021 | 0.050* | |
| C7 | 0.2495 (2) | 0.3348 (2) | 0.65680 (7) | 0.0541 (5) | |
| C1 | 0.3646 (2) | 0.2763 (2) | 0.57188 (6) | 0.0440 (4) | |
| H1 | 0.3678 | 0.3966 | 0.5696 | 0.053* | |
| C2 | 0.2854 (2) | 0.2261 (2) | 0.61907 (6) | 0.0459 (4) | |
| C8 | 0.5294 (2) | 0.2082 (2) | 0.57268 (6) | 0.0486 (4) | |
| H8A | 0.5820 | 0.2392 | 0.6037 | 0.058* | |
| H8B | 0.5252 | 0.0891 | 0.5713 | 0.058* | |
| C9 | 0.7739 (2) | 0.1855 (3) | 0.52294 (8) | 0.0571 (5) | |
| H9 | 0.7533 | 0.0726 | 0.5118 | 0.068* | |
| C4 | 0.1703 (3) | 0.0101 (3) | 0.66772 (10) | 0.0788 (7) | |
| H4 | 0.1439 | −0.1001 | 0.6714 | 0.095* | |
| C6 | 0.1758 (3) | 0.2841 (3) | 0.69932 (8) | 0.0749 (7) | |
| H6 | 0.1540 | 0.3592 | 0.7243 | 0.090* | |
| C10 | 0.8567 (3) | 0.2759 (3) | 0.48204 (9) | 0.0744 (6) | |
| H10A | 0.7901 | 0.2810 | 0.4528 | 0.112* | |
| H10B | 0.9507 | 0.2186 | 0.4745 | 0.112* | |
| H10C | 0.8817 | 0.3855 | 0.4930 | 0.112* | |
| C3 | 0.2444 (2) | 0.0620 (3) | 0.62573 (8) | 0.0607 (5) | |
| H3 | 0.2677 | −0.0143 | 0.6013 | 0.073* | |
| C11 | 0.8699 (3) | 0.1785 (4) | 0.57050 (10) | 0.0913 (8) | |
| H11A | 0.8804 | 0.2873 | 0.5841 | 0.137* | |
| H11B | 0.9711 | 0.1353 | 0.5638 | 0.137* | |
| H11C | 0.8196 | 0.1086 | 0.5939 | 0.137* | |
| C5 | 0.1353 (3) | 0.1218 (3) | 0.70422 (9) | 0.0830 (8) | |
| H5 | 0.0840 | 0.0871 | 0.7323 | 0.100* | |
| O3 | 0.30804 (17) | 0.40215 (16) | 0.44986 (5) | 0.0589 (4) | |
| C13 | 0.3104 (2) | 0.2771 (2) | 0.37008 (6) | 0.0439 (4) | |
| C12 | 0.3761 (2) | 0.3002 (2) | 0.42202 (6) | 0.0467 (4) | |
| C14 | 0.3905 (2) | 0.1855 (2) | 0.33582 (7) | 0.0547 (5) | |
| H14 | 0.4842 | 0.1360 | 0.3453 | 0.066* | |
| C18 | 0.1687 (2) | 0.3448 (2) | 0.35524 (7) | 0.0529 (5) | |
| H18 | 0.1117 | 0.4052 | 0.3778 | 0.064* | |
| C16 | 0.1925 (2) | 0.2349 (3) | 0.27234 (7) | 0.0578 (5) | |
| C15 | 0.3330 (3) | 0.1666 (3) | 0.28760 (7) | 0.0609 (5) | |
| H15 | 0.3901 | 0.1067 | 0.2650 | 0.073* | |
| C17 | 0.1113 (2) | 0.3231 (3) | 0.30701 (7) | 0.0607 (5) | |
| H17 | 0.0157 | 0.3690 | 0.2978 | 0.073* | |
| C19 | 0.1321 (4) | 0.2139 (4) | 0.21933 (9) | 0.0890 (8) | |
| H19A | 0.2018 | 0.2667 | 0.1971 | 0.133* | |
| H19B | 0.0308 | 0.2629 | 0.2158 | 0.133* | |
| H19C | 0.1254 | 0.0989 | 0.2115 | 0.133* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.1200 (5) | 0.0570 (3) | 0.0746 (4) | −0.0060 (3) | 0.0327 (3) | −0.0103 (3) |
| O1 | 0.0601 (8) | 0.0804 (10) | 0.0435 (7) | −0.0112 (7) | −0.0052 (6) | 0.0058 (7) |
| O2 | 0.0651 (9) | 0.0754 (9) | 0.0446 (7) | 0.0106 (7) | −0.0091 (6) | −0.0056 (7) |
| N1 | 0.0427 (8) | 0.0434 (8) | 0.0400 (7) | 0.0015 (6) | 0.0020 (6) | −0.0029 (6) |
| C7 | 0.0561 (11) | 0.0568 (11) | 0.0498 (10) | 0.0044 (9) | 0.0102 (8) | 0.0035 (9) |
| C1 | 0.0488 (10) | 0.0454 (9) | 0.0378 (8) | 0.0003 (7) | 0.0026 (7) | 0.0009 (7) |
| C2 | 0.0460 (10) | 0.0515 (10) | 0.0404 (9) | 0.0035 (8) | 0.0026 (7) | 0.0067 (8) |
| C8 | 0.0497 (10) | 0.0523 (10) | 0.0440 (9) | 0.0024 (8) | 0.0034 (8) | 0.0064 (8) |
| C9 | 0.0487 (11) | 0.0585 (12) | 0.0643 (12) | 0.0116 (9) | 0.0063 (9) | −0.0040 (9) |
| C4 | 0.0927 (17) | 0.0631 (14) | 0.0818 (16) | −0.0038 (12) | 0.0234 (13) | 0.0242 (12) |
| C6 | 0.0881 (17) | 0.0820 (16) | 0.0561 (12) | 0.0105 (13) | 0.0285 (12) | 0.0043 (11) |
| C10 | 0.0552 (13) | 0.0929 (17) | 0.0761 (15) | 0.0067 (12) | 0.0194 (11) | −0.0026 (13) |
| C3 | 0.0708 (13) | 0.0549 (12) | 0.0570 (12) | −0.0008 (10) | 0.0103 (10) | 0.0072 (9) |
| C11 | 0.0580 (14) | 0.131 (2) | 0.0845 (17) | 0.0235 (15) | −0.0098 (12) | 0.0098 (16) |
| C5 | 0.0928 (18) | 0.0889 (18) | 0.0691 (15) | 0.0057 (14) | 0.0357 (13) | 0.0274 (13) |
| O3 | 0.0889 (10) | 0.0460 (7) | 0.0420 (7) | 0.0038 (7) | 0.0029 (6) | −0.0067 (6) |
| C13 | 0.0538 (10) | 0.0406 (9) | 0.0375 (8) | −0.0040 (7) | 0.0015 (7) | 0.0012 (7) |
| C12 | 0.0616 (11) | 0.0399 (9) | 0.0388 (9) | −0.0050 (8) | 0.0023 (8) | −0.0008 (7) |
| C14 | 0.0567 (11) | 0.0613 (12) | 0.0460 (10) | 0.0066 (9) | −0.0016 (8) | −0.0078 (9) |
| C18 | 0.0578 (12) | 0.0537 (11) | 0.0475 (10) | 0.0037 (9) | 0.0053 (8) | 0.0015 (8) |
| C16 | 0.0710 (13) | 0.0594 (12) | 0.0426 (10) | −0.0094 (10) | −0.0061 (9) | 0.0016 (9) |
| C15 | 0.0716 (14) | 0.0698 (13) | 0.0414 (10) | 0.0016 (11) | 0.0039 (9) | −0.0116 (9) |
| C17 | 0.0597 (12) | 0.0664 (13) | 0.0555 (11) | 0.0027 (10) | −0.0051 (9) | 0.0076 (10) |
| C19 | 0.114 (2) | 0.0984 (19) | 0.0531 (13) | −0.0079 (16) | −0.0230 (13) | −0.0049 (13) |
Geometric parameters (Å, °)
| Cl1—C7 | 1.741 (2) | C10—H10A | 0.9600 |
| O1—C1 | 1.415 (2) | C10—H10B | 0.9600 |
| O1—H101 | 0.8200 | C10—H10C | 0.9600 |
| O2—C12 | 1.248 (2) | C3—H3 | 0.9300 |
| N1—C8 | 1.490 (2) | C11—H11A | 0.9600 |
| N1—C9 | 1.507 (2) | C11—H11B | 0.9600 |
| N1—H102 | 0.9000 | C11—H11C | 0.9600 |
| N1—H103 | 0.9000 | C5—H5 | 0.9300 |
| C7—C6 | 1.386 (3) | O3—C12 | 1.271 (2) |
| C7—C2 | 1.387 (3) | C13—C14 | 1.383 (2) |
| C1—C2 | 1.513 (2) | C13—C18 | 1.386 (3) |
| C1—C8 | 1.521 (2) | C13—C12 | 1.504 (2) |
| C1—H1 | 0.9800 | C14—C15 | 1.384 (3) |
| C2—C3 | 1.392 (3) | C14—H14 | 0.9300 |
| C8—H8A | 0.9700 | C18—C17 | 1.386 (3) |
| C8—H8B | 0.9700 | C18—H18 | 0.9300 |
| C9—C11 | 1.504 (3) | C16—C17 | 1.381 (3) |
| C9—C10 | 1.517 (3) | C16—C15 | 1.382 (3) |
| C9—H9 | 0.9800 | C16—C19 | 1.513 (3) |
| C4—C5 | 1.376 (4) | C15—H15 | 0.9300 |
| C4—C3 | 1.379 (3) | C17—H17 | 0.9300 |
| C4—H4 | 0.9300 | C19—H19A | 0.9600 |
| C6—C5 | 1.371 (4) | C19—H19B | 0.9600 |
| C6—H6 | 0.9300 | C19—H19C | 0.9600 |
| C1—O1—H101 | 109.5 | H10A—C10—H10C | 109.5 |
| C8—N1—C9 | 115.19 (14) | H10B—C10—H10C | 109.5 |
| C8—N1—H102 | 108.5 | C4—C3—C2 | 121.5 (2) |
| C9—N1—H102 | 108.5 | C4—C3—H3 | 119.2 |
| C8—N1—H103 | 108.5 | C2—C3—H3 | 119.2 |
| C9—N1—H103 | 108.5 | C9—C11—H11A | 109.5 |
| H102—N1—H103 | 107.5 | C9—C11—H11B | 109.5 |
| C6—C7—C2 | 122.1 (2) | H11A—C11—H11B | 109.5 |
| C6—C7—Cl1 | 117.75 (17) | C9—C11—H11C | 109.5 |
| C2—C7—Cl1 | 120.19 (14) | H11A—C11—H11C | 109.5 |
| O1—C1—C2 | 107.74 (14) | H11B—C11—H11C | 109.5 |
| O1—C1—C8 | 110.88 (15) | C6—C5—C4 | 120.3 (2) |
| C2—C1—C8 | 109.32 (14) | C6—C5—H5 | 119.8 |
| O1—C1—H1 | 109.6 | C4—C5—H5 | 119.8 |
| C2—C1—H1 | 109.6 | C14—C13—C18 | 118.17 (16) |
| C8—C1—H1 | 109.6 | C14—C13—C12 | 120.33 (16) |
| C7—C2—C3 | 117.01 (17) | C18—C13—C12 | 121.50 (16) |
| C7—C2—C1 | 123.81 (17) | O2—C12—O3 | 124.06 (16) |
| C3—C2—C1 | 119.18 (17) | O2—C12—C13 | 118.45 (16) |
| N1—C8—C1 | 112.01 (14) | O3—C12—C13 | 117.49 (16) |
| N1—C8—H8A | 109.2 | C13—C14—C15 | 120.84 (18) |
| C1—C8—H8A | 109.2 | C13—C14—H14 | 119.6 |
| N1—C8—H8B | 109.2 | C15—C14—H14 | 119.6 |
| C1—C8—H8B | 109.2 | C13—C18—C17 | 120.41 (18) |
| H8A—C8—H8B | 107.9 | C13—C18—H18 | 119.8 |
| C11—C9—N1 | 111.64 (17) | C17—C18—H18 | 119.8 |
| C11—C9—C10 | 112.2 (2) | C17—C16—C15 | 117.54 (17) |
| N1—C9—C10 | 107.65 (16) | C17—C16—C19 | 121.9 (2) |
| C11—C9—H9 | 108.4 | C15—C16—C19 | 120.6 (2) |
| N1—C9—H9 | 108.4 | C16—C15—C14 | 121.36 (19) |
| C10—C9—H9 | 108.4 | C16—C15—H15 | 119.3 |
| C5—C4—C3 | 119.8 (2) | C14—C15—H15 | 119.3 |
| C5—C4—H4 | 120.1 | C16—C17—C18 | 121.64 (19) |
| C3—C4—H4 | 120.1 | C16—C17—H17 | 119.2 |
| C5—C6—C7 | 119.2 (2) | C18—C17—H17 | 119.2 |
| C5—C6—H6 | 120.4 | C16—C19—H19A | 109.5 |
| C7—C6—H6 | 120.4 | C16—C19—H19B | 109.5 |
| C9—C10—H10A | 109.5 | H19A—C19—H19B | 109.5 |
| C9—C10—H10B | 109.5 | C16—C19—H19C | 109.5 |
| H10A—C10—H10B | 109.5 | H19A—C19—H19C | 109.5 |
| C9—C10—H10C | 109.5 | H19B—C19—H19C | 109.5 |
| C6—C7—C2—C3 | −0.2 (3) | C1—C2—C3—C4 | −179.13 (19) |
| Cl1—C7—C2—C3 | 178.83 (15) | C7—C6—C5—C4 | 1.2 (4) |
| C6—C7—C2—C1 | 179.50 (19) | C3—C4—C5—C6 | −0.9 (4) |
| Cl1—C7—C2—C1 | −1.4 (3) | C14—C13—C12—O2 | −9.8 (3) |
| O1—C1—C2—C7 | −129.79 (18) | C18—C13—C12—O2 | 169.70 (18) |
| C8—C1—C2—C7 | 109.6 (2) | C14—C13—C12—O3 | 171.21 (17) |
| O1—C1—C2—C3 | 49.9 (2) | C18—C13—C12—O3 | −9.3 (3) |
| C8—C1—C2—C3 | −70.6 (2) | C18—C13—C14—C15 | 2.1 (3) |
| C9—N1—C8—C1 | −169.34 (15) | C12—C13—C14—C15 | −178.45 (18) |
| O1—C1—C8—N1 | 68.35 (19) | C14—C13—C18—C17 | −1.1 (3) |
| C2—C1—C8—N1 | −173.02 (14) | C12—C13—C18—C17 | 179.36 (17) |
| C8—N1—C9—C11 | −50.6 (2) | C17—C16—C15—C14 | 0.0 (3) |
| C8—N1—C9—C10 | −174.10 (16) | C19—C16—C15—C14 | 179.6 (2) |
| C2—C7—C6—C5 | −0.7 (4) | C13—C14—C15—C16 | −1.5 (3) |
| Cl1—C7—C6—C5 | −179.8 (2) | C15—C16—C17—C18 | 0.9 (3) |
| C5—C4—C3—C2 | −0.1 (4) | C19—C16—C17—C18 | −178.6 (2) |
| C7—C2—C3—C4 | 0.6 (3) | C13—C18—C17—C16 | −0.3 (3) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H101···O3 | 0.82 | 1.88 | 2.6986 (18) | 173 |
| N1—H103···O2 | 0.90 | 1.91 | 2.7835 (18) | 164 |
| N1—H102···O3i | 0.90 | 1.89 | 2.7824 (19) | 174 |
Symmetry codes: (i) −x+1, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: RK2225).
References
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Feng, H., Tang, Z., Xie, L.-J. & Xing, B.-T. (2010). Acta Cryst. E66, o391. [DOI] [PMC free article] [PubMed]
- Higashi, T. (1995). ABSCOR Rigaku Corporation, Tokyo, Japan.
- Rigaku/MSC (2006). PROCESS-AUTO Rigaku/MSC, The Woodlands, Texas, USA.
- Rigaku/MSC (2007). CrystalStructure Rigaku/MSC, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Takwale, M. G. & Pant, L. M. (1971). Acta Cryst. B27, 1152–1158.
- Tang, Z., Xu, M., Zhang, H.-C. & Feng, H. (2009b). Acta Cryst. E65, o1670. [DOI] [PMC free article] [PubMed]
- Tang, Z., Xu, M., Zheng, G.-R. & Feng, H. (2009a). Acta Cryst. E65, o1501. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810033878/rk2225sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810033878/rk2225Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report