Abstract
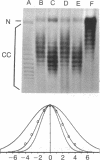
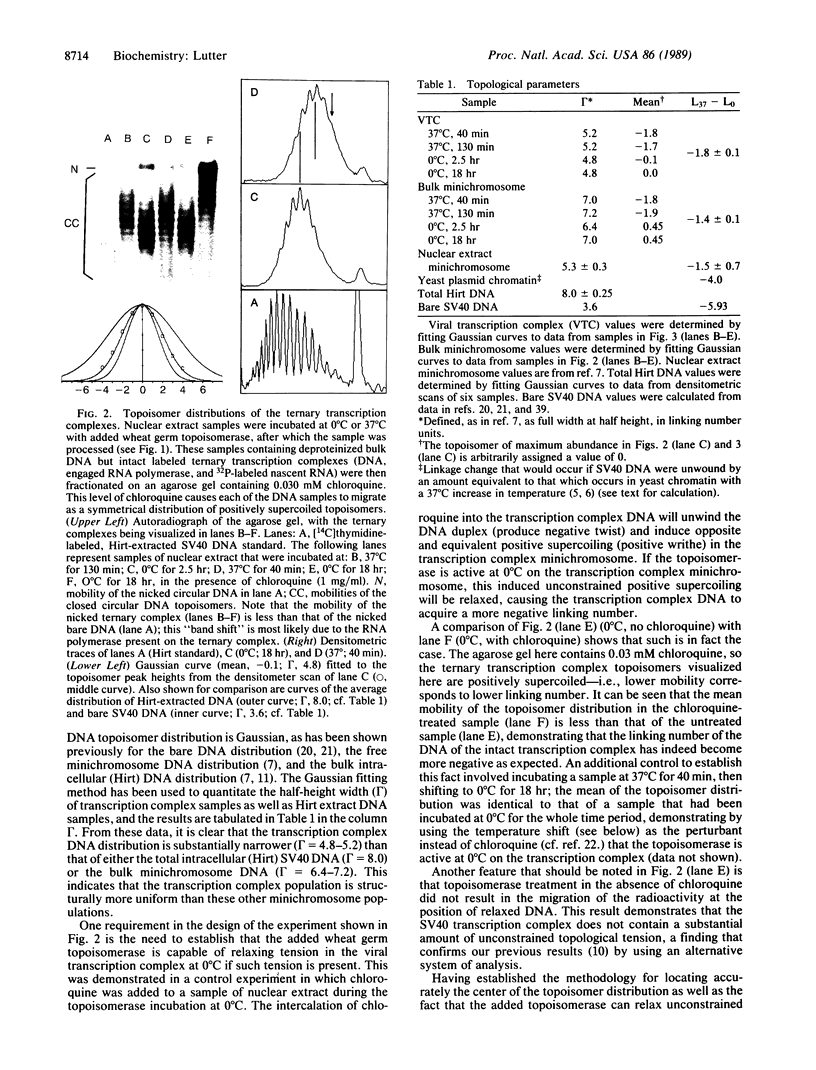
Two long-standing questions in the control of eukaryotic gene expression have been how the structure of transcribing chromatin compares with that of nontranscribing chromatin and how chromatin structure differs among various eukaryotic organisms. We have addressed aspects of these two questions by characterizing the rotational flexibility of the DNA of the simian virus 40 (SV40) transcription complex. When transcription complex samples are incubated with topoisomerase at 0 degrees C or 37 degrees C, the DNA of the 37 degrees C sample is unwound by 1.8 turns relative to that of the 0 degrees C sample. This amount of unwinding is similar to that observed for bulk, untranscribed SV40 minichromosome DNA, indicating that the chromatin structure of a transcribed gene resembles that of a nontranscribed gene in the degree of constraint that it imposes on its DNA. However, this amount of unwinding differs substantially from the value observed for yeast plasmid chromatin DNA, suggesting that yeast chromatin differs significantly from mammalian chromatin in this fundamental property.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrose C., McLaughlin R., Bina M. The flexibility and topology of simian virus 40 DNA in minichromosomes. Nucleic Acids Res. 1987 May 11;15(9):3703–3721. doi: 10.1093/nar/15.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certa U., Colavito-Shepanski M., Grunstein M. Yeast may not contain histone H1: the only known 'histone H1-like' protein in Saccharomyces cerevisiae is a mitochondrial protein. Nucleic Acids Res. 1984 Nov 12;12(21):7975–7985. doi: 10.1093/nar/12.21.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. S., Hsu M. T. Evidence for variation of supercoil densities among simian virus 40 nucleoprotein complexes and for higher supercoil density in replicating complexes. J Virol. 1984 Jul;51(1):14–19. doi: 10.1128/jvi.51.1.14-19.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choder M., Aloni Y. In vitro transcribed SV40 minichromosomes, as the bulk minichromosomes, have a low level of unconstrained negative supercoils. Nucleic Acids Res. 1988 Feb 11;16(3):895–905. doi: 10.1093/nar/16.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choder M., Aloni Y. RNA polymerase II allows unwinding and rewinding of the DNA and thus maintains a constant length of the transcription bubble. J Biol Chem. 1988 Sep 15;263(26):12994–13002. [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito F., Sinden R. R. Supercoiling in prokaryotic and eukaryotic DNA: changes in response to topological perturbation of plasmids in E. coli and SV40 in vitro, in nuclei and in CV-1 cells. Nucleic Acids Res. 1987 Jul 10;15(13):5105–5124. doi: 10.1093/nar/15.13.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Gariglio P., Mousset S. Isolation and partial characterization of a nuclear RNA polymerase - SV40 DNA complex. FEBS Lett. 1975 Aug 1;56(1):149–155. doi: 10.1016/0014-5793(75)80130-x. [DOI] [PubMed] [Google Scholar]
- Hadlock K. G., Quasney M. W., Lutter L. C. Immunoprecipitation of the simian virus 40 late transcription complex with antibody against T-antigen. J Biol Chem. 1987 Nov 15;262(32):15527–15537. [PubMed] [Google Scholar]
- Keller W., Müller U., Eicken I., Wendel I., Zentgraf H. Biochemical and ultrastructural analysis of SV40 chromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):227–244. doi: 10.1101/sqb.1978.042.01.025. [DOI] [PubMed] [Google Scholar]
- Lee K. P., Baxter H. J., Guillemette J. G., Lawford H. G., Lewis P. N. Structural studies on yeast nucleosomes. Can J Biochem. 1982 Mar;60(3):379–388. doi: 10.1139/o82-045. [DOI] [PubMed] [Google Scholar]
- Llopis R., Perrin F., Bellard F., Gariglio P. Quantitation of transcribing native simian virus 40 minichromosomes extracted from CV1 cells late in infection. J Virol. 1981 Apr;38(1):82–90. doi: 10.1128/jvi.38.1.82-90.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopis R., Stark G. R. Two deletions within genes for simian virus 40 structural proteins VP2 and VP3 lead to formation of abnormal transcriptional complexes. J Virol. 1981 Apr;38(1):91–103. doi: 10.1128/jvi.38.1.91-103.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Hereford L. Yeast chromatin is uniformly digested by DNase-I. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4285–4288. doi: 10.1073/pnas.76.9.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardian J. K., Isenberg I. Yeast inner histones and the evolutionary conservation of histone-histone interactions. Biochemistry. 1978 Sep 5;17(18):3825–3833. doi: 10.1021/bi00611a023. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- McMurray C. T., van Holde K. E. Binding of ethidium bromide causes dissociation of the nucleosome core particle. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8472–8476. doi: 10.1073/pnas.83.22.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Cantor C. R. Effect of trypsinization and histone H5 addition on DNA twist and topology in reconstituted minichromosomes. Nucleic Acids Res. 1986 Apr 25;14(8):3293–3310. doi: 10.1093/nar/14.8.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Cantor C. R. Nucleosome core particles suppress the thermal untwisting of core DNA and adjacent linker DNA. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4653–4657. doi: 10.1073/pnas.82.14.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Pederson D. S., Dean A., Simpson R. T. Yeast nucleosomes allow thermal untwisting of DNA. Nucleic Acids Res. 1987 Dec 23;15(24):10311–10330. doi: 10.1093/nar/15.24.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. A. Histone acetylation in baker's yeast. Maintenance of the hyperacetylated configuration in log phase protoplasts. J Biol Chem. 1982 Feb 25;257(4):1565–1568. [PubMed] [Google Scholar]
- Petryniak B., Lutter L. C. Topological characterization of the simian virus 40 transcription complex. Cell. 1987 Jan 30;48(2):289–295. doi: 10.1016/0092-8674(87)90432-6. [DOI] [PubMed] [Google Scholar]
- Proffitt J. H., Davie J. R., Swinton D., Hattman S. 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol Cell Biol. 1984 May;4(5):985–988. doi: 10.1128/mcb.4.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Saavedra R. A., Huberman J. A. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell. 1986 Apr 11;45(1):65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- Saavedra R. A., Huberman J. A. DNA topoisomerases in eukaryotes. Nature. 1985 Sep 5;317(6032):22–22. doi: 10.1038/317022c0. [DOI] [PubMed] [Google Scholar]
- Shure M., Pulleyblank D. E., Vinograd J. The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res. 1977;4(5):1183–1205. doi: 10.1093/nar/4.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. M., Andrésson O. S. DNA sequences of yeast H3 and H4 histone genes from two non-allelic gene sets encode identical H3 and H4 proteins. J Mol Biol. 1983 Sep 25;169(3):663–690. doi: 10.1016/s0022-2836(83)80164-8. [DOI] [PubMed] [Google Scholar]
- Szent-Gyorgyi C., Isenberg I. The organization of oligonucleosomes in yeast. Nucleic Acids Res. 1983 Jun 11;11(11):3717–3736. doi: 10.1093/nar/11.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weiss E., Regnier E., Oudet P. Restriction enzyme accessibility and RNA polymerase localization on transcriptionally active SV40 minichromosomes isolated late in infection. Virology. 1987 Jul;159(1):84–93. doi: 10.1016/0042-6822(87)90350-3. [DOI] [PubMed] [Google Scholar]
- Yaniv M., Cereghini S. Structure of transcriptionally active chromatin. CRC Crit Rev Biochem. 1986;21(1):1–26. doi: 10.3109/10409238609113607. [DOI] [PubMed] [Google Scholar]