Summary
Using single particle electron cryomicroscopy, several helices in the membrane-spanning region of RyR1, including an inner transmembrane helix, a short pore helix, and a helix parallel to the membrane on the cytoplasmic side, have been clearly resolved. Our model places a highly conserved glycine (G4934) at the hinge position of the bent inner helix and two rings of negative charges at the luminal and cytoplasmic mouths of the pore. The kinked inner helix closely resembles the inner helix of the open MthK channel, suggesting that kinking alone does not open RyR1, as proposed for K+ channels.
Introduction
The Ca2+ release channel (also known as the ryanodine receptor, RyR) belongs to a family of intracellular, integral membrane Ca2+ channels and is the largest known ion channel. In mammals, three different RyR isoforms have been identified. They are encoded by distinct genes and share ~70% sequence homology (Rossi and Sorrentino, 2002). This sequence homology accounts for the many functional similarities between RyRs. RyR1 is found primarily in skeletal muscle tissue and mediates the gated release of Ca2+ from sarcoplasmic reticulum (SR) stores into the cytoplasm during excitation-contraction (E-C) coupling. RyR1 has been subjected to the most detailed structure-function analysis of the RyRs due to its high concentration in the SR membrane and the relatively simple procedure for its purification from skeletal muscle. The functional channel is composed of four subunits, each ~565 kDa, constituting a single homotetrameric cation-selective channel pore in the SR membrane (Lai et al., 1989). Mutations in this channel are associated with several human muscular genetic diseases, such as malignant hyperthermia (MH), a pharmacogenetic disorder, and central core disease (CCD), an autosomal dominant myopathy (Guis et al., 2004; Quane et al., 1993).
Extensive efforts have been made to study the 3D structure of the detergent-solubilized RyR1 by using electron microscopy of both single particles (Radermacher et al., 1994; Serysheva et al., 1995, 2005) and 2D crystals (Yin et al., 2005). Though these structural determinations were done at low resolution, they have provided a wealth of information concerning conformational changes associated with opening and localization of binding sites of various cellular effectors (Liu et al., 2004; Orlova et al., 1996; Samso and Wagenknecht, 2002; Wagenknecht et al., 1997). However, substantial questions remain, and the detailed molecular mechanism that underlies RyR1 gating has not yet been resolved. We now report a 9.6 Å resolution structure of RyR1 in a closed conformation, in which a number of membrane-associated α helices can be directly observed. To our knowledge, this represents the first single particle reconstruction at subnanometer resolution providing direct and unambiguous visualization of α helices in the membrane-spanning region.
Results
3D Structure
RyR1 was purified to homogeneity from rabbit skeletal muscle, and a standard [3H]ryanodine binding assay was performed to confirm its functional and structural integrity (Chu et al., 1990; Meissner and el-Hashem, 1992). Figure 1A shows a typical image of ice-embedded RyR1 recorded on a 4k × 4k CCD camera. The image contrast is relatively high, and the particles are randomly oriented within the vitreous ice, producing particle views in a wide range of orientations. The final reconstruction is shown in Figures 1B and 1C. This density map exhibits an immense amount of structural detail, which, lacking crystal structures of any RyR domain, must be interpreted through visual analysis combined with computational tools designed to interpret subnanometer resolution maps (Jiang et al., 2001). At this resolution, only α helices that are sufficiently separated from adjacent densities can be unambiguously identified (Chiu et al., 2005). This manuscript presents a functional interpretation of the structural features seen in the membrane-spanning region of this reconstruction.
Figure 1. Image and Structure of RyR1.
(A) 200 kV electron image of RyR1 embedded in ice recorded at 60,000× magnification with a Gatan 4k × 4k CCD camera on a JEM2010F electron cryomicroscope.
(B and C) Stereoview of the 9.6 Å resolution density map of RyR1 viewed from (B) the cytoplasmic side along the 4-fold axis and (C) the lumenal side in an oblique orientation. The bar denotes 500 Å scale in (A) and 100 Å scale in (B) and (C).
α Helices in the Membrane-Spanning Region
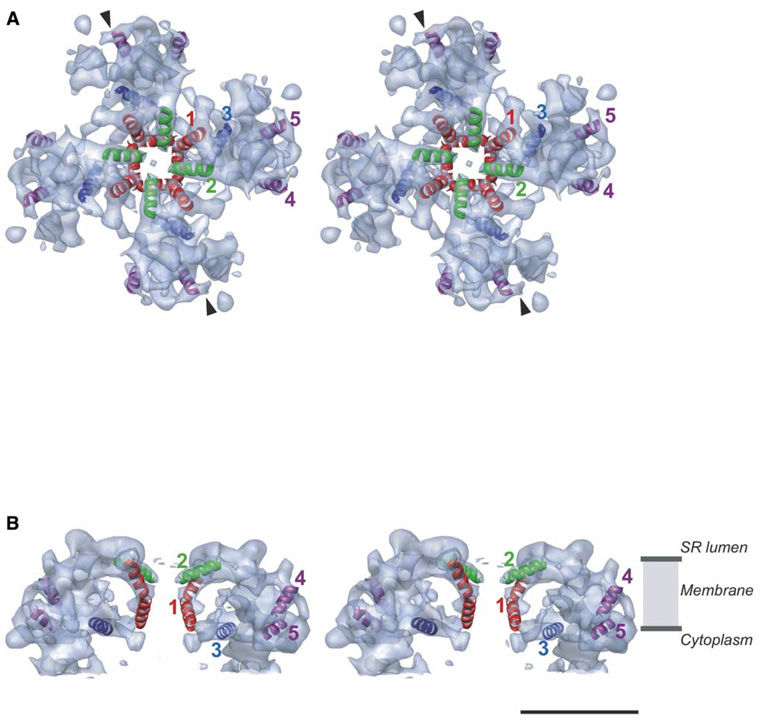
Five rod-like densities have been visually and quantitatively identified as helices in the membrane-spanning portion of the channel (Figures 2A and 2B). It is conceivable that additional helices exist in this region but cannot be unambiguously identified at this resolution. A group of two central rod-like densities, labeled 1 and 2, in each subunit lies near the 4-fold axis of the channel (Figures 2A and 2B and Figure 4A). Putative helix 1 is ~45 Å long. The lower half of this helix is noticeably kinked and bends away from the 4-fold central axis. The apparent tilt of helix 1 with respect to the membrane normal is ~30° above, and ~50° below, the kink. The helices form a large, funnel-like cavity with an apparent diameter of ~30 Å at the luminal side of RyR1, possibly serving as the channel entrance. The funnel tapers toward the cytoplasm, where it has an apparent diameter of ~15 Å. Putative helix 2 is ~22 Å long and has an apparent tilt of ~50° with respect to the membrane normal (Figures 2A and 2B). It points toward the channel’s 4-fold axis to form an apparent opening of ~7 Å with the corresponding helices from the other three subunits. Thus, the putative Ca2+ permeation pathway would run parallel to the channel’s central 4-fold axis, and the lumenal mouth of the channel is lined by four copies of helix 2. A central cavity of ~15 Å diameter is found below the ring formed by four copies of helix 2 (Figure 4A). However, at this resolution, the specific size of any apparent opening or cavity depends on the isosurface display threshold.
Figure 2. Putative Helices in the RyR1 Membrane-Spanning Region.
(A) Stereoview of the membrane-spanning region viewed from the SR luminal side along the 4-fold axis. The densities attributable to α helices are annotated as color ribbons: red, the inner helix; green, the pore helix; blue and purple, the other helices. Arrows denote the direction of the slice shown in Figure 4A.
(B) Side view (view normal to the 4-fold axis) of two of the four subunits of RyR1 with annotated helices showing the relative positions of putative α helices within the SR membrane. A high contour level is used in (A) and (B) to emphasize the strongest densities interpreted as α helices. The bar denotes 50 Å scale.
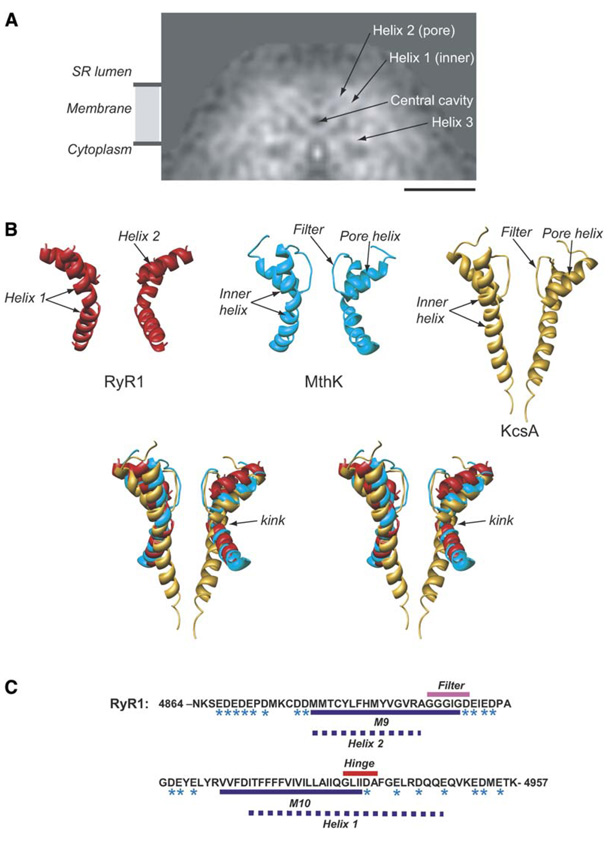
Figure 4. Interpretation of α Helices around the Putative Pore.
(A) A slice of density through RyR1 parallel to the 4-fold axis in the direction indicated by the arrows in (Figure 2A).
(B) Relative arrangement of pore and inner helices from two opposing subunits of the cryoEM map of RyR1 (red). X-ray structures of the MthK channel, 1LNQ (cyan), and of the KcsA channel, 1BL8 (gold), are shown. Two subunits of the RyR1, MthK, and KcsA pore-forming regions are superimposed and are oriented with the cytoplasmic side facing down.
(C) Sequence of the putative pore-forming region of RyR1 (GI: 134134). Residues with negative charges are marked with an asterisk. The dashed lines denote our prediction for the pore-forming helices of RyR1: residues M4879–A4893 correspond to the inner, pore-lining helix (helix 1); residues I4918– E4948 correspond to the P helix (helix 2). The bar in (A) denotes 50 Å scale.
Helix 3 (~22 Å) lies parallel to the plane of the membrane surface (Figures 2A and 2B), likely at the interface between the lipid bilayer and the cytosol. Two rod-like densities shown as purple helical ribbons in Figures 2A and 2B are designated as helices 4 and 5. These two small helices (~18 Å) have various inclination angles with respect to the membrane.
RyR1 must span the membrane an even number of times since both its N terminus and C terminus lie on the cytoplasmic side of the membrane (Grunwald and Meissner, 1995; Marty et al., 1994). The most recent model suggests that a RyR1 monomer spans the membrane with six or eight transmembrane helices (Du et al., 2004). While it would be possible to speculate on this important question based simply on the volume of the transmembrane region, our structure suggests a more complicated arrangement than a simple arrangement of membrane-spanning helices. Helices 1 and 2 are found in the central section of the membrane-spanning region. These helices presumably coincide with the hydrophobic core of the lipid bilayer (Figure 2B). Sections of the density map cut through different levels normal to the channel axis in the membrane-spanning region (Figure 3) demonstrate the high-density peaks composing the transmembrane structure. However, only five peaks of high density can be traced at successive levels to give a 3D picture of rod-like densities that we interpret as α helices. When isolated and visualized as isosurfaces, none of the other identified high-density peaks in the membrane-spanning region of our density map have either the length or the characteristic shape of an α helix as expected at this resolution (Chiu et al., 2005). It is quite possible that this large membrane-spanning region has a mixed α/β structure. Aside from helix 1, none of the identified helices in our map have either sufficient length or the proper orientation to span the membrane.
Figure 3. Cross-Sections through the Membrane-Spanning Region Parallel to the Membrane Plane.
(A) Side view of the membrane-spanning region displayed at the threshold level corresponding to a molecular mass of 2.2 MDa. The levels of sections are indicated with red dashed lines and are numbered beginning from the luminal side.
(B) Sections of density map normal to the 4-fold channel axis at the levels indicated in (A). The threshold for continuous contour lines and color coding has been chosen to highlight the density peaks composing the membrane-spanning region. The positions of the five rod-like densities in the four selected sections in the membrane-spanning region are indicated with corresponding numbers. The bar denotes 50 Å scale.
Sequence Assignment of the Observed α Helices
Comparing our cryoEM structure to the crystal structures of a number of cation channels (Chang et al., 1998; Doyle et al., 1998; Jiang et al., 2002a, 2003b; Miyazawa et al., 2003), we find that the arrangement of α helices in our structure strongly resembles the pore structure of the MthK channel (1LNQ) (Jiang et al., 2002a). For comparison, we also show the corresponding helices from the KcsA channel (1BL8) (Doyle et al., 1998). This comparison clearly demonstrates striking similarity to the MthK channel and relatively poor similarity to the KcsA channel (Figure 4B). This conclusion is further strengthened by the fact that the configuration of the helices in our structure was assigned completely without reference to any of the K+ channel structures. This structural similarity supports the assignment of helix 1 as the inner, pore-lining helix and helix 2 as the pore helix, associated with the channel’s selectivity filter. The kinking of the inner helix in the MthK channel has been suggested to open this channel (Jiang et al., 2002b); however, our structure shows a similar kinked inner helix in the closed conformation of RyR1. While it is possible that subtle structural changes in RyR1 occur upon detergent solubilization, preventing it from being in a fully closed state, previous studies showing sub-stantial conformational changes in the presence of activators would argue against this possibility (Orlova et al., 1996). It seems likely that kinking of the inner helix is not a sufficient conformational change to cause RyR1 channel opening.
Using the nomenclature of the 12 transmembrane helix model, it has been suggested that the ion permeation pathway is formed by the transmembrane helices M9 and M10 (Zorzato et al., 1990). Helix 1 is likely to correspond to M10, proposed to be the inner, pore-lining helix (Du et al., 2004; Takeshima et al., 1989). The length of helix 1 (~45 Å) is consistent with 30 amino acid residues (Figure 4C). In K+ channels, the sequence GXXXXA has been proposed to form the gating hinge in the inner, pore-lining helix (Jiang et al., 2002b). A similar sequence is found in RyR1 at residues G4934–A4939 (Welch et al., 2004) in the center of a putative helical domain (Figure 4C). This allows us to assign G4934 as the kink in helix 1 (Figures 4B and 4C), with I4918–E4948 representing the entire helix.
Helix 2 is likely to correspond to M9, proposed to be the P loop-associated pore helix that enters the channel from the luminal side of the membrane (Gao et al., 2000; Zhao et al., 1999). Our assignment of the M9 sequence to helix 2 in our map suggests that the sequence G(4894)GGIGD(4899), located in the luminal loop between M9 and M10 (Figure 4C), is analogous to the selectivity filter in K+ channels (TVGYGD) (Jiang et al., 2002b). Mutations in the 4894–4899 sequence of RyR1 alter the conductance of the channel (Gao et al., 2000; Zhao et al., 1999), supporting the assignment of this sequence as the selectivity filter.
Functional Interpretation of the Observed α Helices
Given the arrangement of helices 1 and 2 in the 9.6 Å structure of RyR1, there are likely to be negatively charged rings formed by acidic residues on both the luminal and cytosolic sides of the channel pore structure. The sequence (Figure 4C, charged residues indicated by an asterisk) indeed supports this proposition. The presence of negative charges at both pore mouths is a common feature of cation-selective channels. A high density of negative charges is likely to attract the permeant cations and repel intracellular anions from the immediate vicinity of the pore.
Helix 3 is oriented parallel to the plane of the membrane on the cytoplasmic side of the membrane-spanning region (Figures 2A and 2B). We have identified two possible candidates for this helix. If this sequence is N-terminal to the pore helices, it might represent the helix previously designated as 4a/4b (amino acids 4300–4363) (Du et al., 2004), which has been suggested to be either a transmembrane hairpin or a helix that does not span the membrane but associates with the cytosolic membrane leaflet. Deleting this putative helix did not abolish either dihydropyridine receptor (DHPR) activation of RyR1 (orthograde coupling) or the ability of RyR1 to increase DHPR Ca2+ channel activity (retro-grade coupling). However, cells expressing RyR1 with this region deleted had larger L-type currents and Ca2+ transients and were more sensitive to voltage-gated release than cells expressing with wild-type RyR1, suggesting that this sequence may inhibit channel opening. Another possible assignment of helix 3 would be a sequence at the carboxyl-terminal tail of RyR1, connected to the inner helix on the cytoplasmic side of the SR membrane. Since this is the most likely region to be involved in channel gating, this helix might contribute to a gating ring. While our kinked structure in the inner helix suggests that kinking alone does not open RyR1, a gating ring of some sort may still play a role in channel opening. RyR1 can be opened either by the movement of DHPR in the t-tubule membrane, mechanically triggering a change in RyR1 conformation, or by Ca2+ binding. Ca2+-induced expansion of a cytoplasmic gating ring is thought to drive kinking of the inner helix, in turn opening the MthK channel (Jiang et al., 2002b). A similar gating ring mechanism has also been proposed for the cyclic nucleotide-gated channel, HCN (Zagotta et al., 2003). A detailed understanding of the mechanism of channel gating will require comparable structural studies of RyR1 under additional physiological conditions.
Comparison with Previous Structures of RyR1
Recently, a 13.6 Å resolution (based on the 0.5 Fourier shell correlation criterion) cryoEM structure of RyR1 (Samso et al., 2005), comparable to our previously published 14 Å resolution structure (Serysheva et al., 2005), was reported. There are some substantial differences in the conclusions drawn between the present work and that of Samso and coworkers (Samso et al., 2005). The interpretation of their map suggested, based on simple volumetric constraints, that the membrane-spanning region contains at least six transmembrane helices. This statement is consistent with an earlier structural estimate at even lower resolution (Serysheva et al., 1995), but it remains speculative since this number of α helices has not been unambiguously observed in the structure. This argument should not be taken as experimental evidence that the number of transmembrane helices has been determined.
To interpret the density map of the pore region, they relied on the assumption that the RyR1 structure was similar to the KcsA channel (Samso et al., 2005). By visually fitting the KcsA pore helices into their structure, they identified higher-density regions that were then interpreted as α helices. The density map reported here (Figures 1B and 1C) at 9.6 Å resolution permitted identification of five α helices per monomer (Figures 2A and 2B) by a combination of visual inspection of the map itself and the quantitative analysis provided by the SSEhunter program (Baker et al., 2005; Jiang et al., 2001). Figure S1 (see the Supplemental Data available with this article online) shows a view of the central slice of our map in an orientation roughly comparable to that in Figures 4A and 4B in Samso et al. (2005). Our density map clearly exhibits many more structural details in the membrane-spanning region. Although the inner, pore-lining α helices resolved in our density map form a continuous network toward the cytoplasmic region through distinct bridging densities, at current resolution these bridging densities were visualized at much lower display threshold levels than any other identified α helices, and no secondary structure elements could be unambiguously assigned to these densities by using SSEhunter (Baker et al., 2005; Jiang et al., 2001). Therefore, our map does not support the presence of secondary structure elements in the inner branch, as schematized in Samso et al. (2005). We also found no evidence that the membrane-spanning region is more resolved than the cytoplasmic region in our density map as reported in the map of Samso et al. (2005). It should be pointed out that estimation of resolution in specific regions is notoriously susceptible to masking and other artifacts, leading to local overestimates of resolution. To confirm the validity of such measurements, the resolution of the remaining portion of the structure (i.e., the cytoplasmic region) must be determined, and it must be worse than the mean resolution for the overall structure.
Though the resolution of our reconstruction is 9.6 Å, we still consider unambiguous assignment of α helices to be marginal (Jiang et al., 2001). In our approach, all of the reported helices (Figures 2A and 2B) were independently identified both by visual inspection and through use of SSEhunter (Baker et al., 2005; Jiang et al., 2001) without reference to any of the K+ channel structures. This method for identifying putative helices in subnanometer resolution cryoEM maps has been used successfully in a number of previous studies (Jiang et al., 2003a; Schmid et al., 2004; Zhou et al., 2001). The identified α helices in the cryoEM density maps were either confirmed by subsequent determination of a crystal structure of the assembly or by matching them with those in the crystal structures of individual components or homologs (Chiu et al., 2005; Nakagawa et al., 2003).
Conclusions
The 3D structure of RyR1 in a closed conformation has been determined by single particle electron cryo-microscopy at 9.6 Å resolution. This resolution is measured by using the 0.5 cutoff (Böttcher et al., 1997; Ludtke et al., 2004) of the Fourier shell correlation (FSC). Based on the 0.33 FSC criterion, our map would have a resolution of ~8 Å. Interpreting secondary structure elements at this resolution must be performed cautiously. We have identified five densities in the membrane-spanning region with the characteristic features of α helices expected at this resolution (Chiu et al., 2005). At higher resolution, additional helices in this region are likely to be detected. The observation that the pore helices of RyR1 closely match those of MthK strongly supports the reliability and the interpretation of our map. One puzzling issue is that the RyR1 structure was determined in a putatively closed conformation, while that of MthK was in a putatively open conformation. These observations raise the possibility that the observed kinking of the inner helix cannot solely be responsible for the opening of the RyR1 pore. Additional structures of these ion channels in well-defined conformational states at subnanometer resolutions will need to be determined to unambiguously determine the detailed mechanisms of pore opening.
Experimental Procedures
Channel Purification
RyR1 was solubilized with CHAPS from SR membranes of rabbit fast twitch skeletal muscle (Callaway et al., 1994) and was purified according to the procedure established previously (Serysheva et al., 1999). The identity and structural integrity of the protein preparations was confirmed by SDS-PAGE and Western blotting with RyR1-specific antibodies. Functional integrity of the purified channels was verified by using a standard [3H]ryanodine binding assay (data not shown) (Chu et al., 1990; Holmberg and Williams, 1990; Meissner, 1986; Meissner and el-Hashem, 1992; Pessah and Zimanyi, 1991).
Cryo-Specimen Preparation and Imaging
Purified channels, to which 1 mM EGTA was added to maintain them in their closed conformation (Serysheva et al., 1995; Smith et al., 1988), were plunge frozen on Quantifoil holey grids (Quantifoil Micro Tools GmbH, Jena). Images were recorded at 60,000× magnification on a JEM2010F electron microscope with a Gatan liquid nitrogen cryoholder and a Gatan 4k × 4k CCD camera (Booth et al., 2004). The specimen dose was ~15 electrons/Å2 per image frame.
Image Processing and 3D Reconstruction
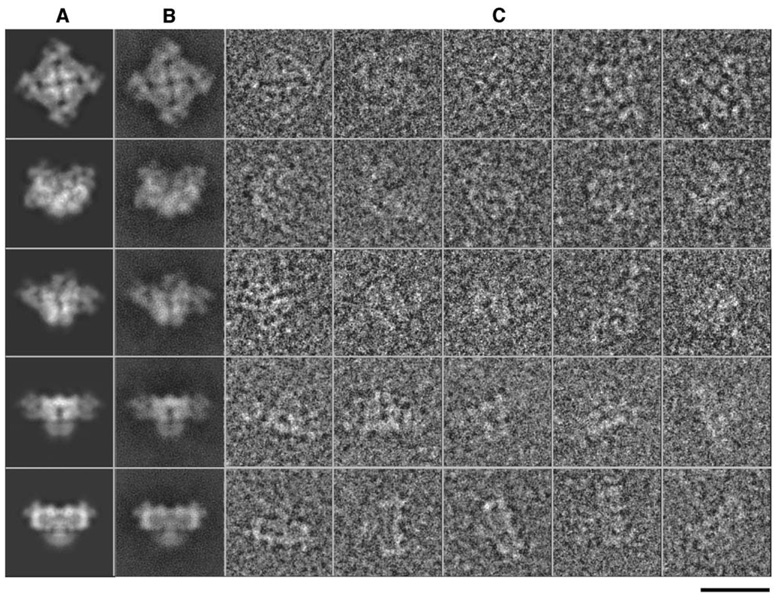
Particles were selected from individual CCD frames, and contrast transfer function parameters were determined by using standard methods in EMAN (Ludtke et al., 2004). Unlike our previous studies on a continuous carbon substrate (Serysheva et al., 2005), ice-embedded particles have a uniform distribution of orientations (Figure 5), providing isotropic resolution. Refinement of the structure is a reference-based iterative process, which has been described previously (Ludtke et al., 2004). Briefly, projections of an initial 3D model, which need not bear any resemblance to the final structure, are generated by using a uniform distribution of orientations, with angular sampling sufficient to obtain a structure of the desired resolution. Individual raw particles are classified by aligning and comparing each to each projection image producing a set of particles associated with each projection. Each of these sets of particles is then iteratively aligned to each other to produce a class average. The orientations of these class averages are known by reference to the projections used to classify the particles, allowing them to be reconstructed, producing a new 3D model. This model is then used as a reference for the next cycle of refinement. Iterative reconstruction with 28,036 particles from 869 CCD frames was continued until convergence was achieved. The reprojections of the final 3D reconstruction agree well with the class averages and the corresponding raw particle images, a key indicator of a successful bias-free reconstruction (Figure 6). The Fourier shell correlation coefficient (van Heel, 1987) was computed between two in-dependent reconstructions based on even and odd halves of the particle data. The final resolution at convergence is 9.6 Å (Figure 7) based on the 0.5 threshold (Böttcher et al., 1997; Ludtke et al., 2004) of the Fourier shell correlation. To evaluate model bias, the structure was rerefined from randomly generated starting models, and the results were compared. The observed helices remained in these other reconstructions. So, while this resolution is not adequate to clearly observe all helices, the five presented here are self-consistent.
Figure 5. Distribution of Particle Orientations over an Asymmetric Unit Used in the Reconstruction.
Brighter dots denote a larger number of particles. This shows a fairly uniform distribution of particle orientations.
Figure 6. Results from Image Processing and 3D Reconstruction.
(A–C) (A) Selected reprojections of the final 3D structure, (B) corresponding class averages, and (C) raw particle images. Their apparent match is a key indication of the self-consistency of the reconstruction. The bar denotes 250 Å scale.
Figure 7. Fourier Shell Correlation between Reconstructions from Even and Odd Halves of the Data Set.
A threshold of 0.5 is used to assess the resolution of the reconstruction, which is 9.6 Å.
Map Visualization and Interpretation
The 3D density map was visualized by using Chimera (Goddard et al., 2005) and AMIRA (Mercury Computer Systems). Secondary structure elements were detected by using the improved version of helixhunter (Jiang et al., 2001) called SSEhunter (Baker et al., 2005), which is available in the AIRS package distributed with EMAN (Ludtke et al., 1999). Helices were manually placed in the reconstruction (Figure 2). After placement, helices were extracted for comparison with the pore and inner helices of MthK and KcsA (Figure 4B) without additional repositioning. The strong agreement between this known structure and our independently analyzed reconstruction is further validation of the 3D density map and the helix identification.
Supplementary Material
Acknowledgments
We thank Drs. Matthew Baker, Wen Jiang, and Tao Ju for kindly providing the SSEhunter program and Matthew Dougherty for assistance in computer graphics. This research has been supported by grants from National Institutes of Health (P41RR02250, P01GM99116, R01AR44864, R01GM072804, and RO1AR41729) and by a Muscular Dystrophy Association grant.
Footnotes
Supplemental Data
Supplemental Data including a figure showing rod-like densities in the central slice of the membrane-spanning region are available at http://www.structure.org/cgi/content/full/13/8/1203/DC1/.
References
- Baker ML, Jiang W, Chiu W. Analysis of intermediate resolution structures. Biophys. J. 2005;88:23. [Google Scholar]
- Booth CR, Jiang W, Baker ML, Hong Zhou Z, Ludtke SJ, Chiu W. A 9 Å single particle reconstruction from CCD coucaptured images on a 200 kV electron cryomicroscope. J. Struct. Biol. 2004;147:116–127. doi: 10.1016/j.jsb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Callaway C, Seryshev A, Wang JP, Slavik KJ, Needleman DH, Cantu C, 3rd, Wu Y, Jayaraman T, Marks AR, Hamilton SL. Localization of the high and low affinity [3H]ryanodine binding sites on the skeletal muscle Ca2+ release channel. J. Biol. Chem. 1994;269:15876–15884. [PubMed] [Google Scholar]
- Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science. 1998;282:2220–2226. doi: 10.1126/science.282.5397.2220. [DOI] [PubMed] [Google Scholar]
- Chiu W, Baker ML, Jiang W, Dougherty M, Schmid MF. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure (Camb) 2005;13:363–372. doi: 10.1016/j.str.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Chu A, Diaz-Munoz M, Hawkes MJ, Brush K, Hamilton SL. Ryanodine as a probe for the functional state of the skeletal muscle sarcoplasmic reticulum calcium release channel. Mol. Pharmacol. 1990;37:735–741. [PubMed] [Google Scholar]
- Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Du GG, Avila G, Sharma P, Khanna VK, Dirksen RT, MacLennan DH. Role of the sequence surrounding predicted transmembrane helix M4 in membrane association and function of the Ca(2+) release channel of skeletal muscle sarcoplasmic reticulum (ryanodine receptor isoform 1) J. Biol. Chem. 2004;279:37566–37574. doi: 10.1074/jbc.M406637200. [DOI] [PubMed] [Google Scholar]
- Gao L, Balshaw D, Xu L, Tripathy A, Xin C, Meissner G. Evidence for a role of the lumenal M3–M4 loop in skeletal muscle Ca(2+) release channel (ryanodine receptor) activity and conductance. Biophys. J. 2000;79:828–840. doi: 10.1016/S0006-3495(00)76339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure (Camb) 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Grunwald R, Meissner G. Lumenal sites and C terminus accessibility of the skeletal muscle calcium release channel (ryanodine receptor) J. Biol. Chem. 1995;270:11338–11347. doi: 10.1074/jbc.270.19.11338. [DOI] [PubMed] [Google Scholar]
- Guis S, Figarella-Branger D, Monnier N, Bendahan D, Kozak-Ribbens G, Mattei JP, Lunardi J, Cozzone PJ, Pellissier JF. Multiminicore disease in a family susceptible to malignant hyperthermia: histology, in vitro contracture tests, and genetic characterization. Arch. Neurol. 2004;61:106–113. doi: 10.1001/archneur.61.1.106. [DOI] [PubMed] [Google Scholar]
- Holmberg SR, Williams AJ. The cardiac sarcoplasmic reticulum calcium-release channel: modulation of ryanodine binding and single-channel activity. Biochim. Biophys. Acta. 1990;1022:187–193. doi: 10.1016/0005-2736(90)90113-3. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Ludtke SJ, Chiu W. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 2001;308:1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002a;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. The open pore conformation of potassium channels. Nature. 2002b;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- Jiang W, Li Z, Zhang Z, Baker ML, Prevelige PE, Chiu W. Coat protein fold and maturation transition of bacterio-phage P22 seen at sub-nanometer resolutions. Nat. Struct. Biol. 2003a;10:131–135. doi: 10.1038/nsb891. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003b;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- Lai FA, Misra M, Xu L, Smith HA, Meissner G. The ryanodine receptor-Ca2+ release channel complex of skeletal muscle sarcoplasmic reticulum. Evidence for a cooperatively coupled, negatively charged homotetramer. J. Biol. Chem. 1989;264:16776–16785. [PubMed] [Google Scholar]
- Liu Z, Zhang J, Wang R, Wayne Chen SR, Wagenknecht T. Location of divergent region 2 on the three-dimensional structure of cardiac muscle ryanodine receptor/calcium release channel. J. Mol. Biol. 2004;338:533–545. doi: 10.1016/j.jmb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Seeing GroEL at 6 Å resolution by single particle electron cryomicroscopy. Structure (Camb) 2004;12:1129–1136. doi: 10.1016/j.str.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Marty I, Villaz M, Arlaud G, Bally I, Ronjat M. Transmembrane orientation of the N-terminal and C-terminal ends of the ryanodine receptor in the sarcoplasmic reticulum of rabbit skeletal muscle. Biochem. J. 1994;298:743–749. doi: 10.1042/bj2980743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner G. Ryanodine activation and inhibition of the Ca2+ release channel of sarcoplasmic reticulum. J. Biol. Chem. 1986;261:6300–6306. [PubMed] [Google Scholar]
- Meissner G, el-Hashem A. Ryanodine as a functional probe of the skeletal muscle sarcoplasmic reticulum Ca2+ release channel. Mol. Cell. Biochem. 1992;114:119–123. doi: 10.1007/BF00240306. [DOI] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Miyazaki N, Taka J, Naitow H, Ogawa A, Fujimoto Z, Mizuno H, Higashi T, Watanabe Y, Omura T, et al. The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure (Camb) 2003;11:1227–1238. doi: 10.1016/j.str.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Orlova EV, Serysheva II, van Heel M, Hamilton SL, Chiu W. Two structural configurations of the skeletal muscle calcium release channel. Nat. Struct. Biol. 1996;3:547–552. doi: 10.1038/nsb0696-547. [DOI] [PubMed] [Google Scholar]
- Pessah IN, Zimanyi I. Characterization of multiple [3H]ryanodine binding sites on the Ca2+ release channel of sarcoplasmic reticulum from skeletal and cardiac muscle: evidence for a sequential mechanism in ryanodine action. Mol. Pharmacol. 1991;39:679–689. [PubMed] [Google Scholar]
- Quane KA, Healy JM, Keating KE, Manning BM, Couch FJ, Palmucci LM, Doriguzzi C, Fagerlund TH, Berg K, Ording H, et al. Mutations in the ryanodine receptor gene in central core disease and malignant hyperthermia. Nat. Genet. 1993;5:51–55. doi: 10.1038/ng0993-51. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Rao V, Grassucci R, Frank J, Timerman AP, Fleischer S, Wagenknecht T. Cryo-electron microscopy and three-dimensional reconstruction of the calcium release channel/ryanodine receptor from skeletal muscle. J. Cell Biol. 1994;127:411–423. doi: 10.1083/jcb.127.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Sorrentino V. Molecular genetics of ryanodine receptors Ca2+-release channels. Cell Calcium. 2002;32:307–319. doi: 10.1016/s0143416002001987. [DOI] [PubMed] [Google Scholar]
- Samso M, Wagenknecht T. Apocalmodulin and Ca2+-calmodulin bind to neighboring locations on the ryanodine receptor. J. Biol. Chem. 2002;277:1349–1353. doi: 10.1074/jbc.M109196200. [DOI] [PubMed] [Google Scholar]
- Samso M, Wagenknecht T, Allen PD. Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat. Struct. Mol. Biol. 2005;12:539–544. doi: 10.1038/nsmb938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MF, Sherman MB, Matsudaira P, Chiu W. Structure of the acrosomal bundle. Nature. 2004;431:104–107. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- Serysheva II, Orlova EV, Chiu W, Sherman MB, Hamilton SL, van Heel M. Electron cryomicroscopy and angular reconstitution used to visualize the skeletal muscle calcium release channel. Nat. Struct. Biol. 1995;2:18–24. doi: 10.1038/nsb0195-18. [DOI] [PubMed] [Google Scholar]
- Serysheva II, Schatz M, van Heel M, Chiu W, Hamilton SL. Structure of the Skeletal Muscle Calcium Release Channel activated with Ca2+ and AMP-PCP. Biophys. J. 1999;77:1936–1944. doi: 10.1016/S0006-3495(99)77035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serysheva II, Hamilton SL, Chiu W, Ludtke SJ. Structure of Ca2+ release channel at 14 Å resolution. J. Mol. Biol. 2005;345:427–431. doi: 10.1016/j.jmb.2004.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Imagawa T, Ma J, Fill M, Campbell KP, Coronado R. Purified ryanodine receptor from rabbit skeletal muscle is the calcium-release channel of sarcoplasmic reticulum. J. Gen. Physiol. 1988;92:1–26. doi: 10.1085/jgp.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H, Nishimura S, Matsumoto T, Ishida H, Kangawa K, Minamino N, Matsuo H, Ueda M, Hanaoka M, Hirose T, et al. Primary structure and expression from complementary DNA of skeletal muscle ryanodine receptor. Nature. 1989;339:439–445. doi: 10.1038/339439a0. [DOI] [PubMed] [Google Scholar]
- van Heel M. Similarity measures between images. Ultramicroscopy. 1987;21:95–100. [Google Scholar]
- Wagenknecht T, Radermacher M, Grassucci R, Berkowitz J, Xin HB, Fleischer S. Locations of calmodulin and FK506-binding protein on the three- dimensional architecture of the skeletal muscle ryanodine receptor. J. Biol. Chem. 1997;272:32463–32471. doi: 10.1074/jbc.272.51.32463. [DOI] [PubMed] [Google Scholar]
- Welch W, Rheault S, West DJ, Williams AJ. A model of the putative pore region of the cardiac ryanodine receptor channel. Biophys. J. 2004;87:2335–2351. doi: 10.1529/biophysj.104.044180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin CC, Han H, Wei R, Lai FA. Two-dimensional crystallization of the ryanodine receptor Ca2+ release channel on lipid membranes. J. Struct. Biol. 2005;149:219–224. doi: 10.1016/j.jsb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Zagotta WN, Olivier NB, Black KD, Young EC, Olson R, Gouaux E. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 2003;425:200–205. doi: 10.1038/nature01922. [DOI] [PubMed] [Google Scholar]
- Zhao M, Li P, Li X, Zhang L, Winkfein RJ, Chen SR. Molecular identification of the ryanodine receptor pore-forming segment. J. Biol. Chem. 1999;274:25971–25974. doi: 10.1074/jbc.274.37.25971. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Baker ML, Jiang W, Dougherty M, Jakana J, Dong G, Lu G, Chiu W. Electron cryomicroscopy and bioinformatics suggest protein fold models for rice dwarf virus. Nat. Struct. Biol. 2001;8:868–873. doi: 10.1038/nsb1001-868. [DOI] [PubMed] [Google Scholar]
- Zorzato F, Fujii J, Otsu K, Phillips M, Green NM, Lai FA, Meissner G, MacLennan DH. Molecular cloning of cDNA encoding human and rabbit forms of the Ca2+ release channel (ryanodine receptor) of skeletal muscle sarcoplasmic reticulum. J. Biol. Chem. 1990;265:2244–2256. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.