Abstract
Arp2/3 complex plays a central role in the de novo nucleation of filamentous actin as branches on existing filaments. To form a new actin filament the complex must bind ATP, protein activators (e.g. Wiskott-Aldrich syndrome proteins, WASp) and the side of an actin filament. Amide Hydrogen/Deuterium exchange (HDX) coupled with mass spectrometry (MS) was used to examine the structural and dynamic properties of the mammalian Arp2/3 complex in the presence of both ATP and the activating peptide segment from WASp. Changes in the rate of hydrogen exchange indicate that ATP binding causes conformational rearrangements of Arp2 and Arp3 that are transmitted allosterically to the ArpC1, ArpC2, ArpC4 and ArpC5 subunits. These data are consistent with the closure of nucleotide-binding cleft of Arp3 upon ATP binding, resulting in structural rearrangements that propagate throughout the complex. Binding of the VCA domain of WASp to ATP-Arp2/3 further modulates the rates of hydrogen exchange in these subunits, indicating that a global conformational reorganization is occurring. These effects may include the direct binding of activators to Arp3, Arp2 and ARPC1; alterations in the relative orientations of Arp2 and Arp3; and the long-range transmission of activator-dependent signals to segments proposed to be involved in binding the F-actin mother filament.
Keywords: Arp2/3, actin, cytoskeleton, amide hydrogen/deuterium exchange, dynamics
INTRODUCTION
The spatial and temporal coordination of actin cytoskeletal assembly and disassembly is essential for motility, cytokinesis, membrane trafficking and the establishment and maintenance of cell morphology 1. Arp2/3 complex is responsible for the de novo nucleation of actin filaments, forming characteristic “Y” branches off the side of existing filaments2. The Arp2/3 complex is composed of seven subunits including two actin related proteins, Arp2 and Arp3 that both bind ATP and are thought to form a nucleation site for the addition of new actin monomers 3. The five additional protein subunits, denoted Arp complex (ARPC) 1–5, are essential for a full activity 4; 5; 6. Branch formation by Arp2/3 complex requires ATP and association with the side of an actin filament and members of the WASp/Scar (Wiskott-Aldrich Syndrome Protein/Suppressor of cyclic AMP receptor) family of nucleation promoting factors (NPFs) 7; 8. A three-dimensional reconstruction of the actin filament branch junction shows the Arp2 and Arp3 subunits arranged like consecutive subunits in an actin filament forming the base for the daughter filament 9. Arp2 and Arp3 must move 3 nm closer together from their positions in inactive Arp2/3 complex to achieve their positions in the branch junction.
Binding of Mg2+-ATP to Arp2/3 complex increases the fluorescence resonance energy transfer (FRET) between fluorescent proteins fused to ARPC1 and ARPC3 indicating that the fluorescent proteins are closer together 10. Point mutations in the nucleotide binding pocket of Arp3 reduced FRET, suggesting that nucleotide binding contributes to a structural change in Arp3 10. In contrast, high resolution crystal structures suggest that nucleotide binding favors closure of the nucleotide binding cleft of Arp3 without large conformational changes in the complex 11; 12.
WASp family proteins promote nucleation by Arp2/3 complex by means of a conserved C-terminal segment called the VCA domain (reviewed in Welch and Mullins, 2002 13). The VCA region is the minimal segment required for activation 7; 13; 14; 15, consisting of a verprolin-homology sequence that binds and may recruit actin monomers to Arp2/3 complex 16, a central hydrophobic region for activation, and an acidic tail region for high-affinity binding to the complex. WASp binding increases FRET within the nucleotide-bound Arp2/3 complex 10 and favors a more compact organization of Arp2/3 complex in reconstructions of electron micrographs (EM) of single particles 17; 18. Importantly, ATP and WASp cooperatively activate Arp2/3 complex. ATP binding to both Arps is required for the nucleation of new actin filaments 17; 19; 20 and is coupled to NPF binding, such that binding of either ligand increases the affinity of the other (reviewed in 21). Homology modeling 22, cross-linking 23, EM 18; 24, and small angle x-ray scattering (SAXS) 25, have all been used to map putative WASp binding regions on Arp2/3 complex; however, structural evidence at the amino acid level is still needed to fully define the binding sites, associated structural rearrangements and the mechanism of Arp2/3 activation.
While high-resolution structures exist for the inactive complex, complementary approaches are needed to answer many unresolved questions about mechanisms. Most or all of subdomains 1 and 2 of Arp2 are disordered in crystal structures 11; 12; 26 leaving questions about the detailed interactions between Arp2 and Arp3 that might be important for function. In particular, the complex with bound activator has not been crystallized to establish the location of the binding site and how activators affect inter and intra-subunit dynamics at the atomic level. Oxidative footprinting and mass spectrometry experiments probed the effects of ATP and WASp binding on the conformation of the complex in solution at the amino acid level 27. Measured changes in reactivity/surface accessibility of amino acid side chains are consistent with the crystallographic observations that ATP binding results in conformational alterations in Arp2 and Arp3 without significant rearrangements of the other five subunits 11; 12. Footprinting further suggested that binding of WASp to the ATP-Arp2/3 complex results in rearrangements of Arp2 and Arp3 leading to an actin-dimer-like compact conformational state 27, without the involvement of the other subunits. These data, however, are limited to those solvent accessible amino acid side-chains present on the protein surface, so subtle conformational changes or those involving backbone contributions may have been missed.
A complementary approach is to combine amide hydrogen/deuterium exchange (HDX) with mass spectrometry to identify subtle structural changes induced by ligand binding or interactions between proteins 28; 29; 30. Some hydrogens within a protein exchange continuously with protons of solvent water (reviewed in Englander, 2006 30) and can be exchanged with deuterons (2H) when dissolved in deuterium oxide (D2O) at neutral pH. Because amide protons are present at every amino acid, except proline, this technique offers the potential to map structural changes along the entire length of the polypeptide chain. The rate of HDX of amide hydrogens varies by many orders of magnitude depending on their environment 29; 30; 31, including differences in solvent accessibility and the involvement in hydrogen bonding that helps stabilize secondary and tertiary structure. Thus, ligand binding might alter the HDX rate of a protein or peptide segment directly due to steric blockage (i.e., shielding from solvent) or indirectly arising from a conformational rearrangement induced by ligand binding. Generally, HDX alone cannot distinguish between these two mechanisms. Changes in exchange rates can be mapped onto existing structures and complimentary biochemical and biophysical methods such as mutagenesis, crosslinking, and molecular modeling can be used to distinguish between direct and indirect binding effects.
In this work, we utilized amide hydrogen/deuterium exchange combined with mass spectrometry to probe structural changes of the Arp2/3 complex associated with binding ATP and WASp-VCA. We first examined the global effects of Mg2+-ATP and WASp-VCA binding, revealing those subunits that are responsive to ligand binding. We then characterized specific regions showing differing degrees of HDX within the complex using liquid chromatography coupled to high resolution Fourier Transform Ion Cyclotron Resonance (FT-ICR) mass spectrometry. These results provide insights into structural and dynamic changes involved in the mechanism of WASp activation of the Arp2/3 complex and allow for a comparison of existing models for Arp2/3 activation.
RESULTS
Global HDX of the Basal and Activated Arp2/3 Complex
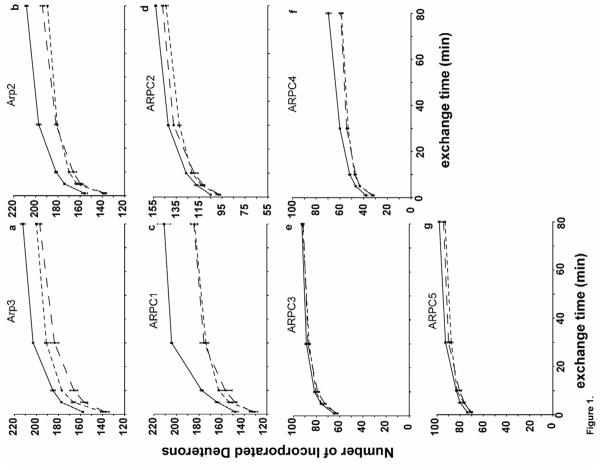
We compared the time course of HDX of bovine thymus Arp2/3 complex without ATP and with ATP ±WASp-VCA. In general, proton exchange was faster in solvent accessible regions than buried regions. Most of the ARPC4 subunit in the Arp2/3 complex is buried from solvent by other subunits 11; 12; 26, so only modest exchange rates were observed for ARPC4 relative to the other subunits (Figure 1). This behavior verified that the Arp2/3 complex was intact and stable during the HDX experiments.
Figure 1. Time course of HDX of apo-Arp2/3 complex (solid line), ATP-Arp2/3 complex (short-dashed line), and ATP/WASp-Arp2/3 complex (long-dashed line).
The number of hydrogens exchanged determined by the mass increase for each individual subunit as a function of exchange time. a, Arp3; b, Arp2; c, ARPC1; d, ARPC2; e, ARPC3; f, ARPC4; g, ARPC5.
Time courses of deuterium exchange clearly showed that bound ATP reduced the number of incorporated deuterons into all subunits of Arp2/3 complex except ARPC3 (Figure 1). Bound ATP had the greatest impact on Arp2, Arp3 and ARPC1, followed by ARPC2, ARPC4, and ARPC5. Even the small difference in deuterium incorporation in ARPC5 was consistent and significant, given the standard deviations calculated from triplicate samples.
Surprisingly, addition of WASp-VCA to the ATP-bound Arp2/3 complex caused only a slight difference in global deuterium incorporation (Figure 1). Compared to the ATP-bound complex, only Arp3 exhibits a further global reduction in deuterium incorporation, while Arp2 and ARPC2 exchanged slightly more deuterium (Figure 1).
Region-specific changes in HDX in specific regions of the inactive and active complex
Ligand binding may influence regional deuterium incorporation without changing global deuterium incorporation owing to compensating positive and negative changes. Therefore, to obtain a more detailed understanding of the structural changes that occur upon ligand binding to Arp2/3 complex, we investigated the deuterium incorporation of individual peptide segments at the 80 minute time point when the differences in deuterons exchanged were largest (Figure 1). Apo-, ATP-, and ATP/WASp-Arp2/3 complexes were subjected to HDX for 80 minutes, quenched with pH 2.5 phosphate buffer in a chilled ice bath, and then digested for 87 seconds on a pepsin column. The resulting peptides were separated by HPLC followed by mass analysis.
A 7 Tesla FT-ICR MS equipped with a standard electrospray source detected a large number of overlapping peptides produced by pepsin digestion including 1745 of the 1981 residues in the complex, with the following coverages: Arp3 (93%), Arp2 (90%), ARPC1 (90%), ARPC2 (93%), ARPC3 (79%), ARPC4 (85%) and ARPC5 (88%). During the HDX experiments, these numbers were reduced to 82% of Arp3, 67% of Arp2, 69% of ARPC1, 79% of ARPC2, 65% of ARPC3, 35% of ARPC4, and 67% of ARPC5, or 69%, due to the broadening of some mass peaks caused by partial deuterium incorporation, and the inability to differentiate between similar masses at the same retention time. Figure 2a compares the deuterium exchange into individual peptides with and without ATP while Figure 2b compares deuterium exchange into individual peptides of ATP-Arp2/3 with and without WASp-VCA. Peptides with significantly different deuterium incorporation levels were mapped onto the composite structural model based on nucleotide-bound bovine Arp2/3 described above (Figures 3–5).
Figure 2. The changes in deuterium incorporation in peptic peptides in a) ATP-bound vs. apo-Arp2/3 complex and b) ATP- vs. ATP+WASp-Arp2/3 complex.
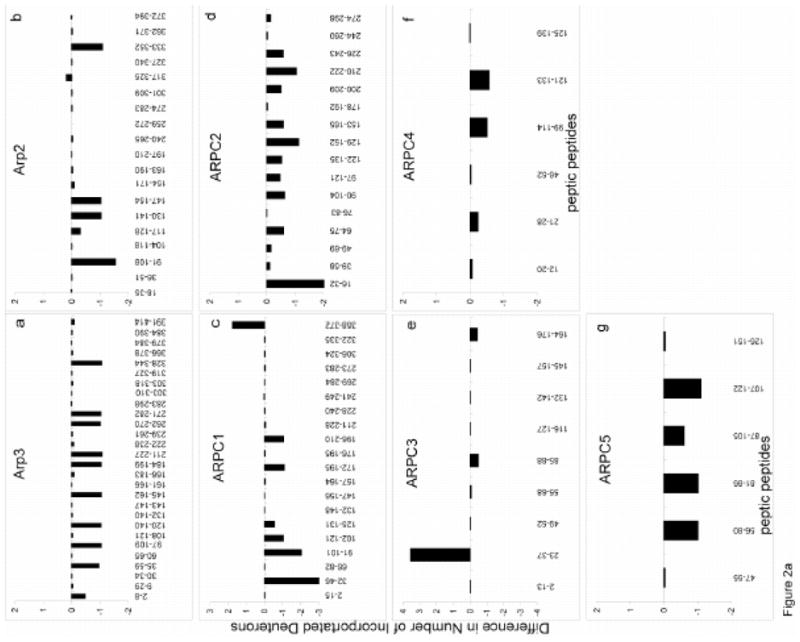
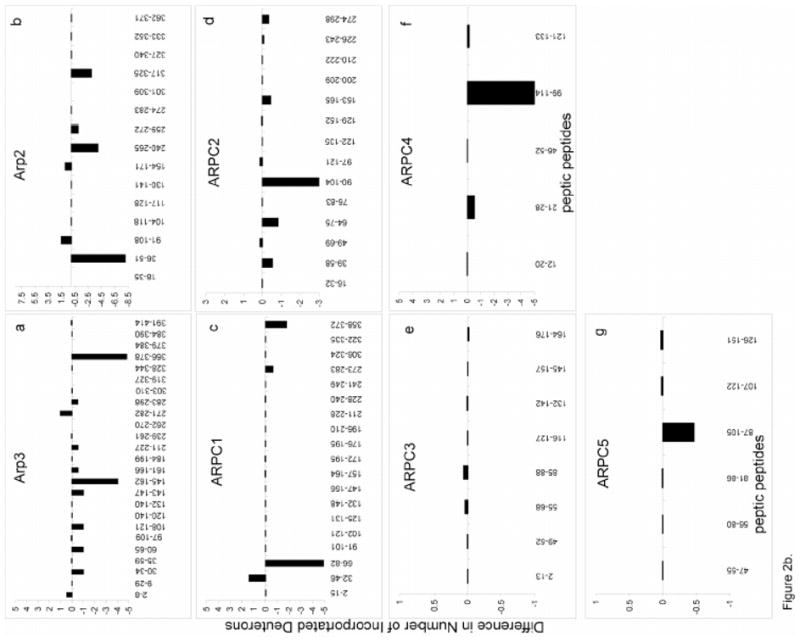
Peptides from each subunit are plotted against the difference in deuterium incorporation following an 80 minute HDX. Differences in deuterium incorporation were determined by the mass difference in individual peptides between a, the apo- and the ATP-bound complex and b, the ATP- and ATP/WASp-bound where negative numbers indicate enhanced protection when ligand is bound, while positive numbers indicate decreased protection. a, Arp3; b, Arp2; c, ARPC1; d, ARPC2; e, ARPC3; f, ARPC4; g, ARPC5.
Figure 3. Mapping the local HDX changes on a ribbon model of the Arp3-Arp2 dimer.
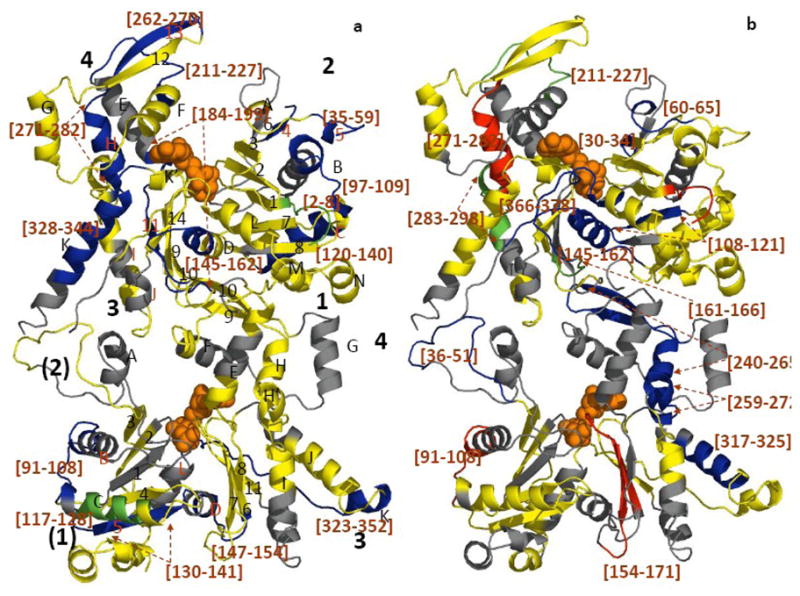
a) compares deuterium exchange into individual peptic peptides with and without ATP; b) compares deuterium exchange into individual peptides of ATP-Arp2/3 complex with and without WASp-VCA.) Differences in deuterium incorporation were determined by the mass difference in individual peptides between a) the apo- and the ATP-bound complex and b) the ATP- and ATP/WASp-bound complex. Subdomains of Arp2 and Arp3 are labeled 1–4. Arp3 (top) is based on the crystal structure (1TYQ) and the Arp2 (bottom) is from the composite structure of 1TYQ with modeled subdomains 1 and 2. α-helices are labeled with capital letters and β-sheets are labeled with numbers in a). Segments of the polypeptide chains cleaved by pepsin are labeled by degree of protection: red >+0.5 Da; yellow, −0.5 Da to 0.5 Da; green, −0.5 to −1 Da; blue <−1 Da; and grey, missing peptides. Peptides with significant changes in deuterium incorporation are labeled (brackets).
Figure 5. Mapping the local HDX changes on the model of the ATP-Arp2/3 complex with and without WASp-VCA.
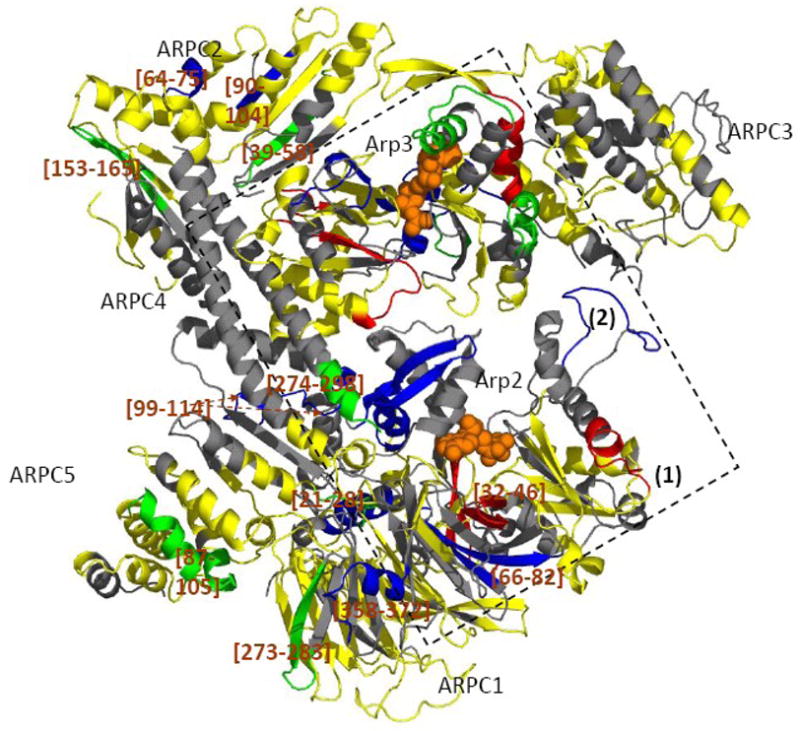
Differences in deuterium incorporation were determined by the mass difference in individual peptic peptides between the ATP- and the ATP/WASp-bound complex. The subdomains 1 and 2 of Arp2 are from the composite model. Peptidic peptides are colored as described in Figure 3 and 4. The square indicates possible WASp binding sites.
Influence of ATP binding on H/D Exchange of Arp3 and Arp2
ATP binding reduced deuterium incorporation (i.e. protection) in peptides [184–199], [211–227] and [328–344] of Arp3 (Figures 2a and 3a), which all contact ATP directly in the crystal structure 11. Notably, ATP also protected peptides distant fromthe nucleotide binding cleft from exchange, including Arp3 peptides [2–8], [35–59], [97–109], [120–140], [145–162], and [262–282] (Figure 3a).
ATP binding also reduced deuterium incorporation into peptides [91–108], [117–128], [130–141], [147–154], and [333–352] of Arp2 (Figure 3a). The first three peptides belong to subdomains 1 and 2 of Arp2 that are disordered in the crystal structure. Marginal protections were noted in the nucleotide-binding cleft (Figure 2a), although the difference is just below our defined significance level as indicated by yellow coding (Figure 3a), suggesting that ATP binding to Arp2 does not induce an obvious cleft-closure as seen in Arp3 (Figures 3a and 4). In addition, no significant differences were observed in the region between Arp3 and Arp 2 upon nucleotide binding (Figure 3a, yellow), indicating that ATP is not sufficient to bring Arp2 and Arp3 into a dimer-like conformation.
Figure 4. Mapping the local HDX changes on the model of the Arp2/3 complex with and without ATP.
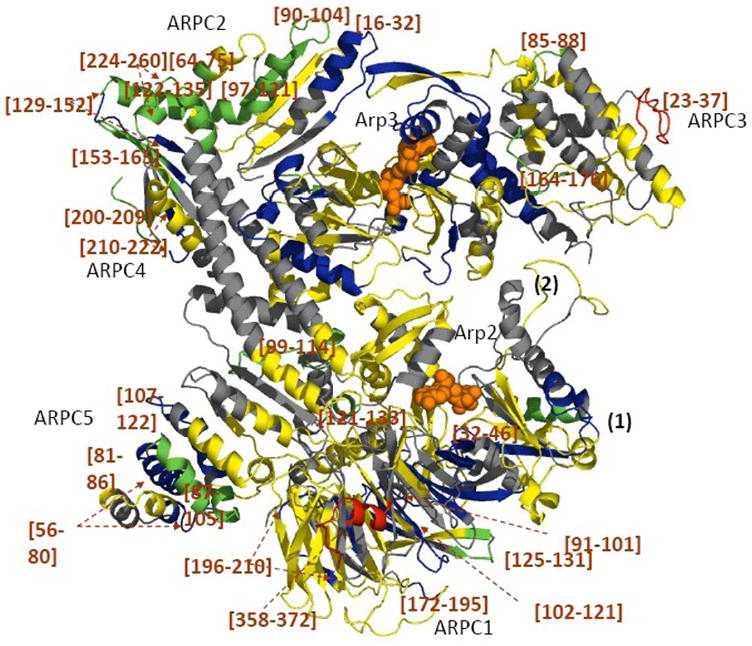
Differences in deuterium incorporation were determined by the mass difference in individual peptic peptides between the apo- and the ATP-bound complex. The subdomains 1 and 2 of Arp2 are from the composite model. Peptic peptides are colored as described in Figure 3.
Influence of ATP binding on H/D Exchange of Other Subunits
ATP bindingnot only reduced deuterium incorporation levels in its direct binding partners, Arp3 and Arp2, but also in the other subunits. ATP binding reduced deuterium exchange into peptides [147–154] and [333–352] of Arp2 together with peptides ARPC1 [91–121], a long loop positioned parallel to loop Arp2 [333–352], and loop ARPC1 [125–131], which is in close proximity to Arp2 [147–154] (Figure 4). ATP binding also protected ARPC1 [32–46], a loop distant from Arp2, suggesting that the effects of ATP binding are communicated throughout this entire subunit. ATP binding to the Arps also reduces deuterium exchange in five beta strands of ARPC1, only one of which is at the Arp2-ARPC1 interface (Figure 4). At the same time, a C-terminal extension of ARPC1 [358–372] became more flexible and displayed an increase in deuterium incorporation. ATP binding to the Arps predominantly reduced deuterium exchange in the two homologous globular domains of ARPC2. Most of the peptides from the long C-terminal helix of ARPC2 were not recovered. The effects of ATP on ARPC1 suggest that the closure of the nucleotide binding cleft of Arp3 makes the adjacent domains of ARPC1 more compact and/or less dynamic (Figure 4). ATP binding to the Arps reduced deuterium exchange more in ARPC5 than APRC4. On the other hand, ATP binding to the Arps significantly increased deuterium incorporation on the loop ARPC3 [23–37], indicating this conformational adjustment increased the solvent accessibility and/or dynamics of this loop.
Arp3 and Arp2 H/D Exchange upon ATP and WASp-VCA Binding
The addition of WASp-VCA to ATP-bound Arp2/3 complex significantly reduced the rates of deuterium incorporation in [145–162] and [366–378] (Figures 2B and 3B). These two α-helices are in the center of this subunit, close to the nucleotide binding site. It is notable that ATP binding alone protected [145–162], but not [366–378]. WASp-VCA binding modestly reduced deuterium incorporation in Arp3 peptides [30–34], [60–65], [108–121], [143–147], [161–166], [211–227] and [283–298]. Notably, WASp-VCA binding increased deuterium incorporation in Arp3 peptides [2–8] and [271–282], the opposite of the effect of ATP binding alone.
The most significant effect of WASp-VCA binding in Arp2 was protection of 8 amide hydrogens in the peptide [36–51] from the “DNase binding loop”. This peptide in subdomain 2 of Arp2 does not appear in electron density maps of Arp2/3 complex. Based on the composite structure of Arp2 with modeled subdomains 1 and 2 (Figure 3), this loop is located within or near the interface between Arp3 and Arp2. WASp-VCA binding also reduces deuterium exchange of Arp2 peptide [240–265], which forms aH, and strands β9 and β10 in subdomain 4, which also interface with Arp3. Reduced deuterium exchange was also observed in peptides [259–272] comprising a portion of αH and αH′, and [317–325] within αJ. WASp-VCA binding protects additional amide hydrogens in Arp2 peptides [91–108] consisting of the loop αB-αC in subdomain 1, and [154–171] consisting of β6, β7 and a portion of loop β7-β8 in subdomain 3 (Figure 2b, 3b).
H/D Exchange of Other Subunits upon ATP and WASp-VCA Binding
WASp-VCA binding to ATP-Arp2/3 complex changes deuterium incorporation in all of the other subunits (Figure 2b, 5). WASp-VCA protected 5 amide protons in ARPC1 peptide [66–82] and 2 amide protons in peptide [358–372], but allowed 1 additional amide proton to exchange in ARPC1 peptide [32–46]. WASp-VCA decreased deuterium incorporation in peptides [39–58], [64–75], [90–104], [153–165] and [274–298] of ARPC2 and peptide [99–114] of ARPC4. WASp-VCA slightly protected peptide [87–105] of ARPC5 and caused mostly subtle increases in deuterium exchange in most peptides of ARPC3.
DISCUSSION
High resolution structures of proteins, particularly when bound to macromolecular or small molecule ligands, greatly enhance our understanding of function by identifying catalytic sites, binding domains, and allosteric reorganizations important for regulation. When high resolution structures are not available, alternative and complementary approaches are needed to identify binding interactions. Alternative approaches are particularly important for large multi-component assemblies that may be heterogeneous or otherwise incompatible with atomic resolution analysis. Footprinting by means of hydrogen/deuterium exchange coupled to mass spectrometry provides detailed information about changes in structure and dynamics resulting directly from ligand binding and molecular rearrangements, even in disordered regions of proteins and in polymeric and multi-component systems, such as microtubules 32, actin filaments 33 and viral capsids 34. To our knowledge, the seven-subunit 220 kDa Arp2/3 complex is one of the highest complexity assemblies studied by HDX-MS.
In the present work, we combined measurements of HDX in whole subunits and numerous peptides to assess how ATP and WASp-VCA binding influence the structure and dynamics of Arp2/3 complex. Analysis of both global and local HDX was required to reveal offsetting changes in HDX that resulted in minimal net changes within subunits. We mapped exchange data onto a model of bovine Arp2/3 complex based on crystal structure (1TYQ) to begin to distinguish between direct effects of ligand binding and long-range allosteric effects.
ATP binding induces modest conformational changes in Arp2/3 complex
Multiple crystal structures show that ATP binding in the nucleotide cleft of Arp3 induces local changes in the cleft and a rigid body movement of subdomains 3 and 4 of Arp3 and ARPC3 relative to the rest of the complex that closes the cleft, but does not cause major rearrangements of the complex12. In solution, oxidative footprinting showed that ATP binding protects residues within the nucleotide binding cleft consistent with partial cleft closure 27. Oxidative footprinting also identified ATP-dependent protections in subdomains 3 and 4 of Arp2, but not subdomains 1 and 227. In the present work, HDX revealed that ATP binding results in protections in both Arp3 and Arp2 that are consistent with previous structural data, but these studies also provide new insights into ATP-dependent structural and dynamic alterations.
Global effects of ATP binding on the Arp2/3 complex
ATP binding to Arp3 and Arp2 causes changes in HDX that extend far beyond the nucleotide binding clefts of these subunits to include all five other subunits. ATP binding resulted in mass differences of 3% (13 Da) in Arp3, 5% (19 Da) in Arp2, 8% (27 Da) in ARPC1 and 7% (11Da) in ARPC4 (Figure 1), showing that ATP binding initiates modest conformational changes that propagate throughout Arp2/3 complex and influence its global dynamics. Crystal structures also revealed that ATP binding causes modest changes in conformations throughout the complex 11, whereas solution FRET experiments suggested that nucleotide binding induces large-scale conformational changes that bring ARPC1 and ARPC3 into closer proximity 10. Oxidative footprinting experiments did not detect these conformational adjustments outside the Arps, likely because they have a greater impact on the polypeptide backbone than on the surface accessibility of side chains in ARPC1 and ARPC4, which are the features exploited in oxidative footprinting.
Local effects of ATP binding to Arp3
Remarkably, ATP binding gave only marginal protection to the peptides containing the amino acid residues that form 9 hydrogen bonds with the triphosphate of ATP 11 in our HDX experiments (Figure 2a). These residues are Asp-11, Thr-14, Tyr-16 and Lys-18 in the P1 loop, Asp-172 and Gly173 in the P2 loop and Gly-324 in the P3 loop of the Arp 3 subunit. Main chain amide nitrogens of the glycine residues form these hydrogen bonds, so it is expected that ATP would protect these peptides. Perhaps compensating positive and negative HDX changes within the same peptide masked site-specific changes. Interestingly, ATP binding decreased deuterium incorporation into two peptides adjacent to the hydrogen bonding sites, peptide [328–334] comprising αK and loop αK-αK′ (p3 loop), situated directly under the adenine ring. ATP binding also protected peptide [184–189], which contains the N-terminal end of αE that is close to the edge of the adenine ring (Figure 3a) as well as Arp3 β11 and loop αD-β9, which are located in the interface between subdomains 1 and 3, possibly because this subunit is more rigid and/or assumes a more compact conformation upon ATP binding.
Effects of ATP binding on distal regions of Arp3
ATP binding protects many peptides in regions of Arp3 far from the nucleotide cleft, suggesting a significant change in structure or dynamics associated with ATP binding as expected from the cleft-closure observed in crystals 11. All of the distal Arp3 peptides protected by ATP binding are in areas that contact other subunits, including β13 and loop β12–β13 in subdomain 4 as well as the DNase binding loop between β4 and β5 [35–59]. ATP binding also influences HDX of parts of Arp3 that contact adjacent subunits, including loop αB-αC in subdomain 2 in the interface region between Arp3 and ARPC2 and the αK peptides and the loop [164–176] in the interface with ARPC3 (Figure 3a). These data are consistent with molecular dynamics simulations showing that surrounding subunits exert significant effects on the structure and dynamics of Arp3 35. In particular, these calculations suggest that the DNase binding loop of Arp3 plays a role in stabilizing the nucleotide-binding cleft and that nucleotide bound to Arp3 may stabilize by contacts between this loop and ARPC2.
Effects of ATP binding on Arp2
ATP binding protected regions of Arp2 distant from the nucleotide-binding cleft of Arp2, but not residues in the cleft. Protected regions were the modeled loop between αB-αC [91–108], β5 strand [130–141], loop αD-β6 [147–154] within subdomain 1, αK and loop αK-αL [333–352] in subdomain 3 (Figure 3a). All of these features were disordered in crystals of Arp2/3 complex 11. Due to incomplete coverage in the nucleotide-binding cleft, our data do not exclude the possibility that Arp2 may experience an ATP-induced cleft closure similar to Arp3, (Figures 3a and 4).
Effects of ATP binding on ARPC1
ATP binding to the Arps resulted in significant protection of ARPC1 at its interface with Arp2 (Figure 4). This interaction between Arp2 and ARPC1 induces protections in nine additional, β-sheets in ARPC1 (color-coded in blue, Figure 4), suggesting that this interaction increases the rigidity of this subunit.
Effects of ATP binding on ARPC2
ATP binding to Arps reduces deuterium exchange on peptide ARPC2 [16–32] in the interface with Arp3, suggesting a coupling between Arp3 and ARPC2. As a consequence of this interaction, ARPC2 may assume a more rigid conformation with reduced fluctuations, consistent with the large portion of the subunit, which shows protection, e.g. segments 90–165 and 200–209. Given that ARPC2 interacts with the side of actin filaments such small ATP-induced structural and dynamic changes contribute to stabilizing the active conformation of the complex.
Effects of ATP binding on ARPC3
Although ATP binding to Arps has only a negligible effect on global exchange of ARPC3, peptide [23–37] incorporates a higher level of deuterium (Figure 2a). HDX decreased in other peptides, resulting in little net change.
Effects of ATP binding on ARPC5
ATP binding to Arps reduced deuterium exchange in four of the seven helices of ARPC5 including peptides [56–80], [81–86], [87–105] and [107–122] (Figures 2a and 4, green/blue). This effect of ATP binding was greater on regions of ARPC5 more peripheral to the Arps than more proximal regions. Changes initiating in the Arps may spread to ARPC5 through its extended N-terminus which interacts with subdomain 4 of Arp2 and two helices that interface with ARPC4 and a loop in ARPC1 (Figure 4).
Effects of ATP binding on sites that interact with mother filaments
Analysis of phylogenetically conserved residues 22, experiments with a reconstituted dimer of ARPC2 and ARPC4 4 and a structural model based on reconstruction of branch junctions from EM tomograms 9 identified an extensive interface of Arp2/3 complex with the side of an actin filament. Subunits ARPC2 and ARPC4 form much of this interface, but parts of each of the other 5 subunits (including the pointed ends of both Arps) appear to interact with the side of the filament. Remarkably, ATP binding to the Arps influences HDX on many parts of this interface. These results of HDX show that ATP binding to Arps influences many features of the interface of Arp2/3 complex with the mother actin filament and provide clues about why bound ATP is required for actin filament branch formation.
WASp-VCA binding sites on Arp2/3 complex
The absence of high resolution structures limits our understanding of the binding sites for WASp-VCA on Arp2/3 complex. The stoichiometry is not even certain. The V-motif binds to an actin subunit 16. VCA can be chemically crosslinked to Arp3, Arp2 and ARPC1 36; 37, so it is assumed that the C- and A-motifs extend over long distances over the surface of Arp2/3 complex. NMR studies show that CA binds within 25 Å of ARPC3 23. A model based on SAXS data proposed that C-motif binds to the base of Arp2 and the A-motif bind near Arp3 and ARPC3 25. Our HDX data provide some new clues about the binding sites.
VCA binding influences HDX on all seven subunits
Although global HDX data indicate that the effects of WASp-VCA binding to the ATP-Arp2/3 complex are confined to Arp3, Arp2 and ARPC2, analysis of peptides shows that all 7 subunits respond to WASp-VCA binding with altered rates of deuterium incorporation in various regions (Figure 2b). The fact that WASp-VCA addition to the ATP-bound complex increases the protection in Arp3 and decreases the protection in ARPC2 indicates that subunits rearrange upon WASp-VCA binding (Figure 1). VCA binding tightens the Arp3 nucleotide binding cleft and loosens the structure of ARPC2 relative to the ATP-bound state. WASp-VCA binding increases the affinity of both Arp2 and Arp3 for ATP but does not alter the stoichiometry of bound nucleotides 19. Thus, the additional protections brought about by VCA binding are not the consequence of altered backbone solvent accessibility due to further nucleotide binding, but reflect changes the conformation or dynamics of each subunit.
Location of potential VCA binding sites on Arp3
WASp-VCA binding reduces HDX in many parts of Arp3, so this data alone does not pinpoint the VCA binding site. Some of the changes may explain the ability of VCA to enhance nucleotide binding 19; 20. VCA protects loop αK-αL (peptide Arp3 [366–378]), lying just above the nucleotide (Figure 3b) as well as neighboring peptides aD, loop αD-β9 (peptide Arp3 [145–162]) and β7 below the bound ATP, indicating altered solvent accessibility or structural change in this region. VCA binding reduces HDX around one site (A-2) on the surface of Arp3 that was predicted to bind the A-motif based on phylogenetic conservation, electrostatics and a hydrophobic pocket binding pocket for binding the tryptophan one residue removed from the C-terminus of VCA 22. The A-2 site on Arp3 involved A150, W153, R161, F379, M383, and L384. WASp-VCA binding significantly protected residues in the A-2 site on peptide [145–162]) in our HDX experiment. WASp-VCA binding did not protect F379, M383 or L384, but did protect the neighboring peptide [366–378] in the loop αK-αL. Protection of these two peptides was markedly stronger than other peptides in Arp3 (Figure 2b, part a). The other potential binding site on Arp3 (A-1) involves K228, M327, F328, R329, R333, R334, R337, K340, and R341 22. Our HDX detected all of these residues in sites A-1, except for K228, but VCA binding did not protect any of them. Thus A-2 is a strong candidate to bind the A motif of WASp-VCA. A model based on SAXS data proposed that the A-motif extends toward the interface between Arp3 and ARPC3 25, consistant with NMR and crosslinking studies 23 but on the opposite side from the A-2 site. VCA did not influence HDX in this region, but coverage was incomplete.
Location of potential VCA binding sites on Arp2
WASp-VCA binding influences HDX in many parts of Arp2, but the region that seems most likely to interact directly with VCA is the peptide [36–51] comprising β3-αA loop in subdomain 2 (Figure 3b), corresponding to the DNase binding loop of actin. The β3-αA loop and rest of subdomain 2 of Arp2 are disordered in all available crystal structures, so it is remarkable how strongly VCA reduces HDX. Phylogenetic conservation of residues in this loop and the location near the potential A-1 and A-2 sites on Arp3, suggested that this region might interact with the C-motif of VCA 22. Alternatively, VCA binding may also promote the interaction of the β3-αA loop with Arp3, consistent with VCA-dependent protection of the Arp2-Arp3 interface region. The groove between subdomains 1 and 3 of Arp2 is another potential binding site for the C-motif, because the residues are phylogentically conserved 22. VCA binding protects these residues and a model based SAXS illustrates how the VC motif is the right size to extend from the V binding site on an actin subunit to a potential C binding site at the base of Arp2 25. Most other peptides affected by VCA binding are buried in interfaces with other subunits.
Location of potential VCA binding sites on ARPC1
VCA can be chemically crosslinked to ARPC1 36; 37 and VCA protects several peptides from HDX (Figure 5). The large WASp-VCA-dependent protection in peptide [66–82] makes it a likely candidate for WASp binding while modest protections in peptides [273–283] and [358–372] and the decreased protection in [32–46] are more likely the result of conformational changes.
Influence of ATP and WASp-VCA binding on activation of Arp2/3 complex
A number of approaches have contributed to our understanding of how binding of nucleotide and activators influence the structure and activity of Arp2/3 complex. The head-to-tail arrangement of the two Arps in crystal structures of inactive Arp2/3 complex is inappropriate to initiate an actin filament branch 11; 12; 26, but the structure suggested how the two Arps might move closer together by about 30 Å to support nucleation 26. A FRET analysis 10 and electron micrographs of single Arp2/3 complexes 18 supported the concept that WASp-VCA binding favors rearrangement of the complex. A 3D reconstruction of electron tomograms of actin filament branches confirmed that Arp2 and Arp3 reorganize to form the first two subunits in the branch 9, but did not reveal the internal details of the large conformational change. Although binding of ATP and VCA are required for branch formation, this ternary complex is inactive until it binds to the side of an actin filament 15; 38; 39. Therefore ATP, VCA, actin monomers and actin filaments all cooperate to activate Arp2/3 complex. For example, actin filaments promote binding of WASP-VCA to Arp2/3 complex 15 and VCA promotes binding of ATP to the Arps 19.
As suggested above, the ATP-dependent movements in some parts of subunits ARPC2, ARPC5 and ARPC4 may have the effect of promoting a conformation suitable for mother actin filament binding. WASp-VCA binding may induce further structural alterations that are more compatible with the fully activated conformation associated with mother filament engagement. Thus, some regions of the subunits mentioned above may need to undergo additional conformational changes. As predicted, WASp-VCA binding enhanced the protection of peptides ARPC2 [153–165] and ARPC5 [87–105]. Together, these studies suggest that all subunits of the complex are involved in the activation of the complex to form branched daughter filaments.
Our HDX results show that the interactions of ATP and WASP-VCA with the Arp2/3 complex alter levels of HDX in all seven subunits, confirming the involvement of the entire complex in activation. The effects of ATP and WASp-VCA binding on subunits ARPC1, ARPC2, ARPC4 and ARPC5 (Figures 4 and 5) are consistent with coupling between these ligands and association of the ternary complex of Arp2/3 complex-VCA and an actin monomer with the side of an actin filament during branch formation. Consistent with cryoEM and biochemical data, our HDX data support a model in which ATP- and WASp-VCA binding to Arp2/3 complex promotes reorganization of Arp2 and Arp3 as well as all other subunits. Importantly, we have identified specific peptide segments that respond to ligand binding, and these data provide a potential mechanistic basis for the coupling between activator binding, F-actin binding and de novo nucleation. This work provides the foundation for future HDX experiments with the Arp2/3 complex in the presence of ATP, WASP, actin monomers, and actin filaments that should provide additional structural and dynamic detail relevant to the formation of actin filament branches.
MATERIALS AND METHODS
Arp2/3 complex and WASp-VCA
Nucleotide-free Arp2/3 complex was purified from bovine calf thymus 26. Actin polymerization assays40 verified that the activity of the complex in the deuterated buffer used for HDX is comparable to protonated buffer (data not shown). The VCA region of bovine N-WASp was prepared 41. Pepsin and adenosine 5′-triphosphate disodium salt (ATP) (99%) were purchased from Sigma-Aldrich. Deuterium oxide (99.9% deuterium) was obtained from Cambridge Isotope Laboratories. Poros 20AL media and trifluoroacetic acid (TFA) were purchased from Applied Biosystems. Acetonitrile was purchased from Fisher Scientific. All other reagents were of the highest purity available.
HDX/MS experiments
Prior to the HDX reaction, apo-Arp2/3 complex, ATP-Arp2/3 complex, and ATP/WASp-VCA-Arp2/3 complex were incubated in H-buffer (20 mM Tris, 50 mM NaCl, and 1 mM DTT) supplemented with 6.25 mM Mg2+-ATP or both 6.25 mM Mg2+-ATP and 0.16 mM WASp-VCA, at 25°C for 30 minutes. The concentration of Arp2/3 complex was 40 μM in all experiments. To ensure identical sample handling conditions, prior to HDX, apo-Arp2/3 complex was incubated in H-buffer alone. To prepare the deuterated buffer, H-buffer was subjected to several cycles of drying and re-dissolving in D2O. HDX was initiated by diluting the reaction mixtures 20-fold into deuterated aqueous buffer at 25°C. Given the equilibrium dissociation constant (Kd) of ~0.9 μM for WASp-VCA binding 15; 19; 37 these conditions ensure ~90% saturation. After 1, 5, 10, 30, and 80 minutes of reaction, 20 μL aliquots of samples were mixed with equal volumes of pre-chilled quenching buffer (0.5 M phosphate, pH 2.5). The quenched samples were subjected to immediate LC-ESI MS analysis for global HDX experiments, or were treated with an additional on-line pepsin digestion at 0°C and subsequent LC-ESI FT ICR MS analysis for local HDX experiments. All global HDX experiments were performed in triplicate. The maximum measured standard deviation was ±4.0 Da for Arp3, 3.2 Da for Arp2, 6.2 Da for ARPC1, 3.5 Da for ARPC2, 2.1 Da for ARPC3, 2.3 Da for ARPC4 and 2.1 Da for ARPC5. These standard deviations were small given the mass accuracy of LTQ mass spectrometer (±3 Da). The deviations in our triplicate results are smaller than the difference in deuterium incorporation in all subunits upon ATP or ATP/Wasp binding.
LC MS Analysis of individual Arp2/3 complex subunits
The extent of deuterium incorporation into the individual subunits of Arp2/3 complex was determined by LC-ESI MS. A Shimadzu HPLC, with two LC-10AD pumps, was used to generate a fast gradient with 50 μL/minute flow rate. Solvent A was 5% acetonitrile in H2O and 0.05% TFA while solvent B consisted of 95% acetonitrile in H2O and 0.05% TFA. To minimize back exchange, the solvent peek tubing, injector, and column were submerged in an ice bath. Five microliters of quenched complex was loaded onto a 1.0 × 50 mm C3 (MicroTech Scientific, Vista, CA) column. After desalting with 5% solvent B for 5 minutes, the individual subunits were eluted with a 6 minute gradient composed of 15% to 55% B. The effluent was directly delivered into a LTQ mass spectrometer (Thermo Electron Corporation) for mass analysis. The amount of deuterium in each subunit was determined from the mass difference between nondeuterated and deuterated samples without adjustment for deuterium gain or loss during LC-ESI MS analysis. The effect of deuterium gain or loss was not taken into account in this work because the HDX behavior of the three forms of Arp2/3 complex was examined under identical conditions.
LC-ESI-MS and LC-ESI-MS/MS analysis of proteolytic peptides
To identify proteolytic peptides, undeuterated Arp2/3 complex was quenched with phosphate buffer (pH 2.5) in an ice bath, as described above, containing 1 μL of 0.1M TCEP. A 20 μL aliquot was loaded onto an immobilized pepsin column prepared as described by Wang 42. The protein was digested on the column for about 87 seconds (digestion time is calculated as 87 seconds for the 2.1 × 50 mm column at 200 μL/min loading flow rate). The peptic peptides were captured on a C18 trap column (1 × 8 mm, Michrom Bioresources, Inc.) for about 2.5 minutes. This trap column also served as a desalting column. The desalted peptides were applied to a C18 column (1.0 × 50 mm, Vydac) for separation by a switching valve and eluted with a 5–15% gradient for 2 minutes and 15%–65% for 8 minutes at 50 μL/min and infused into a 7T Varian IonSpec FT-ICR-MS (Varian Inc.) or a LTQ (Thermo Electron Corporation) for ESI-MS or ESI-MS/MS, respectively. Peptides were identified by searching against the bovine Arp2/3 sequence (NP_776651, NP_001095683, NP_001014844, NP_001029885, NP_001029443, NP_001069631, NP_001030524) using a combination of accurate masses and MS/MS. The LTQ mass spectrometer was operated in a data dependent mode with dynamic exclusion containing 3 event scans (a full mass scan, a zoom scan, and an MS/MS scan). SEQUEST was used to search against the Arp2/3 and pepsin sequences to determine the peptic peptides. The extent of deuterium incorporation into proteolytic peptides was determined by FT-ICR MS from the centroid mass difference between deuterated and nondeuterated samples. FT-ICR MS was chosen to monitor subtle differences in deuterium level because it affords extremely high resolution and mass accuracy. The Varian 7T QFT-ICR routinely provides better than 5 ppm mass accuracy with external calibration. Based on a systematic error of about 5%, deuterium incorporation of 0.5 Dalton was considered significant.
Models of subdomains 1 and 2 in Arp2
Models of bovine Arp2 domains 1 and 2, which were disordered in the crystal structure of bovine Arp2/3 (PDB Code 1TYQ), were generated using the online MODELLER server, ModWeb (http://modbase.compbio.ucsf.edu/ModWeb20-html/modweb.html). The program identified the actin structure from Dictyostelium discoideum (PDB Code 1NLV) as having the best homology and coverage (49% sequence identity over amino acids 7 to 388 of Arp2). The Arp2 homology model was structurally aligned with the ordered part of Arp2 from the crystal structure (domains 3 and 4). Most disordered portions in the 1TYQ structure were retained in the homology model of Arp2 (i.e., amino acids K7 to I41, D54 to A142, and K351 to K388) except the loop containing R42 to K53 which was modeled from the structure 1ATN using the CCP4 43 program CHAINSAW 44. The peptide connections between segments derived from different sources (X-ray, MODELLER 45; 46, CHAINSAW) were regularized using the program COOT 47. Metal nucleotide positions were taken directly from the 1TYQ structure.
Acknowledgments
We thank Dr. Jeffrey Bonanno at the Albert Einstein College of Medicine for constructing the composite model of the Arp2/3 complex, including subunits 1 and 2 of Arp2. This work was supported by a Ruth Kirschstein postdoctoral fellowship GM074374-02 to B.J.N. and a NIH research grant GM066311 to T.D.P. and S.C.A. W.D.Z. was supported by a NIH Molecular Biophysics Training Grant GM008572.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Wendy D. Zencheck, Email: wzenchec@aecom.yu.edu.
Hui Xiao, Email: hxiao@aecom.yu.edu.
Bradley J. Nolen, Email: bnolen@uoregon.edu.
Ruth Angeletti, Email: angelett@aecom.yu.edu.
Thomas D. Pollard, Email: thomas.pollard@yale.edu.
Steven C. Almo, Email: almo@aecom.yu.edu.
WORKS CITED
- 1.Millard TH, Sharp SJ, Machesky LM. Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex. Biochem J. 2004;380:1–17. doi: 10.1042/BJ20040176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD, Earnshaw WC. Cell Biology. Saunders; Philadelphia: 2004. updated edition edit. [Google Scholar]
- 3.Kelleher JF, Atkinson SJ, Pollard TD. Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J Cell Biol. 1995;131:385–97. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol Cell. 2001;8:1041–52. doi: 10.1016/s1097-2765(01)00393-8. [DOI] [PubMed] [Google Scholar]
- 5.Machesky LM, Gould KL. The Arp2/3 complex: a multifunctional actin organizer. Curr Opin Cell Biol. 1999;11:117–21. doi: 10.1016/s0955-0674(99)80014-3. [DOI] [PubMed] [Google Scholar]
- 6.Winter DC, Choe EY, Li R. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc Natl Acad Sci USA. 1999;96:7288–93. doi: 10.1073/pnas.96.13.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr Biol. 1998;8:1347–56. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 8.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97:221–31. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 9.Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–95. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–79. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Nolen BJ, Littlefield RS, Pollard TD. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci USA. 2004;101:15627–32. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–57. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welch MD, Mullins RD. Cellular control of actin nucleation. Annu Rev Cell Dev Biol. 2002;18:247–88. doi: 10.1146/annurev.cellbio.18.040202.112133. [DOI] [PubMed] [Google Scholar]
- 14.Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003;10:591–8. doi: 10.1038/nsb952. [DOI] [PubMed] [Google Scholar]
- 15.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 16.Dayel MJ, Mullins RD. Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2004;2:E91. doi: 10.1371/journal.pbio.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin AC, Xu XP, Rouiller I, Kaksonen M, Sun Y, Belmont L, Volkmann N, Hanein D, Welch M, Drubin DG. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J Cell Biol. 2005;168:315–28. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, Grigorieff N, Goode BL. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- 19.Dayel MJ, Holleran EA, Mullins RD. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc Natl Acad Sci USA. 2001;98:14871–6. doi: 10.1073/pnas.261419298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Clainche C, Didry D, Carlier MF, Pantaloni D. Activation of Arp2/3 complex by Wiskott-Aldrich Syndrome protein is linked to enhanced binding of ATP to Arp2. J Biol Chem. 2001;276:46689–92. doi: 10.1074/jbc.C100476200. [DOI] [PubMed] [Google Scholar]
- 21.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–26. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 22.Beltzner CC, Pollard TD. Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. J Mol Biol. 2004;336:551–65. doi: 10.1016/j.jmb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 23.Kreishman-Deitrick M, Goley ED, Burdine L, Denison C, Egile C, Li R, Murali N, Kodadek TJ, Welch MD, Rosen MK. NMR analyses of the activation of the Arp2/3 complex by neuronal Wiskott-Aldrich syndrome protein. Biochemistry. 2005;44:15247–56. doi: 10.1021/bi051065n. [DOI] [PubMed] [Google Scholar]
- 24.Volkmann N, Amann KJ, Stoilova-McPhie S, Egile C, Winter DC, Hazelwood L, Heuser JE, Li R, Pollard TD, Hanein D. Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science. 2001;293:2456–9. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- 25.Boczkowska M, Rebowski G, Petoukhov MV, Hayes DB, Svergun DI, Dominguez R. X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–84. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 27.Kiselar JG, Mahaffy R, Pollard TD, Almo SC, Chance MR. Visualizing Arp2/3 complex activation mediated by binding of ATP and WASp using structural mass spectrometry. Proc Natl Acad Sci USA. 2007;104:1552–7. doi: 10.1073/pnas.0605380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katta V, Chait BT. Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun Mass Spectrom. 1991;5:214–7. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- 29.Maier CS, Deinzer ML. Protein conformations, interactions, and H/D exchange. Methods Enzymol. 2005;402:312–60. doi: 10.1016/S0076-6879(05)02010-0. [DOI] [PubMed] [Google Scholar]
- 30.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–9. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akashi S. Investigation of molecular interaction within biological macromolecular complexes by mass spectrometry. Med Res Rev. 2006;26:339–68. doi: 10.1002/med.20051. [DOI] [PubMed] [Google Scholar]
- 32.Xiao H, Verdier-Pinard P, Fernandez-Fuentes N, Burd B, Angeletti R, Fiser A, Horwitz SB, Orr GA. Insights into the mechanism of microtubule stabilization by Taxol. Proc Natl Acad Sci USA. 2006;103:10166–73. doi: 10.1073/pnas.0603704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chik JK, Schriemer DC. Hydrogen/deuterium exchange mass spectrometry of actin in various biochemical contexts. J Mol Biol. 2003;334:373–85. doi: 10.1016/j.jmb.2003.09.044. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Lane LC, Smith DL. Detecting structural changes in viral capsids by hydrogen exchange and mass spectrometry. Protein Sci. 2001;10:1234–43. doi: 10.1110/ps.100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfaendtner J, Voth GA. Molecular dynamics simulation and coarse-grained analysis of the Arp2/3 complex. Biophys J. 2008;95:5324–33. doi: 10.1529/biophysj.108.143313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zalevsky J, Grigorova I, Mullins RD. Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem. 2001;276:3468–75. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
- 37.Weaver AM, Heuser JE, Karginov AV, Lee WL, Parsons JT, Cooper JA. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr Biol. 2002;12:1270–8. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- 38.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci USA. 1999;96:3739–44. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2008;283:7135–44. doi: 10.1074/jbc.M705894200. [DOI] [PubMed] [Google Scholar]
- 40.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zalevsky J, Lempert L, Kranitz H, Mullins RD. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr Biol. 2001;11:1903–13. doi: 10.1016/s0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
- 42.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–8. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 43.Dodson EJ, Winn M, Ralph A. Collaborative Computational Project, number 4: providing programs for protein crystallography. Methods Enzymol. 1997;277:620–33. doi: 10.1016/s0076-6879(97)77034-4. [DOI] [PubMed] [Google Scholar]
- 44.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–59. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 46.Pieper U, Eswar N, Webb BM, Eramian D, Kelly L, Barkan DT, Carter H, Mankoo P, Karchin R, Marti-Renom MA, Davis FP, Sali A. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]