Abstract
SBASE 8.0 is the eighth release of the SBASE library of protein domain sequences that contains 294 898 annotated structural, functional, ligand-binding and topogenic segments of proteins, cross-referenced to most major sequence databases and sequence pattern collections. The entries are clustered into over 2005 statistically validated domain groups (SBASE-A) and 595 non-validated groups (SBASE-B), provided with several WWW-based search and browsing facilities for online use. A domain-search facility was developed, based on non-parametric pattern recognition methods, including artificial neural networks. SBASE 8.0 is freely available by anonymous ‘ftp’ file transfer from ftp.icgeb.trieste.it. Automated searching of SBASE can be carried out with the WWW servers http://www.icgeb.trieste.it/sbase/ and http://sbase.abc.hu/sbase/.
INTRODUCTION
SBASE is a collection of protein domain sequences designed to facilitate detection of domain homologies based on simple database search (1,2). The central concept of the database is the ‘similarity group’, i.e. a group of domain sequences that have significant BLAST similarities to each other. Validated domain groups i.e. the 2005 groups that satisfy these criteria are deposited in SBASE-A; these are the well-known structural and functional domain types. SBASE-B contains 595 groups that are either (i) less well characterized than the groups of SBASE-A, or (ii) are defined by composition (e.g. glycine-rich), cellular location (e.g. transmembrane, etc.). These groups are sometimes defined in an overlapping manner, e.g. an extracellular domain (SBASE-B) may contain an EGF-module (SBASE-A).
The current release 8.0 of SBASE contains annotated protein sequence segments consistently named by structure, function, biased composition, binding-specificity and/or similarity to other proteins.
The main developments with respect to the previous release (release 7.0) can be summarized as follows:
(i) Release 8.0 contains 294 898 sequence entries, 24% more than release 7.0, divided into 2600 groups (Table 1). SBASE-A contains 2005 domain groups (139 765 domain sequences), SBASE-B contains 595 domain groups (155 133 domain sequences). The groups are further classified on the basis of the BLAST similarity scores. The list of all clusters with at least two members is deposited into a separate database, SBASE-CLUSTERS, identified by the standard name and by the (optional) subclass number included in the SC field of the records.
Table 1. Increase of data in SBASE release 8.0.
| Release |
Date |
Records |
Amino acids |
Size (Mb) |
| 1.0 |
April 2, 1992 |
27 221 |
1 551 445 |
17.2 |
| 2.0 |
February 13, 1993 |
34 518 |
1 922 524 |
24.9 |
| |
|
(+27%) |
(+24%) |
(+45%) |
| 3.0 |
May 28, 1994 |
41 749 |
2 339 538 |
37.3 |
| |
|
(+21%) |
(+22%) |
(+50%) |
| 4.0 |
June 15, 1995 |
61 137 |
3 281 782 |
50 |
| |
|
(+48%) |
(+40%) |
(+34%) |
| 5.0 |
October 6, 1996 |
79 862 |
4 118 506 |
75 |
| |
|
(+30%) |
(+25%) |
(+50%) |
| 6.0 |
October 23, 1998 |
130 703 |
10 457 771 |
115 |
| |
|
(+63%) |
(+154%) |
(+53%) |
| 7.0 |
October 23, 1999 |
237 937 |
18 964 500 |
221 |
| |
|
(+82%) |
(+81%) |
(+92%) |
| 8.0 |
October 23, 2000 |
294 898 |
28 250 878 |
290 |
| (+24%) | (+33%) | (+24%) |
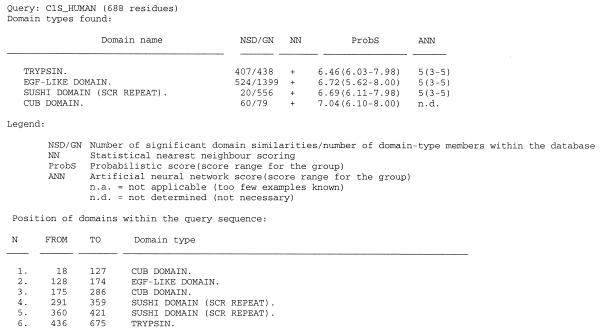
(ii) The search facility of SBASE has been updated so as to include a series of identification steps based on non-parametric pattern recognition. First a simple statistical nearest neighbor scoring is applied, based on internal or within-group group similarities. A sequence segment is considered a member of a given domain group if its similarity parameters, i.e. the number of significant similarities to other members of the group, and the average of the corresponding similarity score (3) are above the threshold levels automatically established for that group, and if it has no sequential overlap with any other domain group. In the second step a probabilistic scoring is applied that takes into consideration the distribution of both the internal and the external similarities (i.e. similarities to non-group members) (4). Thirdly, a neural network identification scheme is applied. A detailed evaluation of the system will be published elsewhere (J.Murvai, K.Vlahovicek, C. Szepesvári and S.Pongor, manuscript in preparation). An output example is shown in Figure 1.
Figure 1.
Results of domain prediction using the SBASE prediction server.
(iii) The domain search facility is now applied to automatically search for new domains, which will allow a faster updating of the database.
DESCRIPTION OF THE DATA
Source and origin of data
SBASE data originate from three main sources: (i) from the SWISS-PROT protein sequence databank (5); (ii) from the Protein Sequence Database of the Protein Identification Resource (PIR International) (6); and (iii) from the literature. The sequences are either translated from nucleotide sequence databases (7,8) or directly keyed in at the protein level. From a total of 294 898 records in SBASE 7.0, 203 479 (69%), 67 826 (23%) and 23 593 (8%) are of eukaryotic, prokaryotic and viral origin, respectively. Domain sizes vary in length between 2 and 3000 amino acids.
Domains included in SBASE are protein sequence segments with known structure and/or function. The main entry classes and representative examples are summarized in Table 1. The boundaries of the domains are determined by homology to domains with known boundaries such as given in the PROT-FAM (9) and in the PFAM databases (10), the INTERPRO resource (11) as well as in the original publications.
Cross-references
SBASE 8.0 has cross-references to several protein and nucleic acid databanks, as well as to the PROSITE (12) PRINTS-S (13), PRODOM (14), BLOCKS (15) and PFAM (10) domain databases, the Protein Structure Data Bank (16) and the database of human Mendelian inheritance (17) (Table 1). In each record, the DR-lines contain the cross-reference data.
Record structure
The format of SBASE 8.0 follows that of the EMBL and SWISS-PROT databases and can be directly formatted under the GCG program package [Wisconsin Package Version 10.0, Genetics Computer Group (GCG), Madison, WI].
DISTRIBUTION AND ACCESS
Distribution
SBASE 8.0 (October 23, 2000) is distributed by anonymous ftp file transfer from ftp.icgeb.trieste.it. The complete database (including the records and list of clusters), is 290 MB; its compressed form is 41 MB.
BLAST search by WWW server
SBASE 8.0 can be searched by the BLAST program using the WWW server http://www.icgeb.trieste.it/sbase/. A related server was created in order to assign SBASE domain homologies on the basis of BLAST searches performed on the SWISS-PROT database and on the PIR International databases (6). This service (available at http://sbase.abc.hu/sbase/ and at domain@abc.hu) returns the best potential domain homologies ranked according to BLAST score.
Access by WWW server
Record retrieval and the above services can be accessed also using the WWW server at http://www.icgeb.trieste.it. At present, cross-references to SBASE-CLUSTERS, EMBL, MEDLINE, MIM, PRINTS, PRODOM, PROSITE and SWISS-PROT can be directly accessed through the WWW server.
Citation
Users of SBASE and of the www servers are asked to cite this article in their publications, e.g. in the following form: ‘The sequence homologies were analyzed searching the SBASE protein domain sequence library release 8.0 via automated electronic mail (WWW) server’.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by EMBnet, the European Molecular Biology Network in the framework of EU grant ERBBIO4-CT96-0030. SBASE was established in 1990 and is maintained collaboratively by the International Center for Genetic Engineering and Biotechnology, Trieste, Italy and the Agricultural Biotechnology Center, Gödöllö, Hungary.
References
- 1.Pongor S., Skerl,V., Cserzo,M., Hatsagi,Z., Simon,G. and Bevilacqua,V. (1992) The SBASE domain library: A collection of annotated protein sequence segments. Protein Eng., 6, 391–395. [DOI] [PubMed] [Google Scholar]
- 2.Murvai J., Vlahovicek,K., Barta,E., Cataletto,B. and Pongor,S. (2000) The SBASE protein domain library, release 7.0: a collection of annotated protein sequence segments. Nucleic Acids Res., 28, 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murvai J., Vlahovicek,K., Barta,E., Parthasarathy,S., Hegyi,H., Pfeiffer,F. and Pongor,S. (1999) The domain-server: direct prediction of protein domain-homologies from BLAST search. Bioinformatics, 15, 343–344. [DOI] [PubMed] [Google Scholar]
- 4.Murvai J., Vlahovicek,K. and Pongor,S. (2000) A simple probabilistic scoring method for protein domain identification. Bioinformatics, in press. [DOI] [PubMed] [Google Scholar]
- 5.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker W.C., Garavelli,J.S., Huang,H., McGarvey,P.B., Orcutt,B.C., Srinivasarao,G.Y., Xiao,C., Yeh,L.S., Ledley,R.S., Janda,J.F. et al. (2000) The protein information resource (PIR). Nucleic Acids Res., 28, 41–44. Updated article in this issue: Nucleic Acids Res. (2001), 29, 29–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker W., van den Broek,A., Camon,E., Hingamp,P., Sterk,P., Stoesser,G. and Tuli,M.A. (2000) The EMBL nucleotide sequence database. Nucleic Acids Res., 28, 19–23. Updated article in this issue: Nucleic Acids Res. (2001), 29, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mewes H.W., Frishman,D., Gruber,C., Geier,B., Haase,D., Kaps,A., Lemcke,K., Mannhaupt,G., Pfeiffer,F., Schuller,C. et al. (2000) MIPS: a database for genomes and protein sequences. Nucleic Acids Res., 28, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Bucher,P., Codani,J.-J., Corpet,F., Croning,M.D.R., Durbin,R. et al. (2000) InterPro – An integrated documentation resource for protein families, domains and functional sites. CCP11 Newsletter, 3. [DOI] [PubMed]
- 12.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attwood T.K., Croning,M.D., Flower,D.R., Lewis,A.P., Mabey,J.E., Scordis,P., Selley,J.N. and Wright,W. (2000) PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res., 28, 225–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2000) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henikoff J.G., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the blocks database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. Updated article in this issue: Nucleic Acids Res. (2001), 29, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler D.L., Chappey,C., Lash,A.E., Leipe,D.D., Madden,T.L., Schuler,G.D., Tatusova,T.A. and Rapp,B.A. (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 28, 10–14. Updated article in this issue: Nucleic Acids Res. (2001), 29, 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]