Abstract
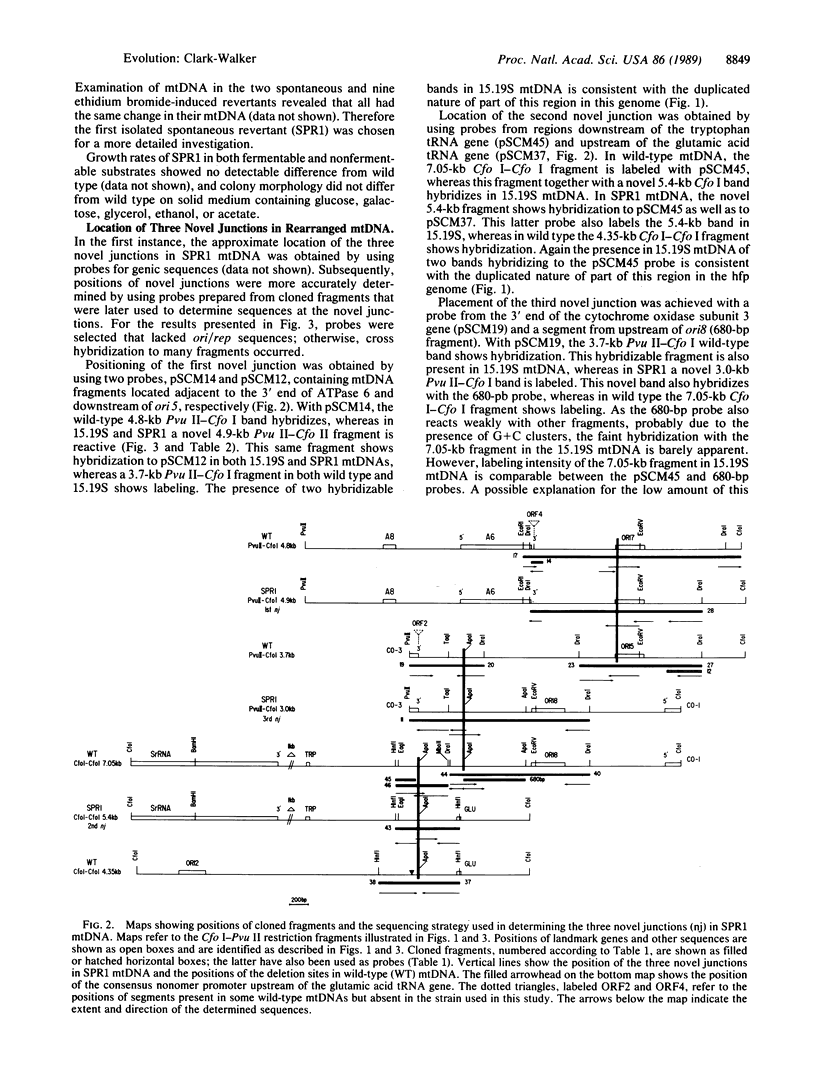
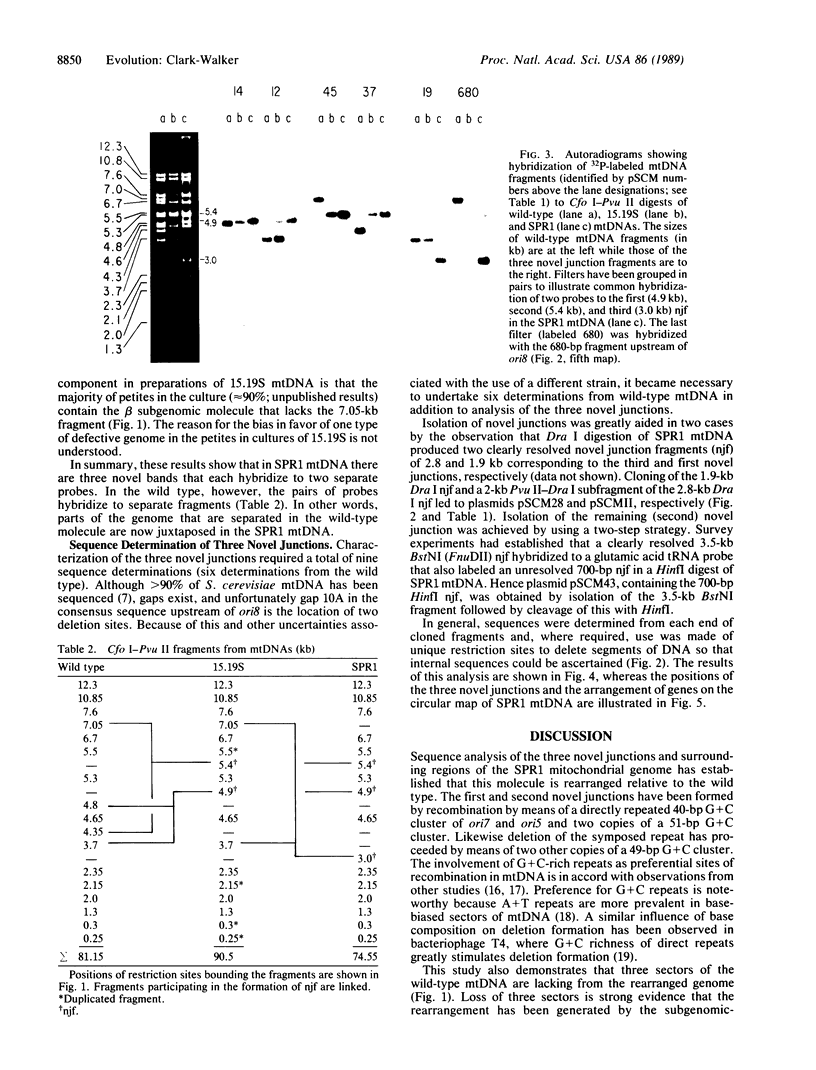
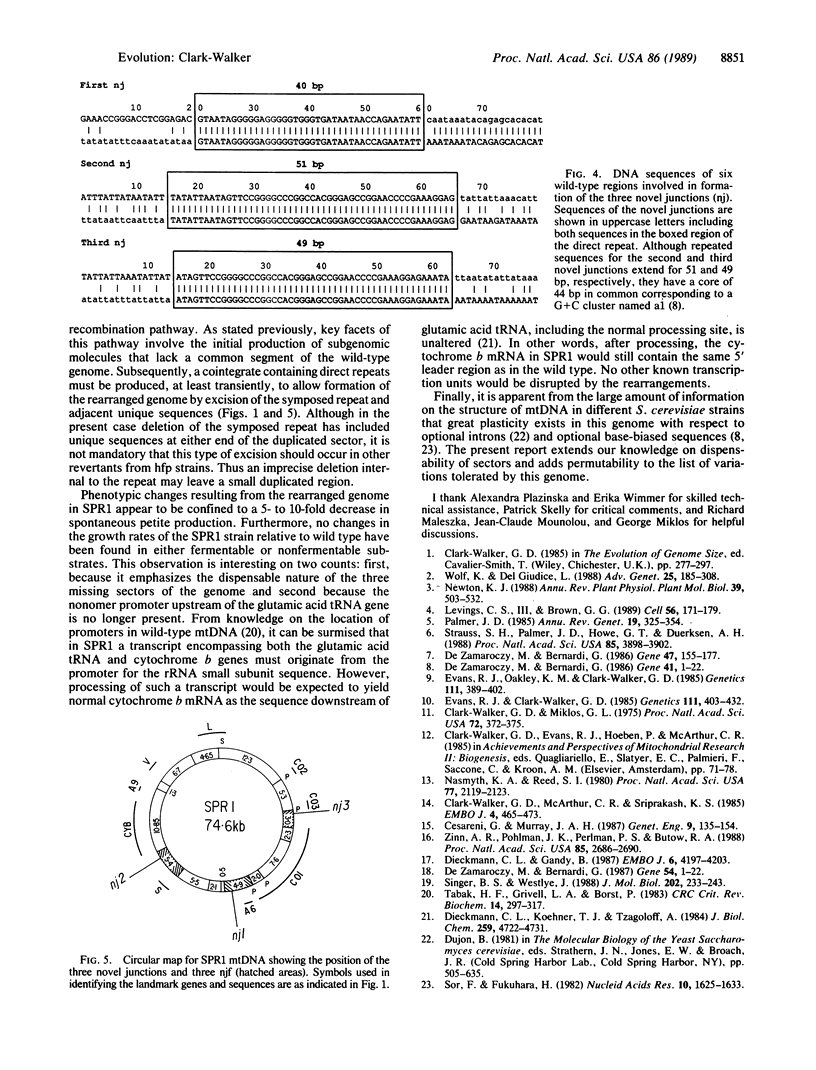
A revertant (SPR1) from a high-frequency petite strain of Saccharomyces cerevisiae has been shown by mapping and sequence analysis to have a rearranged mitochondrial genome. In vivo rearrangement has occurred through a subgenomic-recombination pathway involving the initial formation of subgenomic molecules in nascent petite mutants, recombination between these molecules to form an intermediate with direct repeats, and subsequent excision of the resident or symposed duplication to yield a molecule with three novel junctions and a changed gene order. Sequencing of the novel junctions shows that intramolecular recombination in each case occurs by means of G + C-rich short direct repeats of 40-51 base pairs. Mapping and sequence analysis also reveal that the SPR1 mitochondrial genome lacks three sectors of the wild-type molecule of 4.4, 1.7, and 0.5 kilobases. Each of these sectors occurs in nontemplate, base-biased DNA, that is over 90% A + T. Absence of these sectors together with a rearranged gene order does not appear to affect the phenotype of SPR1, as colony morphology and growth rate on a number of different substrates are not detectably different from the wild type. Lack of phenotypic change suggests that mitochondrial gene expression has not been noticeably disrupted in SPR1 despite deletion of the consensus nonomer promoter upstream from the glutamic acid tRNA gene. Dispensability of DNA sectors and the presence of recombinogenic short, direct repeats are mandatory features of the subgenomic-recombination pathway for creating rearrangements in baker's yeast mtDNA. It is proposed that, in other organisms, organelle genomes containing these elements may undergo rearrangement by the same steps.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clark-Walker G. D., McArthur C. R., Sriprakash K. S. Location of transcriptional control signals and transfer RNA sequences in Torulopsis glabrata mitochondrial DNA. EMBO J. 1985 Feb;4(2):465–473. doi: 10.1002/j.1460-2075.1985.tb03652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D., Miklos G. L. Complementation in cytoplasmic petite mutants of yeast to form respiratory competent cells. Proc Natl Acad Sci U S A. 1975 Jan;72(1):372–375. doi: 10.1073/pnas.72.1.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C. L., Gandy B. Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J. 1987 Dec 20;6(13):4197–4203. doi: 10.1002/j.1460-2075.1987.tb02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C. L., Koerner T. J., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP1, a yeast nuclear gene involved in 5' end processing of cytochrome b pre-mRNA. J Biol Chem. 1984 Apr 25;259(8):4722–4731. [PubMed] [Google Scholar]
- Evans R. J., Clark-Walker G. D. Elevated levels of petite formation in strains of Saccharomyces cerevisiae restored to respiratory competence. II. Organization of mitochondrial genomes in strains having high and moderate frequencies of petite mutant formation. Genetics. 1985 Nov;111(3):403–432. doi: 10.1093/genetics/111.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. J., Oakley K. M., Clark-Walker G. D. Elevated levels of petite formation in strains of Saccharomyces cerevisiae restored to respiratory competence. I. Association of both high and moderate frequencies of petite mutant formation with the presence of aberrant mitochondrial DNA. Genetics. 1985 Nov;111(3):389–402. doi: 10.1093/genetics/111.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings C. S., 3rd, Brown G. G. Molecular biology of plant mitochondria. Cell. 1989 Jan 27;56(2):171–179. doi: 10.1016/0092-8674(89)90890-8. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Singer B. S., Westlye J. Deletion formation in bacteriophage T4. J Mol Biol. 1988 Jul 20;202(2):233–243. doi: 10.1016/0022-2836(88)90454-8. [DOI] [PubMed] [Google Scholar]
- Sor F., Fukuhara H. Nature of an inserted sequence in the mitochondrial gene coding for the 15S ribosomal RNA of yeast. Nucleic Acids Res. 1982 Mar 11;10(5):1625–1633. doi: 10.1093/nar/10.5.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss S. H., Palmer J. D., Howe G. T., Doerksen A. H. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3898–3902. doi: 10.1073/pnas.85.11.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak H. F., Grivell L. A., Borst P. Transcription of mitochondrial DNA. CRC Crit Rev Biochem. 1983;14(4):297–317. doi: 10.3109/10409238309102797. [DOI] [PubMed] [Google Scholar]
- Wolf K., Del Giudice L. The variable mitochondrial genome of ascomycetes: organization, mutational alterations, and expression. Adv Genet. 1988;25:185–308. doi: 10.1016/s0065-2660(08)60460-5. [DOI] [PubMed] [Google Scholar]
- Zinn A. R., Pohlman J. K., Perlman P. S., Butow R. A. In vivo double-strand breaks occur at recombinogenic G + C-rich sequences in the yeast mitochondrial genome. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2686–2690. doi: 10.1073/pnas.85.8.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The AT spacers and the var1 genes from the mitochondrial genomes of Saccharomyces cerevisiae and Torulopsis glabrata: evolutionary origin and mechanism of formation. Gene. 1987;54(1):1–22. doi: 10.1016/0378-1119(87)90342-8. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The GC clusters of the mitochondrial genome of yeast and their evolutionary origin. Gene. 1986;41(1):1–22. doi: 10.1016/0378-1119(86)90262-3. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The primary structure of the mitochondrial genome of Saccharomyces cerevisiae--a review. Gene. 1986;47(2-3):155–177. doi: 10.1016/0378-1119(86)90060-0. [DOI] [PubMed] [Google Scholar]