Abstract
In the title compound, C14H15NO2S, the two aromatic rings enclose a dihedral angle of 70.53 (10)°. A weak intramolecular C—H⋯O hydrogen bond generates an S(6) ring motif. The crystal structure features inversion-related dimers linked by pairs of N—H⋯O hydrogen bonds.
Related literature
For the synthesis, see: Deng & Mani (2006 ▶). For the biological activity of sulfonamides, see: Pandya et al. (2003 ▶); Supuran & Scozzafava (2000 ▶). For the effects of substituents on the crystal structures of and bond lengths in aryl sulfonamides, see: Sharif et al. (2010 ▶); Gowda et al. (2008 ▶, 2009 ▶, 2010 ▶); Nirmala et al. (2009a
▶,b
▶)·For graph-set notation, see: Bernstein et al. (1995 ▶); Etter (1990 ▶).
Experimental
Crystal data
C14H15NO2S
M r = 261.34
Triclinic,
a = 8.6419 (8) Å
b = 8.8016 (8) Å
c = 9.2509 (7) Å
α = 88.187 (4)°
β = 77.010 (4)°
γ = 74.812 (4)°
V = 661.41 (10) Å3
Z = 2
Mo Kα radiation
μ = 0.24 mm−1
T = 296 K
0.28 × 0.17 × 0.08 mm
Data collection
Bruker APEXII CCD diffractometer
11831 measured reflections
3259 independent reflections
2323 reflections with I > 2σ(I)
R int = 0.038
Refinement
R[F 2 > 2σ(F 2)] = 0.044
wR(F 2) = 0.122
S = 1.01
3259 reflections
169 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.24 e Å−3
Δρmin = −0.21 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 1997 ▶); software used to prepare material for publication: WinGX (Farrugia, 1999 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810008329/bt5205sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810008329/bt5205Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O2i | 0.81 (3) | 2.11 (3) | 2.904 (2) | 170 (2) |
| C4—H4⋯O1 | 0.93 | 2.45 | 3.049 (2) | 122 |
Symmetry code: (i) 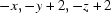 .
.
Acknowledgments
The authors are grateful to Mr Muhammad Hussain of Bana International for providing technical support to the Materials Chemistry Laboratory, Government College University.
supplementary crystallographic information
Comment
Sulfonamides are well known for their antibacterial and enzyme inhibitor properties (Pandya et al., 2003). Aromatic sulfonamides were also reported to inhibit the growth of tumor cells (Supuran & Scozzafava, 2000). In continuation of our studies (Sharif et al., 2010), herein, we report the crystal structure of the title compound.
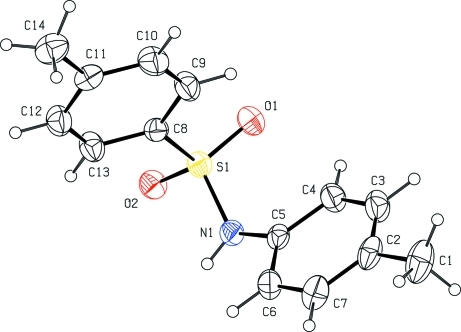
The title molecule (I), (Fig. 1), is bent at the N atom with the C8—SO2—NH—C5 torsion angle of -60.71 (18)°. The dihedral angle between the two aromatic rings is 70.53 (10)°.
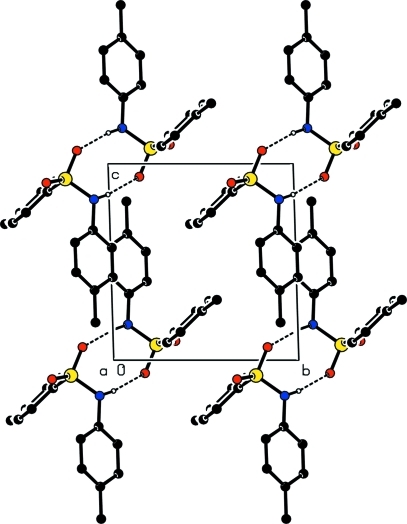
The molecular conformation of the title compound is stabilized by a weak intramolecular C—H···O hydrogen bond, generating an S(6) ring motif (Etter, 1990; Bernstein et al., 1995) (Table 1). In the crystal structure of the title compound, inversion-related molecules are linked into dimers by pairs of N—H···O hydrogen bonds, forming an R22(8) graph-set motif (Table 1 and Fig. 2).
Experimental
The synthesis of the title compound was performed by the procedure reported by Deng & Mani (2006). 4-methyl aniline 0.535 g (5 mmol) was dissolved in 10 ml distilled water and pH of the solution was adjusted to 8 by using (3 M) Na2CO3. p-toluene sulfonyl chloride 0.95 g (5 mmol) was added under continuous stirring at room temperature. pH of the reaction mixture during stirring was maintained between 8-9 with 3 M Na2CO3. When the solution was clear, pH was adjusted to 2-3 using 3 M HCl. The precipitate formed was filtered and recrystallized from methanol.
Refinement
The H atom bonded to N was refined freely. The other H atoms were positioned geometrically and were treated as riding on their parent C atoms, with C—H = 0.93-0.96 Å and Uiso(H) = 1.2Ueq(C) or 1.5Ueq(methyl C).
Figures
Fig. 1.
Perspective view of the title compound with the atoms labelled and displacement ellipsoids depicted at the 30% probability level for all non-H atoms.
Fig. 2.
Partial packing view showing the formation of dimers through N—H···O hydrogen bonds [symmetry code: - x, 2 -y, 2 -z]. For the sake of clarity, H atoms not involved in hydrogen bonding are omitted. Hydrogen bonding is indicated by dashed lines.
Crystal data
| C14H15NO2S | Z = 2 |
| Mr = 261.34 | F(000) = 276 |
| Triclinic, P1 | Dx = 1.312 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 8.6419 (8) Å | Cell parameters from 3233 reflections |
| b = 8.8016 (8) Å | θ = 2.4–25.6° |
| c = 9.2509 (7) Å | µ = 0.24 mm−1 |
| α = 88.187 (4)° | T = 296 K |
| β = 77.010 (4)° | Rod like, light brown |
| γ = 74.812 (4)° | 0.28 × 0.17 × 0.08 mm |
| V = 661.41 (10) Å3 |
Data collection
| Bruker APEXII CCD diffractometer | 2323 reflections with I > 2σ(I) |
| Radiation source: sealed tube | Rint = 0.038 |
| graphite | θmax = 28.4°, θmin = 3.4° |
| φ and ω scans | h = −11→11 |
| 11831 measured reflections | k = −11→11 |
| 3259 independent reflections | l = −11→12 |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.122 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.0642P)2 + 0.0376P] where P = (Fo2 + 2Fc2)/3 |
| 3259 reflections | (Δ/σ)max < 0.001 |
| 169 parameters | Δρmax = 0.24 e Å−3 |
| 0 restraints | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell esds are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement on F2 for ALL reflections except those flagged by the user for potential systematic errors. Weighted R-factors wR and all goodnesses of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The observed criterion of F2 > σ(F2) is used only for calculating -R-factor-obs etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R-factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.18536 (5) | 0.76639 (5) | 0.92202 (4) | 0.0448 (2) | |
| O1 | 0.34628 (15) | 0.66385 (16) | 0.90275 (15) | 0.0579 (4) | |
| O2 | 0.10049 (15) | 0.83328 (15) | 1.06641 (12) | 0.0537 (4) | |
| N1 | 0.1950 (2) | 0.91823 (19) | 0.81866 (16) | 0.0471 (5) | |
| C1 | 0.4564 (3) | 0.9071 (3) | 0.1918 (2) | 0.0860 (9) | |
| C2 | 0.3869 (3) | 0.9072 (3) | 0.3562 (2) | 0.0589 (7) | |
| C3 | 0.4706 (3) | 0.8100 (3) | 0.4474 (2) | 0.0635 (7) | |
| C4 | 0.4115 (2) | 0.8091 (2) | 0.5988 (2) | 0.0570 (7) | |
| C5 | 0.2605 (2) | 0.9077 (2) | 0.66194 (18) | 0.0423 (5) | |
| C6 | 0.1743 (2) | 1.0070 (2) | 0.5727 (2) | 0.0559 (6) | |
| C7 | 0.2379 (3) | 1.0074 (3) | 0.4220 (2) | 0.0661 (8) | |
| C8 | 0.0609 (2) | 0.66738 (19) | 0.85725 (18) | 0.0428 (5) | |
| C9 | 0.1315 (2) | 0.5487 (2) | 0.7513 (2) | 0.0608 (7) | |
| C10 | 0.0316 (3) | 0.4755 (2) | 0.6978 (2) | 0.0668 (8) | |
| C11 | −0.1361 (2) | 0.5162 (2) | 0.7502 (2) | 0.0526 (6) | |
| C12 | −0.2036 (2) | 0.6345 (2) | 0.8566 (2) | 0.0597 (7) | |
| C13 | −0.1067 (2) | 0.7097 (2) | 0.9103 (2) | 0.0566 (7) | |
| C14 | −0.2439 (3) | 0.4335 (3) | 0.6927 (2) | 0.0702 (8) | |
| H1 | 0.118 (3) | 0.994 (3) | 0.843 (2) | 0.068 (7)* | |
| H1A | 0.52890 | 0.97510 | 0.17190 | 0.1290* | |
| H1B | 0.36830 | 0.94410 | 0.14160 | 0.1290* | |
| H1C | 0.51650 | 0.80190 | 0.15690 | 0.1290* | |
| H3 | 0.57180 | 0.74160 | 0.40540 | 0.0760* | |
| H4 | 0.47290 | 0.74260 | 0.65750 | 0.0680* | |
| H6 | 0.07230 | 1.07450 | 0.61420 | 0.0670* | |
| H7 | 0.17880 | 1.07680 | 0.36360 | 0.0790* | |
| H9 | 0.24500 | 0.51800 | 0.71600 | 0.0730* | |
| H10 | 0.07920 | 0.39670 | 0.62450 | 0.0800* | |
| H12 | −0.31690 | 0.66430 | 0.89310 | 0.0720* | |
| H13 | −0.15450 | 0.78950 | 0.98270 | 0.0680* | |
| H14A | −0.27990 | 0.48930 | 0.61030 | 0.1050* | |
| H14B | −0.33800 | 0.43100 | 0.77020 | 0.1050* | |
| H14C | −0.18250 | 0.32780 | 0.66110 | 0.1050* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0441 (3) | 0.0466 (3) | 0.0429 (2) | −0.0104 (2) | −0.0093 (2) | −0.0033 (2) |
| O1 | 0.0451 (7) | 0.0608 (8) | 0.0641 (8) | −0.0050 (6) | −0.0149 (6) | −0.0001 (6) |
| O2 | 0.0579 (8) | 0.0586 (8) | 0.0416 (6) | −0.0118 (6) | −0.0084 (5) | −0.0055 (6) |
| N1 | 0.0467 (9) | 0.0452 (9) | 0.0461 (8) | −0.0127 (8) | −0.0018 (6) | −0.0058 (7) |
| C1 | 0.0891 (17) | 0.131 (2) | 0.0475 (11) | −0.0617 (17) | 0.0057 (10) | −0.0124 (12) |
| C2 | 0.0595 (13) | 0.0780 (14) | 0.0495 (10) | −0.0430 (11) | −0.0023 (9) | −0.0089 (10) |
| C3 | 0.0528 (12) | 0.0689 (13) | 0.0619 (12) | −0.0190 (10) | 0.0074 (9) | −0.0175 (10) |
| C4 | 0.0472 (11) | 0.0590 (12) | 0.0575 (11) | −0.0080 (9) | −0.0032 (8) | −0.0046 (9) |
| C5 | 0.0404 (9) | 0.0443 (9) | 0.0449 (9) | −0.0197 (8) | −0.0036 (7) | −0.0069 (7) |
| C6 | 0.0432 (10) | 0.0656 (12) | 0.0558 (11) | −0.0151 (9) | −0.0042 (8) | 0.0038 (9) |
| C7 | 0.0588 (13) | 0.0898 (16) | 0.0566 (11) | −0.0317 (12) | −0.0141 (9) | 0.0142 (11) |
| C8 | 0.0452 (10) | 0.0391 (9) | 0.0426 (8) | −0.0108 (7) | −0.0070 (7) | 0.0009 (7) |
| C9 | 0.0454 (11) | 0.0571 (12) | 0.0748 (13) | −0.0116 (9) | −0.0022 (9) | −0.0200 (10) |
| C10 | 0.0683 (14) | 0.0542 (12) | 0.0765 (14) | −0.0164 (11) | −0.0098 (11) | −0.0226 (10) |
| C11 | 0.0611 (12) | 0.0487 (10) | 0.0549 (10) | −0.0222 (9) | −0.0189 (9) | 0.0085 (8) |
| C12 | 0.0452 (11) | 0.0654 (13) | 0.0676 (12) | −0.0160 (10) | −0.0080 (9) | −0.0067 (10) |
| C13 | 0.0465 (11) | 0.0610 (12) | 0.0581 (11) | −0.0120 (9) | −0.0032 (8) | −0.0160 (9) |
| C14 | 0.0783 (15) | 0.0678 (14) | 0.0790 (14) | −0.0320 (12) | −0.0324 (12) | 0.0043 (11) |
Geometric parameters (Å, °)
| S1—O1 | 1.4217 (14) | C11—C14 | 1.511 (3) |
| S1—O2 | 1.4324 (12) | C11—C12 | 1.372 (2) |
| S1—N1 | 1.6276 (16) | C12—C13 | 1.373 (3) |
| S1—C8 | 1.7576 (18) | C1—H1A | 0.9600 |
| N1—C5 | 1.429 (2) | C1—H1B | 0.9600 |
| N1—H1 | 0.81 (3) | C1—H1C | 0.9600 |
| C1—C2 | 1.504 (3) | C3—H3 | 0.9300 |
| C2—C7 | 1.376 (4) | C4—H4 | 0.9300 |
| C2—C3 | 1.368 (3) | C6—H6 | 0.9300 |
| C3—C4 | 1.379 (3) | C7—H7 | 0.9300 |
| C4—C5 | 1.376 (3) | C9—H9 | 0.9300 |
| C5—C6 | 1.375 (2) | C10—H10 | 0.9300 |
| C6—C7 | 1.379 (3) | C12—H12 | 0.9300 |
| C8—C13 | 1.374 (3) | C13—H13 | 0.9300 |
| C8—C9 | 1.373 (2) | C14—H14A | 0.9600 |
| C9—C10 | 1.382 (3) | C14—H14B | 0.9600 |
| C10—C11 | 1.374 (3) | C14—H14C | 0.9600 |
| S1···H4 | 3.0400 | H1···O2i | 2.11 (3) |
| O1···C4 | 3.049 (2) | H1A···N1iv | 2.8000 |
| O2···N1i | 2.904 (2) | H1A···C5iv | 3.0100 |
| O2···C14ii | 3.377 (3) | H1B···H7 | 2.4300 |
| O1···H4 | 2.4500 | H1C···H3 | 2.4700 |
| O1···H9 | 2.6100 | H3···H1C | 2.4700 |
| O1···H12iii | 2.8900 | H3···H9vii | 2.5400 |
| O2···H13 | 2.6100 | H4···S1 | 3.0400 |
| O2···H1i | 2.11 (3) | H4···O1 | 2.4500 |
| N1···O2i | 2.904 (2) | H6···H1 | 2.3000 |
| N1···H1Aiv | 2.8000 | H6···C7v | 3.0400 |
| C2···C4iv | 3.481 (3) | H7···H1B | 2.4300 |
| C4···O1 | 3.049 (2) | H9···O1 | 2.6100 |
| C4···C2iv | 3.481 (3) | H9···H3vii | 2.5400 |
| C6···C6v | 3.595 (3) | H10···H14C | 2.4400 |
| C6···C7v | 3.587 (3) | H12···O1viii | 2.8900 |
| C7···C6v | 3.587 (3) | H12···H14B | 2.4500 |
| C14···O2ii | 3.377 (3) | H13···O2 | 2.6100 |
| C2···H14Cvi | 3.0800 | H14B···H12 | 2.4500 |
| C5···H1Aiv | 3.0100 | H14C···H10 | 2.4400 |
| C7···H6v | 3.0400 | H14C···C2vi | 3.0800 |
| H1···H6 | 2.3000 | ||
| O1—S1—O2 | 119.28 (8) | C2—C1—H1A | 109.00 |
| O1—S1—N1 | 108.52 (9) | C2—C1—H1B | 110.00 |
| O1—S1—C8 | 108.38 (8) | C2—C1—H1C | 109.00 |
| O2—S1—N1 | 104.26 (8) | H1A—C1—H1B | 109.00 |
| O2—S1—C8 | 108.41 (8) | H1A—C1—H1C | 109.00 |
| N1—S1—C8 | 107.41 (8) | H1B—C1—H1C | 109.00 |
| S1—N1—C5 | 123.93 (13) | C2—C3—H3 | 118.00 |
| C5—N1—H1 | 112.3 (13) | C4—C3—H3 | 119.00 |
| S1—N1—H1 | 114.3 (16) | C3—C4—H4 | 120.00 |
| C1—C2—C7 | 121.5 (2) | C5—C4—H4 | 120.00 |
| C1—C2—C3 | 121.5 (2) | C5—C6—H6 | 120.00 |
| C3—C2—C7 | 117.01 (18) | C7—C6—H6 | 120.00 |
| C2—C3—C4 | 122.9 (2) | C2—C7—H7 | 119.00 |
| C3—C4—C5 | 119.16 (18) | C6—C7—H7 | 119.00 |
| C4—C5—C6 | 119.03 (16) | C8—C9—H9 | 120.00 |
| N1—C5—C6 | 118.64 (16) | C10—C9—H9 | 120.00 |
| N1—C5—C4 | 122.18 (16) | C9—C10—H10 | 119.00 |
| C5—C6—C7 | 120.51 (18) | C11—C10—H10 | 119.00 |
| C2—C7—C6 | 121.4 (2) | C11—C12—H12 | 119.00 |
| C9—C8—C13 | 119.86 (17) | C13—C12—H12 | 119.00 |
| S1—C8—C9 | 119.76 (14) | C8—C13—H13 | 120.00 |
| S1—C8—C13 | 120.37 (13) | C12—C13—H13 | 120.00 |
| C8—C9—C10 | 119.12 (17) | C11—C14—H14A | 109.00 |
| C9—C10—C11 | 121.65 (16) | C11—C14—H14B | 109.00 |
| C10—C11—C14 | 121.20 (17) | C11—C14—H14C | 110.00 |
| C10—C11—C12 | 118.13 (17) | H14A—C14—H14B | 109.00 |
| C12—C11—C14 | 120.68 (17) | H14A—C14—H14C | 109.00 |
| C11—C12—C13 | 121.15 (17) | H14B—C14—H14C | 109.00 |
| C8—C13—C12 | 120.09 (16) | ||
| O1—S1—N1—C5 | 56.26 (18) | C3—C4—C5—C6 | −1.4 (3) |
| O2—S1—N1—C5 | −175.62 (16) | C3—C4—C5—N1 | −177.08 (19) |
| C8—S1—N1—C5 | −60.71 (18) | C4—C5—C6—C7 | 0.3 (3) |
| O1—S1—C8—C13 | 155.34 (14) | N1—C5—C6—C7 | 176.04 (19) |
| O2—S1—C8—C13 | 24.51 (16) | C5—C6—C7—C2 | 1.3 (3) |
| N1—S1—C8—C13 | −87.60 (15) | S1—C8—C9—C10 | −177.84 (13) |
| O2—S1—C8—C9 | −156.51 (14) | C13—C8—C9—C10 | 1.2 (3) |
| N1—S1—C8—C9 | 91.38 (15) | S1—C8—C13—C12 | 178.40 (14) |
| O1—S1—C8—C9 | −25.69 (16) | C9—C8—C13—C12 | −0.6 (3) |
| S1—N1—C5—C6 | 134.31 (16) | C8—C9—C10—C11 | −1.4 (3) |
| S1—N1—C5—C4 | −50.0 (2) | C9—C10—C11—C12 | 1.0 (3) |
| C3—C2—C7—C6 | −1.5 (4) | C9—C10—C11—C14 | −178.98 (17) |
| C1—C2—C7—C6 | 179.9 (2) | C10—C11—C12—C13 | −0.5 (3) |
| C7—C2—C3—C4 | 0.2 (4) | C14—C11—C12—C13 | 179.57 (17) |
| C1—C2—C3—C4 | 178.9 (2) | C11—C12—C13—C8 | 0.2 (3) |
| C2—C3—C4—C5 | 1.2 (4) |
Symmetry codes: (i) −x, −y+2, −z+2; (ii) −x, −y+1, −z+2; (iii) x+1, y, z; (iv) −x+1, −y+2, −z+1; (v) −x, −y+2, −z+1; (vi) −x, −y+1, −z+1; (vii) −x+1, −y+1, −z+1; (viii) x−1, y, z.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O2i | 0.81 (3) | 2.11 (3) | 2.904 (2) | 170 (2) |
| C4—H4···O1 | 0.93 | 2.45 | 3.049 (2) | 122 |
Symmetry codes: (i) −x, −y+2, −z+2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: BT5205).
References
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl.34, 1555–1573.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Deng, X. & Mani, N. S. (2006). Green Chem 8, 835–838.
- Etter, M. C. (1990). Acc. Chem. Res. 23, 120-126.
- Farrugia, L. J. (1997). J. Appl. Cryst.30, 565.
- Farrugia, L. J. (1999). J. Appl. Cryst.32, 837–838.
- Gowda, B. T., Foro, S., Babitha, K. S. & Fuess, H. (2008). Acta Cryst. E64, o1692. [DOI] [PMC free article] [PubMed]
- Gowda, B. T., Foro, S., Nirmala, P. G. & Fuess, H. (2010). Acta Cryst. E66, o14. [DOI] [PMC free article] [PubMed]
- Gowda, B. T., Foro, S., Nirmala, P. G., Terao, H. & Fuess, H. (2009). Acta Cryst. E65, o800. [DOI] [PMC free article] [PubMed]
- Nirmala, P. G., Gowda, B. T., Foro, S. & Fuess, H. (2009a). Acta Cryst. E65, o3184. [DOI] [PMC free article] [PubMed]
- Nirmala, P. G., Gowda, B. T., Foro, S. & Fuess, H. (2009b). Acta Cryst. E65, o3208. [DOI] [PMC free article] [PubMed]
- Pandya, R., Murashima, T., Tedeschi, L. & Barrett, A. G. M. (2003). J. Org. Chem.68, 8274–8276. [DOI] [PubMed]
- Sharif, S., Akkurt, M., Khan, I. U., Salariya, M. A. & Ahmad, S. (2010). Acta Cryst. E66, o73–o74. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Supuran, C. T. & Scozzafava, A. (2000). J. Enzyme Inhib. Med. Chem.15, 597–610. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablocks global, I. DOI: 10.1107/S1600536810008329/bt5205sup1.cif
Structure factors: contains datablocks I. DOI: 10.1107/S1600536810008329/bt5205Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report