Abstract
The SCF (Skp1–cullin–F-box proteins), also known as CRL (cullin-based RING ligase), is the largest family of E3 ubiquitin ligases that mediate approximately 20% ubiquitinated protein substrates for 26S proteasome degradation. Through promoting timely degradation of many key regulatory proteins, SCF E3 ligase controls numerous cellular processes; its dysfunction contributes to a number of human diseases, including cancer. The RING component of SCF complex consists of 2 family members, RBX1 (RING box protein 1), also known as ROC1 (regulator of cullins), and RBX2/ROC2 (also known as SAG [sensitive to apoptosis gene]), both of which are essential for the catalytic activity of SCF. RBX1 and RBX2 are evolutionarily conserved from yeast to humans and play an essential role during mouse embryonic development. Moreover, RBX1 and RBX2 are both overexpressed in multiple human cancer tissues and required for the growth and survival of cancer cells. In this review, we will discuss the similarities and differences between 2 RING family members, their regulation of SCF E3 ligase activity, and their role in development, cancer cell survival, and skin carcinogenesis, along with a brief discussion of RBX-SCF E3 ligases as the cancer targets and a recently discovered small molecule inhibitor of SCF E3 ligases as a novel class of anticancer drugs.
Keywords: anticancer targets, protein degradation, neddylation, RING box proteins, SCF E3 ubiquitin ligases, ubiquitin-proteasome system
Introduction
Protein modification by ubiquitin or ubiquitin-like proteins (e.g., SUMO and NEDD8) has been widely appreciated as one of the major posttranslational regulatory mechanisms for modulating most aspects of cell physiology.1 The ubiquitin-proteasome system (UPS) targets many regulatory proteins for rapid proteolysis inside all living cells with 2 distinct steps: covalent attachment of multiple ubiquitin molecules to the protein substrates, a process called ubiquitination, followed by substrate degradation by 26S proteasome.2 Ubiquitin is conjugated to target proteins by a stepwise cascade of triplet enzymes: namely, ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3), ultimately transferring to a lysine residue on the substrate.3 Multiple runs of this cascade reaction lead to polyubiquitination. The substrates with a ubiquitin chain on K48 linkage are recognized by 26S proteasomes for targeted degradation, whereas proteins with a ubiquitin chain on K63 linkage can be activated to regulate the NF-κB signaling pathway,4,5 DNA replication and repair,6,7 and intracellular trafficking.8 Furthermore, proteins can be monoubiquitinated, which may change their activity and subcellular localization.9,10
The E3 ubiquitin ligases are considered to be the most important components of the UPS, as they confer a high degree of specificity and selectivity towards their target substrates in cells.11 Tetrameric SCF (Skp1–cullin/Rbx–F-box protein) E3 ubiquitin ligase, also known as CRL (cullin-based RING ligase), is the largest family of E3 ligases that consists of an adaptor protein, SKP1, a scaffold protein, cullin (with 7 family members), a substrate receptor protein, F-box protein (with approximately 70 members), and a catalytic RING component (with 2 members, RBX1 and RBX2).12,13 By promoting a variety of short-lived regulatory proteins for targeted degradation, SCF E3s play the essential role in many biological processes, including cell cycle progression, DNA replication, signal transduction, gene transcription, and development, among others.14,15 In this review, we will focus on 2 RING components of SCF complex, RBX1 and RBX2, and discuss their cloning, biochemical and biological characterization, as well as their regulation of SCF E3 ligase activity and cell growth and survival, with emphasis on their role in human cancer and as potential cancer targets.
RBX1 and RBX2: Gene Cloning and Characterization
Cloning and Gene Structure
RBX1 was initially cloned in 1999 by 4 independent groups via either biochemical purification or yeast 2-hybrid screening.16-19 This protein was designated as RBX1 (RING box protein) or ROC1 (regulator of cullins) and Hrt1 as a Cdc53- associated protein. For simplicity, we will refer to RBX1/ROC1/Hrt1 hereafter as RBX1. Human RBX1 gene consists of 5 exons and 4 introns and encodes 108 amino acids with a molecular weight of about 14 kDa.20 One characteristic feature of the human RBX1 structure is that it contains a RING-H2 finger domain (Cys42-X2-Cys45-X29-Cys75-X1-His77-X2-His80-X2-Cys83-X10-Cys94-X2-Asp97, where X can be any amino acid) located at its carboxyl terminus. Through the defined motif of cysteine and histidine residues, the RING finger domain binds 2 zinc atoms per molecule in a unique cross-brace arrangement: the first and third pairs of cysteine/histidine create the first binding site, whereas the second and fourth pairs of cysteine/histidine form the other21 (Fig. 1). The conserved RING finger domain is required for its ubiquitin ligase activity.22,23
Figure 1.
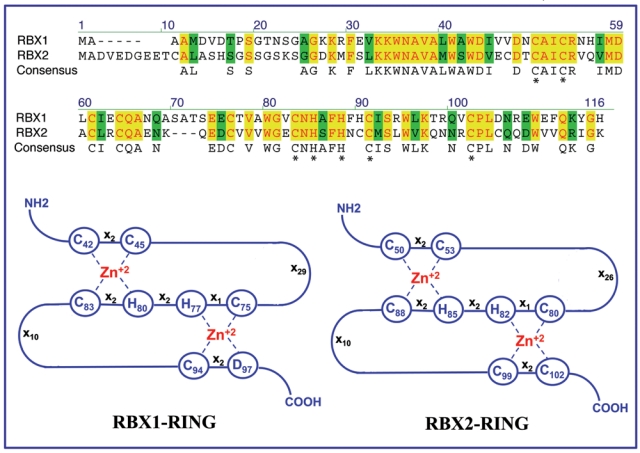
Comparison of amino acid compositions and RING structures of RBX1 and RBX2.
RBX2, also known as SAG (sensitive to apoptosis gene), was originally cloned in our laboratory as a redox-inducible antioxidant protein.24,25 The human SAG gene consists of 4 exons and 3 introns with exon 2 spliced out in the wild-type SAG open reading frame.26 Like RBX1 protein, human or mouse SAG encodes a zinc RING protein that consists of 113 amino acids with a molecular weight of about 14 kDa.26 SAG was subsequently characterized as the second member of the RBX family component of SCF ubiquitin ligase with a RING domain–dependent ligase activity when complexed with cullin-1.27 In addition to wild-type SAG, the SAG gene also encodes a splicing variant (SAG-v) with exon 2 retaining, leading to a frameshift and a truncated protein without the C-terminal RING domain, thus lacking cullin-1 binding and ligase activity.27 For simplicity, RBX2/ROC2/SAG will hereafter be referred to as RBX2.
Although RBX1 and RBX2 only share 53% of overall sequence identity, 7 of 8 cysteine/histidine residues that constitute the C3H2C3 RING finger domain are identical (Fig. 1). Both RBX1 and RBX2 are found to be evolutionarily conserved among many species from yeast to humans.21 Sequence conservation between human and mouse RBXs is very high with 100% identity in RBX1 and 96% identity in RBX2. More importantly, either human RBX1 or RBX2 can rescue the death phenotype derived from deletion of the RBX ortholog Hrt1 in yeast, which contains only one family member,27 indicating they are functionally equivalent at least in yeast.
Transcriptional Regulation and Potential Posttranslational Modification
Although the transcription of both RBX1 and RBX2 are subjected to induction upon mitogen stimulation,23 RBX1 is, in general, constitutively expressed, whereas RBX2 is stress inducible.21,28 The known RBX2 inducers include redox agents,24 nitric oxide,29 ischemia/reoxygenation,30,31 hypoxia,28 chemicals such as neurotoxins and 1-methyl-4-phenylpyridinium,32 heat shock,33 tumor promoter TPA,34 and UV irradiation.35 We recently elucidated the molecular basis for mouse Rbx2 induction by TPA through several AP-1 binding sites at the promoter of mouse Rbx2 gene34 and for human RBX2 induction by hypoxia through a HIF-1 consensus binding site at intron 1 of human RBX2 gene.28 The bioinformatic analysis, using the TRANSFAC database (http://www.gene-regulation.com/pub/databases.html),36 of a 2-kb promoter sequence of the human RBX2 gene revealed the consensus binding sites for many transcription factors, including NF-κB, RAR/RXR, c-MYC, AP-1, SP-1, and p53, among others. Further characterization of the potential regulation of RBX2 expression by these transcription factors would broaden our understanding of how RBX2 expression is regulated at the transcription levels in response to environmental stimuli. Although many transcription factor binding sites were also found in the promoter of human RBX1, little is known about how RBX1 is regulated at the transcriptional level. Still little known is the posttranslational regulation of both RBX1 and RBX2. Both proteins appear to be subject to rapid ubiquitination and degradation upon overexpression, but the binding to cullins confers their stabilization23 (unpublished observation).
Tissue Distribution and Cellular Localization
The tissue expression pattern of human or mouse RBX1 and RBX2 is similar. In humans, both proteins are ubiquitously expressed with much higher expression in the heart, skeletal muscle, and testes and less expression in the brain, lung, kidney, and placenta.24,26 In the mouse, both Rbx1 and Rbx2 are also ubiquitously expressed among different tissues with Rbx1 predominantly expressed in the genital tract (ovary, testis, prostate, and uterus)37 and Rbx2 mainly in the brain.21 Most recently, we found that both Rbx1 and Rbx2 are expressed during mouse embryogenesis and that knockout of either Rbx1 or Rbx2 caused embryonic lethality but at different stages of embryonic development20 (unpublished observation). Finally, both human RBX1 and RBX2 are localized in the cytoplasm and nucleus of cells,24,38 indicating their participation in ubiquitination and degradation of both cytoplasmic and nuclear proteins.
RBX1 and RBX2: The RING Components of SCF E3 Ubiquitin Ligases
The Catalytic Components of SCF E3 Ubiquitin Ligases
It is well known that a functional SCF E3 ligase is composed of 4 components: SKP1, a cullin, an F-box protein, and RBX1 or RBX2.12 Within the SCF complex, cullin is a scaffold component that serves as the assembly center, with its amino terminus binding to the adaptor subunit SKP1 and with an F-box receptor protein (e.g., β-TrCP, SKP2, FBXW7) and the carboxyl terminus binding with RBX1 or RBX2.39 The specificity of SCF E3 ligase is determined by the F-box protein that bridges to SCF components via its F-box domain and to its substrates via its WD40 or LRR domains.40 The ligase activity of SCF E3 is determined by the cullin-RBX complex, which catalyzes the transfer of ubiquitin from RBX-bound E2 to protein substrates.41 By complexing with distinct cullins, RBX family members constitute the catalytic cores of SCF complexes.15 Both RBX1 and RBX2 bind to 6 members of the human cullin family (Cul1-3, Cul4A-B, and Cul5) under overexpressed conditions17 and show in vitro E3 ubiquitin ligase activity when complexed with Cul1.27 Under physiological conditions, RBX1 preferentially interacts with Cul2, whereas RBX2 is selectively associated with Cul5,42 although the binding of RBX1-Cul543 and RBX2-Cul128 can also be detected under certain conditions. It is conceivable that 2 RBX family members, along with 7 cullins, approximately 70 F-box proteins, and many adaptor proteins, can assemble, like modules, into the various SCF complexes. The formation of distinct SCFs in a modular manner facilitates ubiquitination of a myriad of substrates, understandably regulating a vast array of cellular functions.15,44,45
The Regulators of SCF E3 Ubiquitin Ligase Activity
It is known that all cullins are subjected to modification at a specific lysine residue located at their C-terminus with the ubiquitin-like protein NEDD8 in a process called neddylation.46-48 Similar to ubiquitination, neddylation is catalyzed through an enzymatic cascade involving the sequential activity of E1, E2, and E3. The NEDD8 cascade is currently known to contain a single E1, NEDD8-activating enzyme (NAE), 2 E2s, UBE2M (also known as UBC12)49-51 and UBE2F,52 and a few candidate E3s. Two NEDD8 E2 enzymes have a certain degree of selectivity. UBE2M specifically regulates neddylation of RBX1/Cul1-4, whereas UBE2F regulates RBX2/Cul5 neddylation.52 Interestingly, RBX1 was reported in 2 studies, serving as a unique NEDD8 E3 ligase,53,54 whereas SCCRO (DCN1), an evolutionally conserved RBX1-binding protein,55 was found to interact with UBE2M and cullins and likely acts as a NEDD8 E3 ligase.56,57
Cullin neddylation activates SCF activity via several mechanisms. First, it prevents inhibitory binding of CAND1 (cullin-associated neddylation-dissociated 1).53,58-61 The crystal structure analysis reveals an N-terminus to C-terminus and a C-terminus to N-terminus binding of CAND1 and cullin-1.59 NEDD8 conjugation of cul-lins blocks the CAND1 binding and thereby facilitates the formation of functional SCFs.59,61,62 Second, neddylation not only enhances the recruitment of ubiquitin-loaded E2 enzymes to the ligase complex but also potently stimulates ubiquitin chain elongation on the substrates through neddylation-induced conformational change at the cullin-RBX interface that allows the ubiquitin-loaded E2 to move closer to the acceptor lysine residue of the substrate protein.58 Third, NEDD8 has been implicated in the formation of higher order cullin-RBX complexes by promoting dimerization, which was shown to be another mechanism for increasing the catalytic efficiency of some SCFs.63 Owing to the critical involvement of neddylation in the activation of SCFs, a potent small molecule inhibitor of NAE was recently discovered that inhibits SCF ligase activity via deneddylation and demonstrates potent anticancer activity64 (see below).
Substrates of RBX1/RBX2-SCF E3 Ubiquitin Ligases
A protein substrate ubiquitinated by a SCF E3 ubiquitin ligase is recognized by its F-box components. The human genome encodes about 70 F-box proteins40 with the majority of them uncharacterized.65 The 3 best-characterized F-box proteins are SKP2, β-TrCPs, and FBXW7 with detailed descriptions found in several excellent reviews.65-67 Interestingly, one F-box protein could recognize the substrates with opposite biological functions (e.g., SKP2 targets both p27 and cyclin E), whereas the same substrate can also be recognized by different F-box proteins with opposite functions (e.g., cyclin E can be targeted by both oncogenic SKP2 and tumor-suppressive FBXW7),14 indicating a precise regulation exists for degradation of a given substrate, which could be carried out by different F-box proteins in a cellular context and spatial-dependent manner. Recently, over 350 putative substrates of SCF complexes were identified by a global protein stability profiling analysis, and many of them are involved in cell cycle, apoptosis, and signaling pathways.68,69 Future studies are directed to validate these substrates and elucidate the biological relevance of their degradation, eventually leading to a better understanding of how precisely the cullin-RBX1/RBX2–based E3 ligases regulate cellular processes under physiological and pathological conditions.
RBX1 and RBX2: Essential for Development
Both RBX1 and RBX2 are evolutionarily conserved proteins from yeast to human.21 Earlier studies have shown that Hrt1, the only yeast homolog of RBX1/RBX2, is a growth-essential gene whose targeted disruption causes yeast death, which can be rescued by either human RBX117,18 or RBX2.27 In Caenorhabditis elegans, siRNA knockdown of Rbx1 (ZK287.5) induced embryonic death and abnormal meiosis,70 whereas knockdown of Rbx2 (R10A10.2) did not cause any significant phenotypic changes.71 In Drosophila, disruption of Roc1a or Roc1b caused lethality or male sterility, respectively, whereas disruption of Roc2 caused no obvious phenotype changes.72,73 These studies suggest that RBX2 homologs in C. elegans and Drosophila are functionally redundant and that RBX2 loss can be compensated by its family member, RBX1 or ROC1a/1b, during embryonic development. Our recent work showed that both Rbx1 and Rbx2 are expressed during mouse embryogenesis and that the deletion of either Rbx1 or Rbx2 causes embryonic lethality but at different stages of development20 (unpublished observation). Thus, both Rbx1 and Rbx2 play an essential role during mouse embryogenesis. Mechanistic study revealed that Rbx1 disruption induces a significant accumulation of p27, a cyclin-dependent kinase inhibitor, normally not expressed in the E6.5 to E7.5 embryo, thus resulting in reduced cell proliferation. Importantly, early embryonic death induced by Rbx1 disruption can be partially rescued by simultaneous deletion of p27, as evidenced by an extension of embryonic life from E6.5 to E9.5, indicating a causal role of p27 accumulation in early embryonic lethality. Thus, the in vivo physiological function of Rbx1 seems to ensure cell proliferation by targeting p27 for degradation during the early stage of embryonic development.20 We are in the process of defining potential mechanisms by which Rbx2 disruption causes embryonic death at later stages of development. The fact that embryonic lethality can be induced from the disruption of Rbx1 in a Rbx2 wild-type background and vice versa indicates that the in vivo functions of Rbx1 and Rbx2 are nonredundant and that Rbx1 and Rbx2 may target different sets of substrates during embryonic development.
RBX1 and RBX2 in Cancer and as Cancer Drug Targets
RBX1 and RBX2 in Human Cancer and Mouse Skin Carcinogenesis
A previous study has shown that RBX2 mRNA is overexpressed in lung cancer, which correlated with poor patient survival.74 We have shown that RBX2 protein was overexpressed in a subset of colon cancer tissues.75 Using immune staining, we recently found that RBX2 protein is overexpressed in a number of human cancer tissues, including carcinomas of the lung, colon, stomach, and liver,76 whereas RBX1 protein is also overexpressed in carcinomas of the lung, liver, and breast.38 It is unclear, however, whether overexpression of RBX1 or RBX2 is the cause or consequence of human carcinogenesis. To address the role of RBX2 in carcinogenesis, we recently generated a RBX2 transgenic model with targeted expression of human RBX2 in mouse skin epidermal cells driven by a K-14 promoter. Using a DMBA-TPA 2-stage skin carcinogenesis protocol, we found that RBX2 transgenic expression significantly inhibits carcinogenesis at an early stage via promoting degradation of c-Jun, thus inhibiting AP-1 activity, as evidenced by a prolonged latency period for tumor formation and reduced tumor numbers. However, at later stages of carcinogenesis, when tumors have formed, RBX2 transgenic expression increased tumor size, not because of accelerated proliferation, but because of reduced apoptosis, resulting from NFκB activation due to Rbx2-mediated IκB degradation.77 The stage-dependent targeting of c-Jun and IκB was facilitated by the expression of Fbxw7 (which targets c-Jun) but not β-TrCP (which targets IκB) at the early stage and vice versa at the later stage.77 We also tested the role of RBX2 in UVB-induced skin carcinogenesis using the same RBX2 transgenic model and found that RBX2 transgenic expression increased the rate of skin hyperplasia but not the rate of skin carcinogenesis. This appears to be mediated by simultaneous targeting for the degradation of c-Jun/AP-1 (leading to antiproliferation) and p27 (leading to proliferation), implying a more important role of p27 in the blockage of UVB skin hyperplasia.35
RBX1 and RBX2 in Regulation of Cell Survival and Apoptosis
One way to test the potential role of a given gene found overexpressed in human cancer cells is to determine if it is required for the maintenance of cancer cell growth and survival upon its silencing. We used this strategy in the testing of RBX1 and RBX2, both of which are overexpressed in human cancers, and found that siRNA silencing of either RBX1 or RBX2 in multiple cancer cell lines suppressed cancer cell growth in monolayer culture, inhibited their clonal survivals, and blocked their ability to grow in soft agar, a characteristic of the cancer cell phenotype. This is achieved by the induction of apoptosis and senescence for RBX1 and of apoptosis only for RBX2.38,76 Thus, either RBX1 or RBX2 is required for cancer cell proliferation and survival as well as for the maintenance of the cancer cell phenotype. It is likely that cancer cells are addicted to RBX1/RBX2 overexpression and that their knockdown by siRNA induces death, thus providing a sound rationale to target RBX1/RBX2 E3 ligase for anticancer therapy.78
A number of studies by our group and other laboratories have shown that ectopic expression of RBX2 protects tissues or cells from apoptosis induced by a variety of agents, including metal ions and redox compounds,24,79 nitric oxide,29 neurotoxin and 1-methyl-4-phenylpyridinium,32 heat shock,33 UV irradiation,35 and ischemia/hypoxia both in vitro30 and in vivo.31,80 RBX2 overexpression also promotes S-phase entry and cell growth under serum-starved conditions81 and inhibits TPA-induced neoplastic transformation by targeting c-Jun for degradation.34 Likewise, silencing RBX2 expression by antisense or siRNA inhibits tumor cell growth,75 enhances apoptosis induced by etoposide and TRAIL,82 enhances TPA-induced neoplastic transformation,34 and sensitizes cells to radiation.76,83 These cellular functions are mediated through RBX2’s antioxidant activity by scavenging ROS24,29 and through RBX2’s E3 ubiquitin ligase activity by promoting the degradation of p27, c-Jun, procaspase-3, IκBα, HIF-1α, and NOXA.28,34,76,77,81,82 Thus, stress-inducible RBX2 plays an important role in cell survival in response to various stressed conditions.
RBX1 but Not RBX2 Regulates Senescence in a p53/Rb-Independent Manner
Interestingly, our recent studies showed that siRNA silencing of RBX1 but not RBX2 induces senescence, although silencing of either RBX1 or RBX2 induces apoptosis.38,76 Cellular senescence is a tumor-suppressive mechanism to restrict tumor development by arresting the proliferation of potentially tumorigenic cells and killing cancer cells.84,85 Senescence is mainly mediated by 2 tumor suppressor pathways: p53 and pRB.84-86 Interestingly, senescence induced by RBX1 silencing is independent of p53 or pRB. It appears to be the consequence of DNA damage responses38 triggered by the accumulation of CDT1 and ORC1, 2 known substrates of RBX1-SCF E3 ligase87-90 that regulates DNA rereplication91-93 (unpublished observation). In addition to induction of senescence, siRNA silencing of RBX1, but not RBX2, also causes growth arrest at the G2/M phase, which is associated with the accumulation of 14-3-3σ and down-regulation of cyclin B1 and CDC2.38 In the case of apoptosis induction, accumulation of proapoptotic protein PUMA and reduction of antiapoptotic proteins Bcl-2, Mcl-1, and survivin are associated with RBX1 silencing,38 whereas accumulation of proapoptotic protein NOXA is associated with RBX2 silencing.76 Thus, even in the same cancer cell lines, targeting RBX1 or RBX2 could cause accumulation of different sets of substrates that trigger some secondary responses, eventually leading to both overlapping and unique biological consequences.
RBX1 and RBX2 as Cancer Drug Targets
RBX1 and RBX2 possess several ideal features as anticancer targets. First, these are “druggable” enzymes. Second, they are overexpressed in human cancers and are required for cancer cell survival as well as for the maintenance of cancer cell phenotype. Third, normal cells are less sensitive to their silencing, particularly in the case of RBX2,76 conferring potential therapeutic windows.78,94 However, development and optimization of a high throughput screening (HTS) for small molecule inhibitors of RBX1/RBX2-SCF E3 ligase face significant technical hindrances. First, the assay requires the production of multiple active proteins. For example, the screening for inhibitors of RBX-cullin autoubiquitination will require 5 proteins/enzymes, including ubiquitin, E1, E2, RBX1 or RBX2, and one of cullins, whereas for inhibitors of substrate ubiquitination, it requires SKP1, an F-box protein, and a substrate in addition to the above 5 proteins. Second, the follow-up counterscreening to sort out particular E3 inhibitors from those that inhibit E1 or E2 would be an equally daunting task. Thus, although several HTS assays have been developed to screen for E3 inhibitors,95 none of them can be readily adapted for RBX1/2-SCF E3 screening.
Although no specific small molecule inhibitor of SCF E3 ubiquitin ligases has been successfully discovered, the screening for small molecule inhibitors of NAE led to the identification of MLN4924,64 which binds to NAE to create a covalent NEDD8-MLN4924 adduct that blocks NAE enzymatic activity.96 Since all cullins, which are the major known substrates of neddylation,47 are subjected to neddylation for SCF E3 activation,53,58-60 MLN4924, by blocking cullin neddylation, inhibits SCF E3 ligases and causes accumulation of SCF E3 substrates, leading to the induction of apoptosis in a number of human cancer cell models64,97 as well as induction of senescence in a prostate cancer model.98 Furthermore, MLN4924 was well tolerated at various doses and treatment regimens in mice,64 demonstrating a selective killing of cancer cells. Encouragingly, MLN4924 has advanced to several phase I clinical trials for solid tumors and hematological malignancies.99
Conclusion and Perspectives
RBX1 and RBX2 are 2 small RING family proteins that control the activity of entire SCF E3 ubiuqitin ligases, the largest family of E3 ubiquitin ligases that mediate approximately 20% ubiquitinated protein substrates for 26S proteasome degradation.64 The regulation and functions of RBX1 and RBX2 can be summarized in Figure 2. Upon induction by mitogens or other stresses (in the case of RBX2), RBX1 and RBX2 form SCFs with their respective cullins and catalyze cullin neddylation to fully activate SCFs. Continued overexpression of RBX1/2 and activation of SCFs render cancer cells addicted to such growth-promoting environments and become vulnerable to pharmaceutical inhibition of SCF ligase activity (e.g., MLN4924) or to siRNA silencing of RBX1 or RBX2. Thus, RBX1/2-SCF E3s provide a promising anticancer target for selective killing of cancer cells with activated SCFs. Future work should be directed to differentiate overlapping as well as unique functions of each RBX family member by dissecting unique signaling pathways controlled by their respective substrates, which are likely being ubiquitinated and degraded by different combinations of RBX-cullin E3s in a cellular context and spatial-dependent manner. A comprehensive dissection of signaling pathways controlled by individual RBX family members would facilitate selectively targeting one family member for gained specificity and reduced normal tissue toxicity. Given the fact that RBX1 is constitutively expressed in normal tissues and cells, selective targeting stress-inducible RBX2 may render a higher therapeutic window due to less normal cell toxicity.
Figure 2.
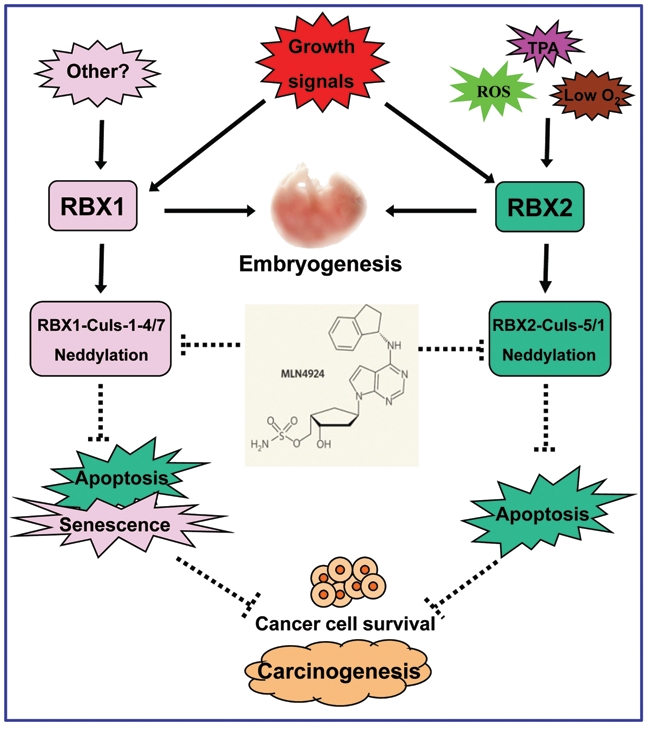
Regulation and biological functions. RBX1 and RBX2 are induced upon growth signals, whereas RBX2 is also induced by various stresses, including ROS, TPA, and hypoxia. While both are required for mouse embryonic development, RBX1 or RBX2 forms SCF E3 ubiquitin ligases by complexing with their respective cullins. Following cullin neddylation, RBX1-SCFs and RBX2-SCFs are activated to promote the growth and survival of cancer cells. MLN4924, a small molecule inhibitor of NAE, blocks cullin neddylation to inactivate SCFs, leading to the induction of apoptosis and senescence, thus acting as a novel class of anticancer drugs.
Acknowledgments
The authors thank Dr. Lijun Jia for stimulating discussion in review outlines.
Footnotes
This work is supported by National Cancer Institute (NCI) grants (CA111554 and CA118762) to Yi Sun.
The author(s) declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1. Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159-80 [DOI] [PubMed] [Google Scholar]
- 2. Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596-613 [DOI] [PubMed] [Google Scholar]
- 3. Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425-79 [DOI] [PubMed] [Google Scholar]
- 4. Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275-86 [DOI] [PubMed] [Google Scholar]
- 5. Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769-96 [DOI] [PubMed] [Google Scholar]
- 6. Bennett EJ, Harper JW. DNA damage: ubiquitin marks the spot. Nat Struct Mol Biol. 2008;15:20-2 [DOI] [PubMed] [Google Scholar]
- 7. Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479-89 [DOI] [PubMed] [Google Scholar]
- 8. Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610-8 [DOI] [PubMed] [Google Scholar]
- 9. Hicke L, Schubert HL, Hill CP. Ubiquitin- binding domains. Nat Rev Mol Cell Biol. 2005;6:610-21 [DOI] [PubMed] [Google Scholar]
- 10. Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8:499-504 [DOI] [PubMed] [Google Scholar]
- 11. Nagy V, Dikic I. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. Biol Chem. 2010;391:163-9 [DOI] [PubMed] [Google Scholar]
- 12. Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399-434 [DOI] [PubMed] [Google Scholar]
- 13. Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133-70 [DOI] [PubMed] [Google Scholar]
- 14. Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369-81 [DOI] [PubMed] [Google Scholar]
- 15. Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9-20 [DOI] [PubMed] [Google Scholar]
- 16. Kamura T, Koepp DM, Conrad MN, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657-61 [DOI] [PubMed] [Google Scholar]
- 17. Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3:535-41 [DOI] [PubMed] [Google Scholar]
- 18. Seol JH, Feldman RMR, Zachariae WZ, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tan P, Fuchs SY, Chen A, et al. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IkBa. Mol Cell. 1999;3:527-33 [DOI] [PubMed] [Google Scholar]
- 20. Tan M, Davis SW, Saunders TL, Zhu Y, Sun Y. RBX1/ROC1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of p27. Proc Natl Acad Sci U S A. 2009;106:6203-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3:635-50 [DOI] [PubMed] [Google Scholar]
- 22. Chen A, Wu K, Fuchs SY, Tan P, Gomez C, Pan ZQ. The conserved RING-H2 finger of ROC1 is required for ubiquitin ligation. J Biol Chem. 2000;275:15432-9 [DOI] [PubMed] [Google Scholar]
- 23. Ohta T, Michel JJ, Xiong Y. Association with cullin partners protects ROC proteins from proteasome-dependent degradation. Oncogene. 1999;18:6758-66 [DOI] [PubMed] [Google Scholar]
- 24. Duan H, Wang Y, Aviram M, et al. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Swaroop M, Bian J, Aviram M, et al. Expression, purification, and biochemical characterization of SAG, a RING finger redox sensitive protein. Free Radicals Biol Med. 1999;27:193-202 [DOI] [PubMed] [Google Scholar]
- 26. Swaroop M, Gosink M, Sun Y. SAG/ROC2/Rbx2/Hrt2, a component of SCF E3 ubiquitin ligase: genomic structure, a splicing variant, and two family pseudogenes. DNA Cell Biol. 2001;20:425-34 [DOI] [PubMed] [Google Scholar]
- 27. Swaroop M, Wang Y, Miller P, et al. Yeast homolog of human SAG/ROC2/Rbx2/Hrt2 is essential for cell growth, but not for germination: chip profiling implicates its role in cell cycle regulation. Oncogene. 2000;19:2855-66 [DOI] [PubMed] [Google Scholar]
- 28. Tan M, Gu Q, He H, Pamarthy D, Semenza GL, Sun Y. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1alpha ubiquitination and degradation. Oncogene. 2008;27:1404-11 [DOI] [PubMed] [Google Scholar]
- 29. Yang ES, Park JW. Regulation of nitric oxide-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Res. 2006;40:279-84 [DOI] [PubMed] [Google Scholar]
- 30. Chanalaris A, Sun Y, Latchman DS, Stephanou A. SAG attenuates apoptotic cell death caused by simulated ischaemia/reoxygenation in rat cardiomyocytes. J Mol Cell Cardiol. 2003;35:257-64 [DOI] [PubMed] [Google Scholar]
- 31. Yang GY, Pang L, Ge HL, et al. Attenuation of ischemia-induced mouse brain injury by SAG, a redox-inducible antioxidant protein. J Cereb Blood Flow Metab. 2001;21:722-33 [DOI] [PubMed] [Google Scholar]
- 32. Kim SY, Kim MY, Mo JS, Park JW, Park HS. SAG protects human neuroblastoma SH-SY5Y cells against 1-methyl-4-phenylpyridinium ion (MPP+)-induced cytotoxicity via the downregulation of ROS generation and JNK signaling. Neurosci Lett. 2007;413:132-6 [DOI] [PubMed] [Google Scholar]
- 33. Lee SJ, Yang ES, Kim SY, Shin SW, Park JW. Regulation of heat shock-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Biol Med. 2008;45:167-76 [DOI] [PubMed] [Google Scholar]
- 34. Gu Q, Tan M, Sun Y. SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 2007;67:3616-25 [DOI] [PubMed] [Google Scholar]
- 35. He H, Gu Q, Zheng M, Normolle D, Sun Y. SAG/ROC2/RBX2 E3 ligase promotes UVB-induced skin hyperplasia, but not skin tumors, by simultaneously targeting c-Jun/AP-1 and p27. Carcinogenesis. 2008;29:858-65 [DOI] [PubMed] [Google Scholar]
- 36. Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perin JP, Seddiqi N, Charbonnier F, et al. Genomic organization and expression of the ubiquitin-proteasome complex-associated protein Rbx1/ROC1/Hrt1. Cell Mol Biol. 1999;45:1131-7 [PubMed] [Google Scholar]
- 38. Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69:4974-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng N, Schulman BA, Song L, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703-9 [DOI] [PubMed] [Google Scholar]
- 40. Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu K, Fuchs SY, Chen A, et al. The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol Cell Biol. 2000;20:1382-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kamura T, Maenaka K, Kotoshiba S, et al. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kamura T, Burian D, Yan Q, et al. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J Biol Chem. 2001;276:29748-53 [DOI] [PubMed] [Google Scholar]
- 44. Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739-51 [DOI] [PubMed] [Google Scholar]
- 45. Jia L, Sun Y. RBX1/ROC1-SCF E3 ubiquitin ligase is required for mouse embryogenesis and cancer cell survival. Cell Div. 2009;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985-97 [DOI] [PubMed] [Google Scholar]
- 47. Rabut G, Peter M. Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:969-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36:802-6 [DOI] [PubMed] [Google Scholar]
- 49. Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036-42 [DOI] [PubMed] [Google Scholar]
- 50. Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17:2208-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Osaka F, Kawasaki H, Aida N, et al. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang DT, Ayrault O, Hunt HW, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301:392-8 [DOI] [PubMed] [Google Scholar]
- 55. Yang X, Zhou J, Sun L, et al. Structural basis for the function of DCN-1 in protein neddylation. J Biol Chem. 2007;282:24490-4 [DOI] [PubMed] [Google Scholar]
- 56. Kim AY, Bommelje CC, Lee BE, et al. SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J Biol Chem. 2008;283:33211-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kurz T, Chou YC, Willems AR, et al. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23-35 [DOI] [PubMed] [Google Scholar]
- 58. Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995-1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Goldenberg SJ, Cascio TC, Shumway SD, et al. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517-28 [DOI] [PubMed] [Google Scholar]
- 60. Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci U S A. 2008;105:12230-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zheng J, Yang X, Harrell JM, et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10:1519-26 [DOI] [PubMed] [Google Scholar]
- 62. Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511-8 [DOI] [PubMed] [Google Scholar]
- 63. Merlet J, Burger J, Gomes JE, Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci. 2009;66:1924-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732-6 [DOI] [PubMed] [Google Scholar]
- 65. Skaar JR, D’Angiolella V, Pagan JK, Pagano M. SnapShot: F box proteins II. Cell. 2009;137:1358, e1351 [DOI] [PubMed] [Google Scholar]
- 66. Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83-93 [DOI] [PubMed] [Google Scholar]
- 67. Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923-9 [DOI] [PubMed] [Google Scholar]
- 69. Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918-23 [DOI] [PubMed] [Google Scholar]
- 70. Sasagawa Y, Urano T, Kohara Y, Takahashi H, Higashitani A. Caenorhabditis elegans RBX1 is essential for meiosis, mitotic chromosomal condensation and segregation, and cytokinesis. Genes Cells. 2003;8:857-72 [DOI] [PubMed] [Google Scholar]
- 71. Moore R, Boyd L. Analysis of RING finger genes required for embryogenesis in C. elegans. Genesis. 2004;38:1-12 [DOI] [PubMed] [Google Scholar]
- 72. Donaldson TD, Noureddine MA, Reynolds PJ, Bradford W, Duronio RJ. Targeted disruption of Drosophila Roc1b reveals functional differences in the Roc subunit of Cullin-dependent E3 ubiquitin ligases. Mol Biol Cell. 2004;15:4892-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Reynolds PJ, Simms JR, Duronio RJ. Identifying determinants of cullin binding specificity among the three functionally different Drosophila melanogaster Roc proteins via domain swapping. PLoS One. 2008;3:e2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sasaki H, Yukiue H, Kobayashi Y, et al. Expression of the sensitive to apoptosis gene, SAG, as a prognostic marker in nonsmall cell lung cancer. Int J Cancer. 2001;95:375-7 [DOI] [PubMed] [Google Scholar]
- 75. Huang Y, Duan H, Sun Y. Elevated expression of SAG/ROC2/Rbx2/Hrt2 in human colon carcinomas: SAG does not induce neoplastic transformation, but its antisense transfection inhibits tumor cell growth. Mol Carcinog. 2001;30:62-70 [PubMed] [Google Scholar]
- 76. Jia L, Yang J, Hao X, et al. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gu Q, Bowden TG, Normolle D, Sun Y. SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage dependent targeting of c-Jun/AP1 and IkB/NF-kB. J Cell Biol. 2007;178:1009-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sun Y. Alteration of SAG mRNA in human cancer cell lines: requirement for the RING finger domain for apoptosis protection. Carcinogenesis. 1999;20:1899-903 [DOI] [PubMed] [Google Scholar]
- 80. Kim DW, Lee SH, Jeong MS, et al. Transduced Tat-SAG fusion protein protects against oxidative stress and brain ischemic insult. Free Radic Biol Med. 2010;48:969-77 [DOI] [PubMed] [Google Scholar]
- 81. Duan H, Tsvetkov LM, Liu Y, et al. Promotion of S-phase entry and cell growth under serum starvation by SAG/ROC2/Rbx2/Hrt2, an E3 ubiquitin ligase component: association with inhibition of p27 accumulation. Mol Carcinog. 2001;30:37-46 [DOI] [PubMed] [Google Scholar]
- 82. Tan M, Gallegos JR, Gu Q, et al. SAG/ROC-SCFbeta-TrCP E3 ubiquitin ligase promotes pro-caspase-3 degradation as a mechanism of apoptosis protection. Neoplasia. 2006;8:1042-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Tan M, Zhu Y, Kovacev J, et al. Disruption of SAG/RBX2/ROC2 induces radiosensitization by increasing ROS levels and blocking NF-kB activation in mouse embryonic stem cells. Free Radic Biol Med. 2010;49:976-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729-40 [DOI] [PubMed] [Google Scholar]
- 85. Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat Rev Cancer. 2008;8:450-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Itahana K, Campisi J, Dimri GP. Mechanisms of cellular senescence in human and mouse cells. Biogerontology. 2004;5:1-10 [DOI] [PubMed] [Google Scholar]
- 87. Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem. 2003;278:30854-8 [DOI] [PubMed] [Google Scholar]
- 88. Mendez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol Cell. 2002;9:481-91 [DOI] [PubMed] [Google Scholar]
- 89. Hu J, Xiong Y. An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J Biol Chem. 2006;281:3753-6 [DOI] [PubMed] [Google Scholar]
- 90. Nishitani H, Sugimoto N, Roukos V, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659-72 [DOI] [PubMed] [Google Scholar]
- 92. Speck C, Chen Z, Li H, Stillman B. ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA. Nat Struct Mol Biol. 2005;12:965-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liontos M, Koutsami M, Sideridou M, et al. Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res. 2007;67:10899-909 [DOI] [PubMed] [Google Scholar]
- 94. Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol Therapy. 2003;2:623-9 [PubMed] [Google Scholar]
- 95. Sun Y. Overview of approaches for screening for ubiquitin ligase inhibitors. Methods Enzymol. 2005;399:654-63 [DOI] [PubMed] [Google Scholar]
- 96. Brownell JE, Sintchak MD, Gavin JM, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102-11 [DOI] [PubMed] [Google Scholar]
- 97. Swords RT, Kelly KR, Smith PG, et al. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796-800 [DOI] [PubMed] [Google Scholar]
- 98. Lin HK, Chen Z, Wang G, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Soucy TA, Smith PG, Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin Cancer Res. 2009;15:3912-6 [DOI] [PubMed] [Google Scholar]


