Abstract
Both transfer RNA (tRNA) and cytochrome c are essential to cellular function: tRNA mediates protein synthesis while cytochrome c is required for oxidative phosphorylation and apoptosis induction. tRNA has recently been implicated as a direct regulator of the well-conserved apoptotic role of cytochrome c. Interaction between these molecules could potentially coordinate biosynthesis, energy production and apoptosis. Here we review the diversity and dynamics of tRNA and how this class of non-coding RNAs may regulate the role of cytochrome c in apoptosis. We comment on unanswered questions in the cell biology of this interaction and how answers may influence our understanding of disease.
Key words: tRNA, cytochrome c, apoptosis, caspase, mitochondria, protein synthesis, cancer
tRNA: The Genetic Interpreter
tRNA interprets the genetic code by physically connecting nucleic acid codons to amino acids. The human nuclear genome codes for over 500 tRNAs. Nuclear tRNAs are transcribed as longer pretRNAs by RNA polymerase III, trimmed at each end, and spliced to remove introns as necessary.1 All mature tRNAs are 73 to 93 ribonucleotides and fold into similar tertiary structures. tRNAs are activated by conjugation to amino acids at a CCA trinucleotide sequence at their 3′ ends. Opposite this end in the folded structure of tRNA, a three nucleotide anti-codon sequence can pair with mRNA codons on the ribosome. tRNA must interact transiently rather than stably with cellular proteins to function in protein synthesis. This unique requirement for tRNAs may allow them more functional versatility than other small non-coding RNAs. This is exemplified by the way in which the human immunodeficiency virus uses tRNA to prime reverse transcription of its RNA genome.2
The process of tRNA maturation involves extensive modification in both the nucleus and cytoplasm. Approximately 100 chemically distinct modifications have been reported; most functionally undefined.1 It was until recently believed that following maturation, nuclear tRNAs localized exclusively to the cytosol. However, new studies have showed that under stress conditions such as nutrient deprivation, mature tRNAs can undergo retrograde translocation to the nucleus.3
The human mitochondrial genome codes for 22 tRNAs. Unlike many other organisms, these include a complete set able to specify all common amino acids. Most mitochondrial tRNA are expected to be present in the matrix where mitochondrial protein synthesis occurs. It was generally thought that human mitochondrial and nuclear tRNA systems are completely separate until a recent paper challenged this idea by showing that human mitochondria can import cytosolic tRNA with the addition of ATP.4
Small tRNA-derived fragments (tRFs) were recently recognized as a major RNA species in human cells. Remarkably, one pre-tRNA-derived fragment was shown to be required for cell proliferation.5 Interestingly, several lines of evidence showed that endonucleolytic cleavage of tRNAs exists in cells under a variety of stress conditions. Particularly, tRNA cleavage occurs during oxidative stress.6,7 tRNA halves are present in human cells, Drosophila cells, and likely those of many other organisms.8,9 These fragments may inhibit protein synthesis as “cleaved tRNAs” which are nicked but otherwise fully folded.10 While intriguing, the importance of tRNA halves on cell physiology and the mechanism and regulation of this process remain largely unproven.
Cytochrome c: An Apoptotic Death Inducer
The most conserved role of cytochrome c is in the electron transport chain powering oxidative phosphorylation. Cytochrome c carries electrons from the mitochondrial inner membrane protein complex III to complex IV and is essential for the generation of the mitochondrial membrane potential (Δψ) that drives the formation of ATP. In human and other vertebrate cells, cytochrome c is also a central apoptotic effecter. In these cells two major apoptotic pathways have been described: the extrinsic pathway and the intrinsic pathway. Cytochrome c release into the cytosol is particularly associated with activation of the intrinsic pathway, which responds to intracellular stimuli such as DNA damage, lineage information and oncogene activation. Once in the cytosol cytochrome c binds the adaptor protein Apaf-1 (apoptotic protease-activating factor 1) and assembles the apoptosome complex,11 causing recruitment and activation of the initiator caspase-9.12,13 The subsequent proteolytic cascade effects the morphological changes that define apoptosis including cellular shrinkage, membrane blebbing, nuclear condensation and fragmentation of cells into apoptotic bodies that can be rapidly cleared.
Formation of the apoptosome is intricately regulated. Prior to cytochrome c binding, Apaf-1 is tightly associated with dATP,14,15 which contacts multiple domains of Apaf-1 and helps keep Apaf-1 in its inactive form. Binding of cytochrome c leads to the hydrolysis of the bound dATP to dADP, and subsequent exchange of the Apaf-1-bound dADP with a free dATP. Apaf-1 can then assemble into a functional heptameric platform. This platform recruits multiple procaspase-9 molecules, leading to their oligomerization and subsequent auto-proteolytic processing. The hydrolysis of dATP is enhanced by the combined action of at least three proteins: the tumor suppressor PHAPI, cellular apoptosis susceptibility protein (CAS) and heat shock protein 70 (Hsp70).16 A range of cellular factors oppose apoptosome formation, including high levels of the cations potassium and calcium, as well as the action of the proteins HSP27, HSP90 and prothymosin-α (ProT).12,13 Low levels of dATP also promote apoptosome formation but high levels of dATP inhibit it.17
The ability of cytochrome c to initiate apoptosis depends on numerous cellular factors including the intracellular redox environment. Oxidized but not reduced cytochrome c activates caspases and promotes apoptosis.18–20 Intracellular glutathione (GSH) generated via the pentose phosphate pathway is necessary to inhibit prop-apoptotic cytochrome c function by maintaining it in a reduced form at least in some neurons and cancer cells.21
Discovery of Interaction Between-tRNA and Cytochrome c
The role of nucleotides in cytochrome c-mediated caspase activation led us to speculate that RNA could be involved in this process. Treatment of mammalian S100 extracts with RNase strongly increased cytochrome c-induced caspase-9 activation, while the addition of RNA to the extracts impaired caspase-9 activation. These results suggested that one or more RNA species inhibit a factor required for caspase-9 activation. Systemic evaluation of the steps leading to caspase-9 activation revealed that RNA-mediated inhibition occurs at the level of cytochrome c. We stabilized RNA-protein complexes inside intact cells with low formaldehyde concentration and then lysed cells in buffer containing the strong detergent Empigen BB to prevent non-specific interaction that might occur during cell lysis.22 This analysis showed that several cytosolic and mitochondrial tRNAs specifically associate with cytochrome c. Microinjection of tRNA blunted the ability of cytochrome c to induce apoptosis, while degradation of tRNA by an RNase that preferentially degrades tRNA, Onconase, enhanced apoptosis via the intrinsic pathway. This finding reveals a direct role for tRNA in regulating apoptosis (Fig. 1).23
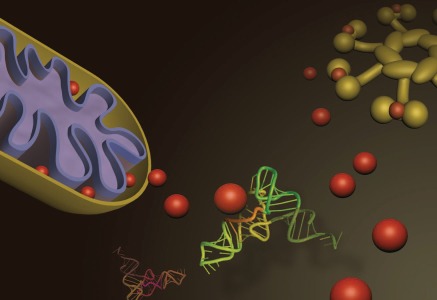
Figure 1.
Cytochrome c (red spheres) is present mostly in the mitochondrial inner membrane space of healthy cells, but translocates to the cytosol when intrinsic apoptotic stimuli are present. Transfer RNA (middle) may interact with cytochrome c and prevent association with Apaf-1, blocking formation of the apoptosome complex (upper right). When active, the apoptosome begins a proteolytic cascade that results in membrane blebbing, nuclear condensation and irreversible cell death.
Interaction Between tRNA and cytochrome c May Inform Our Understanding of Disease
The realization of the Cytochrome c:tRNA interaction may shed important light on diseases associated with genetic mutations in tRNA. Point mutations in human mitochondrial tRNA genes cause a range of neurological, neuromuscular and neurodegenerative syndromes.24,25 It is generally assumed that these result from deficits in mitochondrial protein synthesis. However, protein synthesis defects cannot always be detected.26 The association of mitochondrial tRNA with cytochrome c is a potentially unexplored aspect of pathogenesis in these cases. For example, mitochondrial tRNA mutations might affect cytochrome c binding and alter apoptotic threshold or energy generation via electron transfer. Characterization of the interacting domains of cytochrome c and tRNA should reveal any genotype-phenotype correlations in either molecule resulting from effects on the function of the other.
Inhibition of apoptosis is an essential part of cancer pathogenesis.27 As tRNAs are highly expressed in tumor cells,28 this may represent a mechanism by which tumor cells protect themselves from death. While high tRNA levels are a general requirement of rapid protein synthesis commonly occurring in tumor cells, the overexpression of tRNA in cancer can exceed that of normal cells growing at similar rates, and tRNAs in malignant cells may differ form those in normal cells of the same origin.29–31 The activity of Pol III—mediated tRNA transcription is markedly enhanced by oncogenic cMyc, mTOR and Ras pathways.32–34 The tumor suppressor p53 potently inhibits the activity of Pol III and tRNA synthesis is strongly enhanced in p53 deficient cells.35 Similarly, the tumor suppressor Rb inhibits the activity of Pol III.28,36 Deregulation of RNA Pol III leads to a dramatic increase in the levels of tRNA in tumor cells. The causal effect of high levels of tRNA on cellular transformation was established by a recent study from Robert White's group showing that overexpression of an initiator methionine tRNA by itself can transform 3T3 cells.37
If tumor cells are relianant on high levels of tRNA for both growth and apoptosis resistance, tRNA may be a potential valuable target for tumor therapy. This is supported by the potency of onconase-induced apoptosis in tumor cells.38 Onconase is a member of a growing family of extracellular cytotoxic RNase. While extensive analyses have established that onconase specifically degrades tRNA in cells while leaving mRNA and rRNA intact,39–41 other small RNAs are also possible alternative or complementary targets of onconase.42 Our data shows that onconase disables an apoptosis resistant mechanism downstream of cytochrome c release. If this proves to be the major mechanism by which onconase kills tumor cells, it could provide a rational guide for development and use of related RNase therapeutics.
Conclusions
The identification of the cytochrome c:tRNA interaction reveals a previously unexpected connection between two ancient molecules. This interaction is clearly important in the context of apoptosis, and may represent an evolutionarily conserved connection between metabolism and cell survival. Further characterization of this interaction may illuminate mechanisms operating broadly across eukaryotic life. We hypothesize that focused exploration of tRNA mutation, modification and degradation may lead to a better understanding of some genetic diseases. We also believe that interaction between tRNA and cytochrome c combined with the therapeutic action of onconase provides an onus for exploration of tRNA-based cancer therapy.
Future Directions
Cellular cytochrome c:tRNA interaction is likely affected by relative tRNA affinity, abundance and accessibility toward cytochrome c. While cytochrome c is lysine-rich and highly positvely charged, the inability of rRNAs to serve as effective competitors of cytochrome c:tRNA interaction suggests specific structural features of tRNAs are important.23 Whether cytochrome c recognizes different tRNAs with similar affinity and whether association is affected by aminoacylation or post-transcriptional modification of tRNA nucleotides remain unanswered.
The subcellular localization of cytochrome c:tRNA binding is also enigmatic. Analysis of healthy cells shows that cytochrome c preferentially binds to mitochondrial tRNA. This observation is at odds with the expected sub-mitochondrial localization of cytochrome c in the inter-membrane space and mitochondrial tRNA in the mitochondrial matrix. It is notable that the localization of mitochondrial tRNA has not been visualized by electron microscopy and may be at least partially present in the inner membrane space (IMS). Transport to the IMS may even occur along with cytochrome c as this protein is folded in the matrix before crossing the inner mitochondrial membrane to the IMS. Alternatively, tRNAs may be actively transported between the different compartments of the mitochondria. Cytosolic tRNAs also bind to cytochrome c, although to a lesser extent compared with mitochondrial tRNAs. Combined with the recent finding that cytosolic tRNAs are actively transported into mitochondria, this observation is consistent with a model in which interaction in healthy cells occurs in the inner membrane space. The association of cytochrome c with tRNA in healthy cells may affect the function of cytochrome c in electron transfer. If so, the cytochrome c-tRNA connection may represent a link between protein translation and ATP production. During apoptosis, the association of cytochrome c with cytosolic tRNAs is expected to increase as cytochrome c enters the cytosol. Cytosolic tRNAs level may regulate a threshold of apoptotic sensitivity toward cytochrome c thus coordinating protein synthesis with cellular survival. One corollary of this would be that retrograde nuclear translocation of tRNA as in some stress conditions would lower the a cytochrome c-determined apoptotic threshold.
Acknowledgements
We thank Lili Guo for designing the figure. Supported by NIH grants GM60911 and CA108872 to X.Y.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12316
References
- 1.Hopper AK, Pai DA, Engelke DR. Cellular dynamics of tRNAs and their genes. FEBS Lett. 2010;584:310–317. doi: 10.1016/j.febslet.2009.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleiman L, Jones CP, Musier-Forsyth K. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010;584:359–365. doi: 10.1016/j.febslet.2009.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci USA. 2007;104:8845–8850. doi: 10.1073/pnas.0700765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubio MA, Rinehart JJ, Krett B, Duvezin-Caubet S, Reichert AS, Soll D, et al. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc Natl Acad Sci USA. 2008;105:9186–9191. doi: 10.1073/pnas.0804283105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, et al. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 9.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou H, Li Y, Liu X, Wang X. An APAF-1. cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
- 12.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 13.Schafer ZT, Kornbluth S. The apoptosome: physiological, developmental and pathological modes of regulation. Dev Cell. 2006;10:549–561. doi: 10.1016/j.devcel.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Kim HE, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci USA. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- 16.Kim HE, Jiang X, Du F, Wang X. PHAPI CAS and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol Cell. 2008;30:239–247. doi: 10.1016/j.molcel.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Chandra D, Bratton SB, Person MD, Tian Y, Martin AG, Ayres M, et al. Intracellular nucleotides act as critical prosurvival factors by binding to cytochrome c and inhibiting apoptosome. Cell. 2006;125:1333–1346. doi: 10.1016/j.cell.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Wang AJ, Xu JX. Redox state of cytochrome c regulates cellular ROS and caspase cascade in permeablized cell model. Protein Pept Lett. 2008;15:200–205. doi: 10.2174/092986608783489490. [DOI] [PubMed] [Google Scholar]
- 19.Borutaite V, Brown GC. Mitochondrial regulation of caspase activation by cytochrome oxidase and tetramethylphenylenediamine via cytosolic cytochrome c redox state. J Biol Chem. 2007;282:31124–31130. doi: 10.1074/jbc.M700322200. [DOI] [PubMed] [Google Scholar]
- 20.Suto D, Sato K, Ohba Y, Yoshimura T, Fujii J. Suppression of the pro-apoptotic function of cytochrome c by singlet oxygen via a haem redox state-independent mechanism. Biochem J. 2005;392:399–406. doi: 10.1042/BJ20050580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaughn AE, Deshmukh M. Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nat Cell Biol. 2008;10:1477–1483. doi: 10.1038/ncb1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yong J, Kasmin M, Bachorik JL, Wan L, Dreyfuss G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol Cell. 2010;38(4):551–562. doi: 10.1016/j.molcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, et al. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson NG, Clayton DA. Molecular genetic aspects of human mitochondrial disorders. Annu Rev Genet. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- 25.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 26.Janssen GM, Maassen JA, van Den Ouweland JM. The diabetes-associated 3,243 mutation in the mitochondrial tRNA(Leu(UUR)) gene causes severe mitochondrial dysfunction without a strong decrease in protein synthesis rate. J Biol Chem. 1999;274:29744–29748. doi: 10.1074/jbc.274.42.29744. [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 28.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 29.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA overexpression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White K, Tahaoglu E, Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- 31.Kuchino Y, Borek E. Tumour-specific phenylalanine tRNA contains two supernumerary methylated bases. Nature. 1978;271:126–129. doi: 10.1038/271126a0. [DOI] [PubMed] [Google Scholar]
- 32.Larminie CG, Sutcliffe JE, Tosh K, Winter AG, Felton-Edkins ZA, White RJ. Activation of RNA polymerase III transcription in cells transformed by simian virus 40. Mol Cell Biol. 1999;19:4927–4934. doi: 10.1128/mcb.19.7.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, Follettie M, et al. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem. doi: 10.1074/jbc.M109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomez-Roman N, Felton-Edkins ZA, Kenneth NS, Goodfellow SJ, Athineos D, Zhang J, et al. Activation by c-Myc of transcription by RNA polymerases I, II and III. Biochem Soc Symp. 2006:141–154. doi: 10.1042/bss0730141. [DOI] [PubMed] [Google Scholar]
- 35.Crighton D, Woiwode A, Zhang C, Mandavia N, Morton JP, Warnock LJ, et al. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 2003;22:2810–2820. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White RJ. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 37.Marshall L, Kenneth NS, White RJ. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 38.Costanzi J, Sidransky D, Navon A, Goldsweig H. Ribonucleases as a novel pro-apoptotic anticancer strategy: review of the preclinical and clinical data for ranpirnase. Cancer Invest. 2005;23:643–650. doi: 10.1080/07357900500283143. [DOI] [PubMed] [Google Scholar]
- 39.Iordanov MS, Ryabinina OP, Wong J, Dinh TH, Newton DL, Rybak SM, et al. Molecular determinants of apoptosis induced by the cytotoxic ribonuclease onconase: evidence for cytotoxic mechanisms different from inhibition of protein synthesis. Cancer Res. 2000;60:1983–1994. [PubMed] [Google Scholar]
- 40.Suhasini AN, Sirdeshmukh R. Transfer RNA cleavages by onconase reveal unusual cleavage sites. J Biol Chem. 2006;281:12201–12209. doi: 10.1074/jbc.M504488200. [DOI] [PubMed] [Google Scholar]
- 41.Lin JJ, Newton DL, Mikulski SM, Kung HF, Youle RJ, Rybak SM. Characterization of the mechanism of cellular and cell free protein synthesis inhibition by an anti-tumor ribonuclease. Biochem Biophys Res Commun. 1994;204:156–162. doi: 10.1006/bbrc.1994.2439. [DOI] [PubMed] [Google Scholar]
- 42.Zhao H, Ardelt B, Ardelt W, Shogen K, Darzynkiewicz Z. The cytotoxic ribonuclease onconase targets RNA interference (siRNA) Cell Cycle. 2008;7:3258–3261. doi: 10.4161/cc.7.20.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]