Abstract
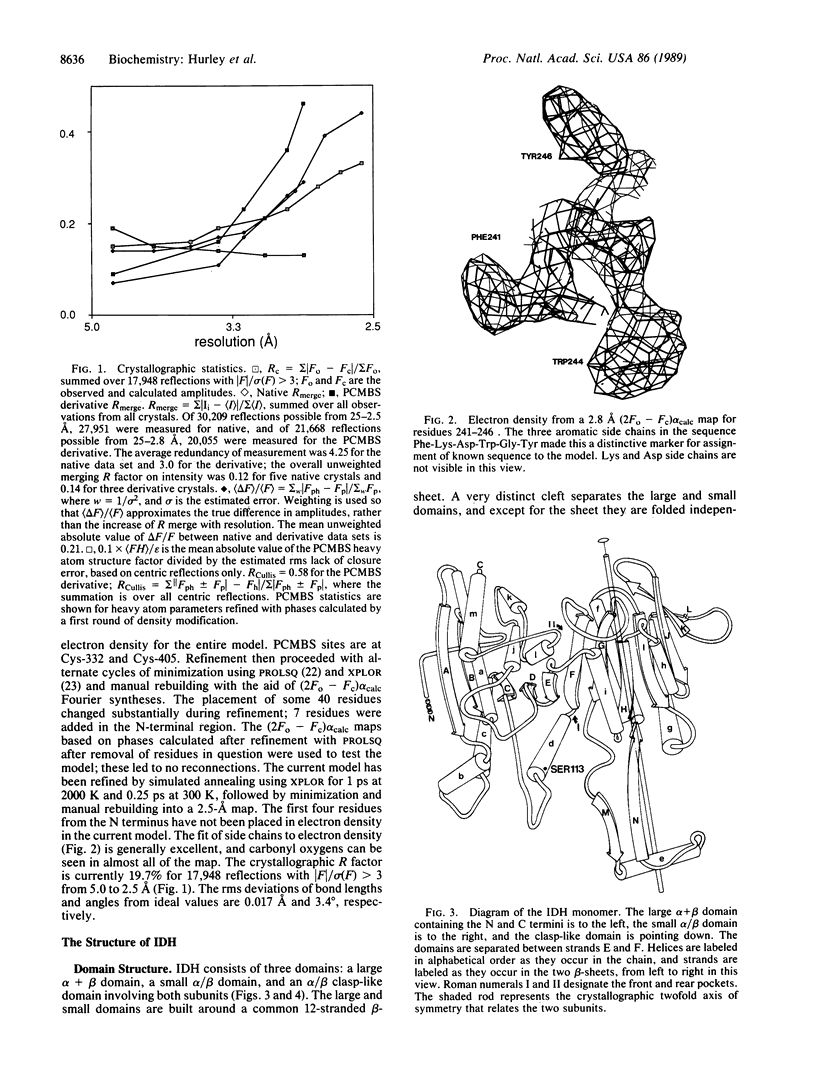
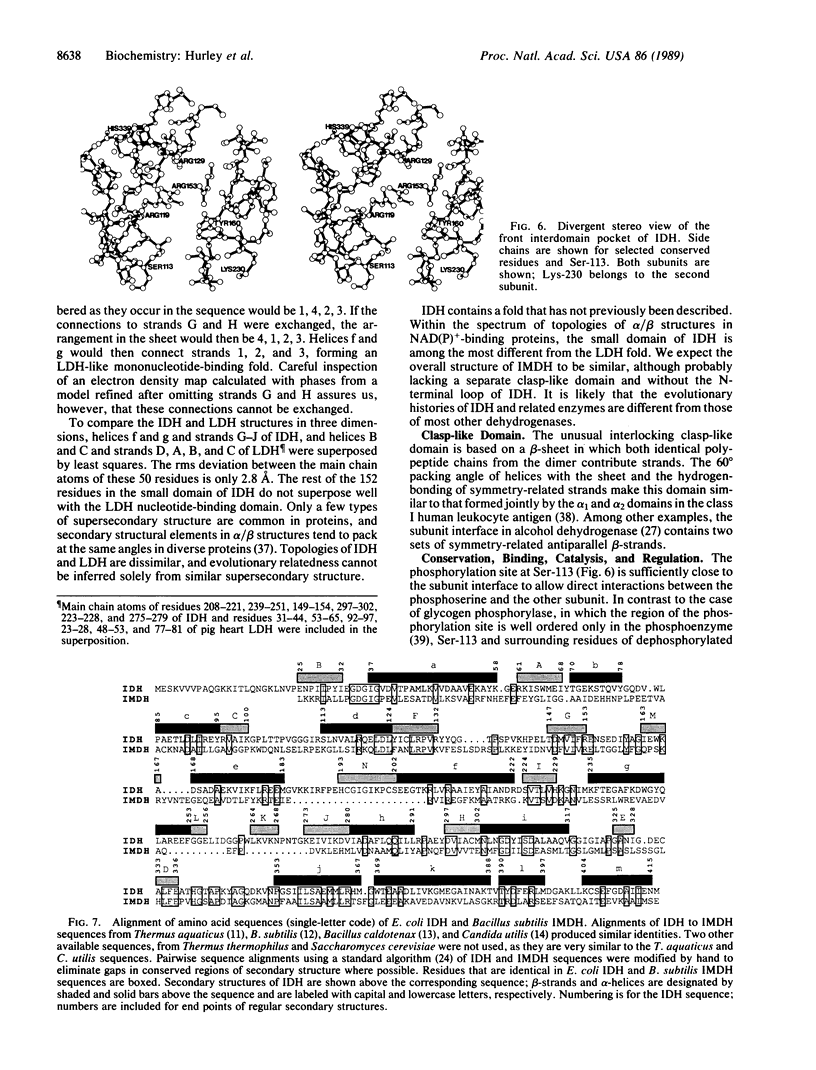
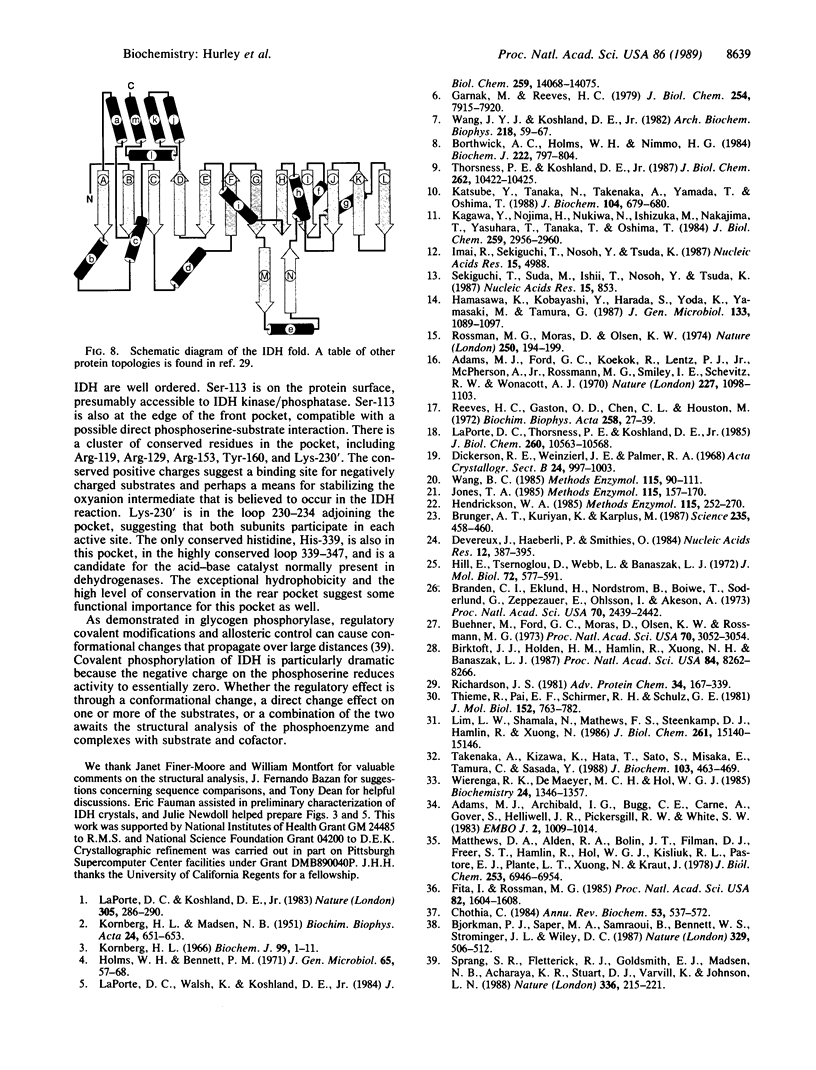
The structure of isocitrate dehydrogenase [threo-DS-isocitrate: NADP+ oxidoreductase (decarboxylating), EC 1.1.1.42] from Escherichia coli has been solved and refined at 2.5 A resolution and is topologically different from that of any other dehydrogenase. This enzyme, a dimer of identical 416-residue subunits, is inactivated by phosphorylation at Ser-113, which lies at the edge of an interdomain pocket that also contains many residues conserved between isocitrate dehydrogenase and isopropylmalate dehydrogenase. Isocitrate dehydrogenase contains an unusual clasp-like domain in which both polypeptide chains in the dimer interlock. Based on the structure of isocitrate dehydrogenase and conservation with isopropylmalate dehydrogenase, we suggest that the active site lies in an interdomain pocket close to the phosphorylation site.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. J., Archibald I. G., Bugg C. E., Carne A., Gover S., Helliwell J. R., Pickersgill R. W., White S. W. The three dimensional structure of sheep liver 6-phosphogluconate dehydrogenase at 2.6 A resolution. EMBO J. 1983;2(6):1009–1014. doi: 10.1002/j.1460-2075.1983.tb01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. J., Ford G. C., Koekoek R., Lentz P. J., McPherson A., Jr, Rossmann M. G., Smiley I. E., Schevitz R. W., Wonacott A. J. Structure of lactate dehydrogenase at 2-8 A resolution. Nature. 1970 Sep 12;227(5263):1098–1103. doi: 10.1038/2271098a0. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Holden H. M., Hamlin R., Xuong N. H., Banaszak L. J. Structure of L-3-hydroxyacyl-coenzyme A dehydrogenase: preliminary chain tracing at 2.8-A resolution. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8262–8266. doi: 10.1073/pnas.84.23.8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987 Oct 8;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Borthwick A. C., Holms W. H., Nimmo H. G. The phosphorylation of Escherichia coli isocitrate dehydrogenase in intact cells. Biochem J. 1984 Sep 15;222(3):797–804. doi: 10.1042/bj2220797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brändén C. I., Eklund H., Nordström B., Boiwe T., Söderlund G., Zeppezauer E., Ohlsson I., Akeson A. Structure of liver alcohol dehydrogenase at 2.9-angstrom resolution. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2439–2442. doi: 10.1073/pnas.70.8.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Buehner M., Ford G. C., Moras D., Olsen K. W., Rossman M. G. D-glyceraldehyde-3-phosphate dehydrogenase: three-dimensional structure and evolutionary significance. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3052–3054. doi: 10.1073/pnas.70.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fita I., Rossmann M. G. The NADPH binding site on beef liver catalase. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1604–1608. doi: 10.1073/pnas.82.6.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnak M., Reeves H. C. Purification and properties of phosphorylated isocitrate dehydrogenase of Escherichia coli. J Biol Chem. 1979 Aug 25;254(16):7915–7920. [PubMed] [Google Scholar]
- Hamasawa K., Kobayashi Y., Harada S., Yoda K., Yamasaki M., Tamura G. Molecular cloning and nucleotide sequence of the 3-isopropylmalate dehydrogenase gene of Candida utilis. J Gen Microbiol. 1987 Apr;133(4):1089–1097. doi: 10.1099/00221287-133-4-1089. [DOI] [PubMed] [Google Scholar]
- Hendrickson W. A. Stereochemically restrained refinement of macromolecular structures. Methods Enzymol. 1985;115:252–270. doi: 10.1016/0076-6879(85)15021-4. [DOI] [PubMed] [Google Scholar]
- Hill E., Tsernoglou D., Webb L., Banaszak L. J. Polypeptide conformation of cytoplasmic malate dehydrogenase from an electron density map at 3.0 angstrom resolution. J Mol Biol. 1972 Dec 30;72(3):577–589. doi: 10.1016/0022-2836(72)90176-3. [DOI] [PubMed] [Google Scholar]
- Holms W. H., Bennett P. M. Regulation of isocitrate dehydrogenase activity in Escherichia coli on adaptation to acetate. J Gen Microbiol. 1971 Jan;65(1):57–68. doi: 10.1099/00221287-65-1-57. [DOI] [PubMed] [Google Scholar]
- Imai R., Sekiguchi T., Nosoh Y., Tsuda K. The nucleotide sequence of 3-isopropylmalate dehydrogenase gene from Bacillus subtilis. Nucleic Acids Res. 1987 Jun 25;15(12):4988–4988. doi: 10.1093/nar/15.12.4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- KORNBERG H. L., MADSEN N. B. Synthesis of C4-dicarboxylic acids from acetate by a glyoxylate bypass of the tricarboxylic acid cycle. Biochim Biophys Acta. 1957 Jun;24(3):651–653. doi: 10.1016/0006-3002(57)90268-8. [DOI] [PubMed] [Google Scholar]
- Kagawa Y., Nojima H., Nukiwa N., Ishizuka M., Nakajima T., Yasuhara T., Tanaka T., Oshima T. High guanine plus cytosine content in the third letter of codons of an extreme thermophile. DNA sequence of the isopropylmalate dehydrogenase of Thermus thermophilus. J Biol Chem. 1984 Mar 10;259(5):2956–2960. [PubMed] [Google Scholar]
- Katsube Y., Tanaka N., Takenaka A., Yamada T., Oshima T. Crystallization and preliminary X-ray data for 3-isopropylmalate dehydrogenase of Thermus thermophilus. J Biochem. 1988 Nov;104(5):679–680. doi: 10.1093/oxfordjournals.jbchem.a122531. [DOI] [PubMed] [Google Scholar]
- Kornberg H. L. The role and control of the glyoxylate cycle in Escherichia coli. Biochem J. 1966 Apr;99(1):1–11. doi: 10.1042/bj0990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPorte D. C., Koshland D. E., Jr Phosphorylation of isocitrate dehydrogenase as a demonstration of enhanced sensitivity in covalent regulation. Nature. 1983 Sep 22;305(5932):286–290. doi: 10.1038/305286a0. [DOI] [PubMed] [Google Scholar]
- LaPorte D. C., Thorsness P. E., Koshland D. E., Jr Compensatory phosphorylation of isocitrate dehydrogenase. A mechanism for adaptation to the intracellular environment. J Biol Chem. 1985 Sep 5;260(19):10563–10568. [PubMed] [Google Scholar]
- LaPorte D. C., Walsh K., Koshland D. E., Jr The branch point effect. Ultrasensitivity and subsensitivity to metabolic control. J Biol Chem. 1984 Nov 25;259(22):14068–14075. [PubMed] [Google Scholar]
- Lim L. W., Shamala N., Mathews F. S., Steenkamp D. J., Hamlin R., Xuong N. H. Three-dimensional structure of the iron-sulfur flavoprotein trimethylamine dehydrogenase at 2.4-A resolution. J Biol Chem. 1986 Nov 15;261(32):15140–15146. [PubMed] [Google Scholar]
- Matthews D. A., Alden R. A., Bolin J. T., Filman D. J., Freer S. T., Hamlin R., Hol W. G., Kisliuk R. L., Pastore E. J., Plante L. T. Dihydrofolate reductase from Lactobacillus casei. X-ray structure of the enzyme methotrexate.NADPH complex. J Biol Chem. 1978 Oct 10;253(19):6946–6954. [PubMed] [Google Scholar]
- Reeves H. C., Daumy G. O., Lin C. C., Houston M. NADP + -specific isocitrate dehydrogenase of Escherichia coli. I. Purification and characterization. Biochim Biophys Acta. 1972 Jan 20;258(1):27–39. doi: 10.1016/0005-2744(72)90964-3. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974 Jul 19;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Sekiguchi T., Suda M., Ishii T., Nosoh Y., Tsuda K. The nucleotide sequence of 3-isopropylmalate dehydrogenase gene from Bacillus caldotenax. Nucleic Acids Res. 1987 Jan 26;15(2):853–853. doi: 10.1093/nar/15.2.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang S. R., Acharya K. R., Goldsmith E. J., Stuart D. I., Varvill K., Fletterick R. J., Madsen N. B., Johnson L. N. Structural changes in glycogen phosphorylase induced by phosphorylation. Nature. 1988 Nov 17;336(6196):215–221. doi: 10.1038/336215a0. [DOI] [PubMed] [Google Scholar]
- Takenaka A., Kizawa K., Hata T., Sato S., Misaka E., Tamura C., Sasada Y. X-ray study of baker's yeast lipoamide dehydrogenase at 4.5 A resolution by molecular replacement method. J Biochem. 1988 Mar;103(3):463–469. doi: 10.1093/oxfordjournals.jbchem.a122293. [DOI] [PubMed] [Google Scholar]
- Thieme R., Pai E. F., Schirmer R. H., Schulz G. E. Three-dimensional structure of glutathione reductase at 2 A resolution. J Mol Biol. 1981 Nov 15;152(4):763–782. doi: 10.1016/0022-2836(81)90126-1. [DOI] [PubMed] [Google Scholar]
- Thorsness P. E., Koshland D. E., Jr Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J Biol Chem. 1987 Aug 5;262(22):10422–10425. [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Wang J. Y., Koshland D. E., Jr The reversible phosphorylation of isocitrate dehydrogenase of Salmonella typhimurium. Arch Biochem Biophys. 1982 Oct 1;218(1):59–67. doi: 10.1016/0003-9861(82)90321-6. [DOI] [PubMed] [Google Scholar]