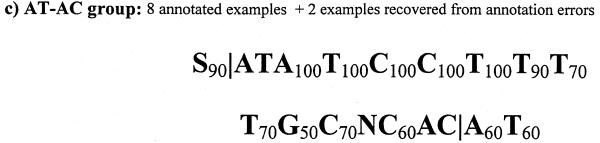Abstract
A database (SpliceDB) of known mammalian splice site sequences has been developed. We extracted 43 337 splice pairs from mammalian divisions of the gene-centered Infogene database, including sites from incomplete or alternatively spliced genes. Known EST sequences supported 22 815 of them. After discarding sequences with putative errors and ambiguous location of splice junctions the verified dataset includes 22 489 entries. Of these, 98.71% contain canonical GT–AG junctions (22 199 entries) and 0.56% have non-canonical GC–AG splice site pairs. The remainder (0.73%) occurs in a lot of small groups (with a maximum size of 0.05%). We especially studied non-canonical splice sites, which comprise 3.73% of GenBank annotated splice pairs. EST alignments allowed us to verify only the exonic part of splice sites. To check the conservative dinucleotides we compared sequences of human non-canonical splice sites with sequences from the high throughput genome sequencing project (HTG). Out of 171 human non-canonical and EST-supported splice pairs, 156 (91.23%) had a clear match in the human HTG. They can be classified after sequence analysis as: 79 GC–AG pairs (of which one was an error that corrected to GC–AG), 61 errors corrected to GT–AG canonical pairs, six AT–AC pairs (of which two were errors corrected to AT–AC), one case was produced from a non-existent intron, seven cases were found in HTG that were deposited to GenBank and finally there were only two other cases left of supported non-canonical splice pairs. The information about verified splice site sequences for canonical and non-canonical sites is presented in SpliceDB with the supporting evidence. We also built weight matrices for the major splice groups, which can be incorporated into gene prediction programs. SpliceDB is available at the computational genomic Web server of the Sanger Centre: http://genomic.sanger.ac.uk/spldb/SpliceDB.html and at http://www.softberry.com/spldb/SpliceDB.html.
INTRODUCTION
The database has been generated as a result of our interest in characterization of observed types of splice sites. New sequences coming every day from genome sequencing projects are mostly annotated by computationally generated information. There is no straightforward procedure to retrieve experimentally supported splice site sequences to study their properties. Currently our knowledge about how the cell is specifying splice sites is not sufficient for accurate and comprehensive computational identification of splice junctions in genomic sequences. Characterization of all known splice sites can help us to increase the quality of gene structure prediction programs. Moreover, many annotated non-canonical splice site sequences may appear in databases as a result of sequencing or annotation errors (1,2). These errors should be found and corrected or discarded in the investigation of splice site characteristics.
EST sequences as an independent source of information were used to verify the annotated splice pairs. This approach has been suggested and exploited by Thanaraj (3), who selected genes without alternative splicing and generated a complex splice site classification system depending on the found EST matches. We extended this approach in using high throughput genome sequencing project (HTG) genomic sequences to verify splice site exonic and intronic composition and applied it to analysis of mammalian genes (4).
Our analysis comprises constitutively as well as alternatively spliced genes. Therefore all kinds of spliced introns of the same gene are included in the database. This is the first public database describing alternative introns supported by ESTs and non-canonical splice junctions.
EST BASED CLASSIFICATION
Every EST similar to any splice construct can be classified depending on quality and type of observed alignment with the annotated gene sequence as (for more details see figure 1b in 4): D-end: EST only covers the left exon; A-end: EST only covers the right exon; B-ends: EST overlaps with a splice junction covering left and right exons with no more than one substitution; Error: EST overlaps with a splice junction covering left and right exons with several mismatches or/and gaps.
When all EST alignments for the same spliced construct have been obtained, every splice site can be classified using the following rules:
(i) if there is some B-end EST, then classify as ‘Supported junction’ (B20) splice pair, otherwise;
(ii) if there is some Error EST, then classify as ‘Error in junction’ (Err) splice pair, otherwise;
(iii) if there is some D-end AND some A-end EST, then classify as ‘Unsupported junction but supported exons’ (5+3) splice pair, otherwise;
(iv) if there is some D-end EST, then classify as ‘Only supported 5′ exon’ (5pr) splice pair, otherwise;
(v) if there is some A-end EST, then classify as ‘Only supported 3′ exon’ (3pr) splice pair, otherwise;
(vi) classify as ‘Completely unsupported’ (Uns) splice pair.
Finally, all splice pairs classified as ‘supported junction’ but with low conservation (identity <95%) within 20 bp. at every side of splice junction were reclassified as ‘Error in junction’.
DATABASE STATUS
This first version of SpliceDB was built using mammalian divisions of the InfoGene database (5), which united information from many GenBank (Release 112) (6) entries describing a particular gene. We obtained 43 337 splice site pairs, of which 22 815 were supported by ESTs. Applying corrections explained in Burset et al. (4) this number was reduced to 22 489 supported and corrected entries. Subdivisions of data in SpliceDB and their content are listed in Table 1.
Table 1. Characteristics of the SpliceDB divisions.
| Sequences of splice
pairs |
Canonical |
Non-canonical |
|
| Mammals | Original from GenBank | 41 722 (96.27%) | 1615 (3.73%) |
| EST supported | 22 374 (98.07%) | 441 (1.93%) | |
| EST supported and corrected | 22 199 (98.71%) | 290 (1.29%) | |
| Human | Original from GenBank | 27 486 (96.55%) | 982 (3.45%) |
| EST supported | 15 384 (98.33%) | 261 (1.67%) | |
| EST supported and corrected | 15 263 (98.89%) | 171 (1.11%) | |
| HTG supported | 156 | ||
More than half (65.69%) of database entries come from human sequences, so we decided to keep separate sets of human splice sites. It may be interesting for scientists working with humans to go directly to these sequences as well as we were able to compare human non-canonical splice sites with HTGs. So, originally we obtained 28 468 human splice site pairs, of which 15 645 (68.57%) were supported by ESTs. After correction procedures 15 434 (68.63% of human entries) of verified splice pairs were presented in the corresponding subdivision.
All subdivisions are subsets of all mammalian annotated splice pair sets and the users can retrieve sequences of any combination of interesting groups.
Mammals or human files are divided at every filter stage into canonical and non-canonical introns. We create three filter stages for every group. The first group is formed by all splice site pairs using original GenBank annotations; the second group comprises pairs supported by ESTs and the third includes pairs supported by ESTs and is automatically corrected, meaning that all ambiguous junctions have been discarded, see Burset et al. (4) for details (Table 1).
In human non-canonical subdivision there is a special file with a subset from non-canonical, EST supported and corrected splice pairs, which are supported by HTG. Analysis of alignment information and possible corrections using HTG have been done manually, studying case-by-case sequence alignments.
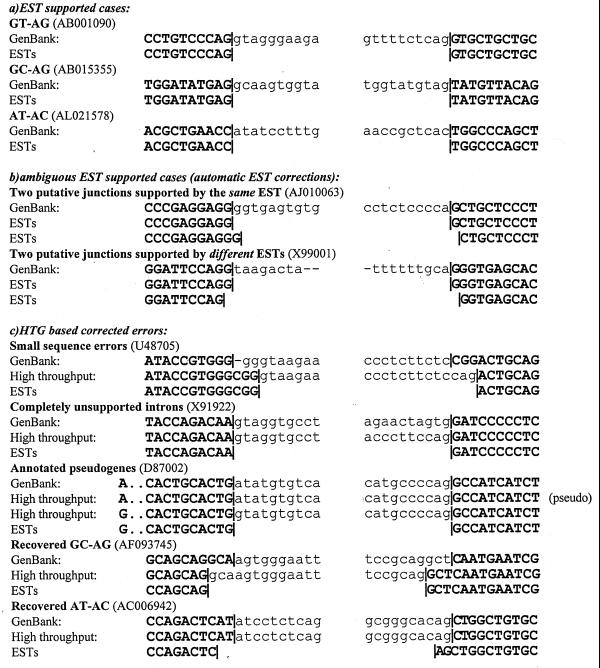
Several examples of EST-supported entries are presented in Figure 1a. As was indicated in Burset et al. (4), we often observe EST-supported sequences with putative errors or, at least, with ambiguity in splice junction position. The same EST may support two splice junctions or several ESTs may support different junctions, as can be seen in Figure 1b.
Figure 1.
Examples of different situations in analysis of annotated splice junctions.
Using HTG sequences allows us to identify a large variety of sequencing and annotation errors. Some entries have only small sequence errors, such as the first example in Figure 1c, which only have a deletion in donor and a substitution in acceptor sites. We recovered here a canonical GT–AG site shifted three positions downstream. Several other cases present completely unsupported introns (Fig. 1c). One very interesting example of errors is annotation of pseudogenes. The functional copy sometimes can be identified in HTG sequences by comparing them with ESTs (assuming that only the functional gene copy will generate EST sequences). In an example of such situation we found a substitution A to G upstream donor site, which helped differentiate the gene functional copy with canonical splice site. The last two examples in Figure 1c cannot be considered as sequence errors. They are examples of wrongly annotated non-canonical sites, which we corrected to typically observed non-canonical pairs.
DATABASE FORMAT
All entries in the database are presented in a tabular format, so every line in any file describes a completely specified splice site pair. We use two kinds of field separators: the different major parts in every entry are separated by the double symbol ‘@@’, and inside the major part the field separator is a typical blank space or tabulator. It allows us to write large sentences inside every major part maintaining clear separation between them. The typical structure of an entry in SpliceDB is:
ID @@ ACCES @@ INTRON @@ DON @@ ACC @@ SEQ_DON @@ SEQ_ACC @@ EST @@ EST_ACCES @@ CORR
ID (database identifier)
This field has always only one word, that is a unique and specific identifier provided to every pair. ID is formed by Infogene (5) entry name, assigned intron number and donor and acceptor positions in the original sequence. All these data are joined using a ‘##’ symbol (i.e. HG_0000731##114##122615##122965).
ACCES (accession number)
This field has always only one word, which refers to one of the original GenBank accession numbers (i.e. AB011399).
INTRON (assigned intron number)
This field has always only one word, that is the intron number assigned to every intron pair in the Infogene database (i.e. 114).
DON (donor number)
This field has always only one word, that is the donor position in the original Infogene entry (i.e. 122615).
ACC (acceptor number)
This field has always only one word, that is the acceptor position in original Infogene entry (i.e. 122965).
SEQ_DON (nucleotide sequence around donor)
The field has always only one word, that is the nucleotide sequence centered in donor conserved dinucleotides, with 40 bp in every side, forming a total sequence of 82 bp (i.e. aacatctgtctctactggaaacctctgcactgaggagcagattgattgataagcaaaaggcttctactgcatttccatcctt).
SEQ_ACC (nucleotide sequence around acceptor)
The field has always only one word, that is the nucleotide sequence centered in acceptor conserved dinucleotides, with 40 bp in every side, forming a total sequence of 82 bp (i.e. aaaaagctcactttttttgttcttcacattttacaggagcagacgcctccgcctagacctgaagcctaccccatccccactc).
EST (EST classification)
This field has always only one word, that is the obtained EST classification [see materials and methods in Burset et al. (4) for details] (i.e. B20).
EST_ACCES (EST accession number supporting classification)
This field has always only one word, that is the accession number of the EST used to support our classification (i.e. gb|N35650|N35650).
CORR (possible corrections)
This field is optional and is specified in free text. All possible corrections are annotated in this field, based on ESTs or in HTGs:
Automatic EST correction in positions [pos1 pos2] using [ESTaccession]. We annotate which positions present ambiguities in addition to the annotated and supported junctions (pos1 and pos2), and the EST accession number that supports alternative junction (ESTaccession).
HTGs [free text]. We provide information about HTGs corresponding to splice junction sequences [for more details see results in Burset et al. (4)].
CONSENSUS AND WEIGHT MATRICES
After analysis of the information presented, we conclude that practically all splice site pairs are limited to three types: GT–AG, GC–AG and AT–AC, and the other kind of introns (if they exist) have a very small frequency (∼0.02% or less).
Alignment of conserved dinucleotides in every type of splice site allows us to observe a certain degree of conservation in surrounding nucleotides, which in practice means a deviation in observed frequencies with respect to expected random distribution. Often this information has been presented in the form of consensus sequences (for every column in aligned sequences we write the most representative nucleotide, or group of nucleotides, indicating this frequency or percent) and as frequency matrices (for every column in aligned sequences we represent the frequency or percentage for every nucleotide, creating a matrix of four rows and as many columns as significant positions). Frequency matrices are more informative and used in gene prediction programs, but a relatively high number of aligned sequences is needed to obtain discriminative matrices.
We present frequence matrices built on verified datasets for GT–AG and GC–AG pair sequences. Because we have a small number of AT–AC cases only consensus sequences are provided for these splice sites (Fig. 2).
Figure 2.
Consensus sequences and weight matrices for major groups of splice site pairs. Frequency matrices have only been calculated for major splice site groups (GT–AG and GC–AG). In the first row we indicated positions with respect to the splice cut point, which is always between –1 and 1. It should be taken into account that negative numbers in donor matrices correspond to exonic regions, but in acceptor matrices positive numbers correspond to exonic regions. In consensus sequences | means cut position (M: A or C, R: A or G, Y: C or T, S: C or G).
SpliceDB is available on the Sanger Centre computational genomic Web server at http://genomic.sanger.ac.uk/spldb/SpliceDB.html and at http://www.softberry.com/spldb/SpliceDB.html.
DISCUSSION
We have applied ESTs and HTG sequences to verify mammalian splice junctions, but there are other model organisms with a lot of genomic data which are interesting to analyze, such as Drosophila melanogaster, Caenorhabditis elegans or Arabidopsis thaliana. We plan to extend our database to nearly all model eukaryotic organisms. Another problem is to install HTG analysis as automatically as possible, because manual intervention is very time consuming (if more accurate).
Observation of practically only three types of splice sites simplifies the problem of their computational identification by gene prediction programs. Consideration of a GC–AG splice pair in the Fgenesh program has been done by Salamov and Solovyev (7), and maintained the accuracy level despite including many more potential splice variants. Addition of AT–AC splice sites (occurring with very low frequency) will probably wait until we accumulate more examples, allowing us to better describe the site characteristics.
Another point to consider is whether to include information about gene structures supported by ESTs. It might be useful for training gene prediction software because it is very sensitive to sequence errors, especially in conserved positions of splice site sequences. Including information about alternative intron positions is very important for developing gene prediction programs that will generate alternative splicing variants.
References
- 1.Penotti, F.E. (1991) Human Pre-mRNA splicing signals. J. Theor. Biol., 150, 385–420. [DOI] [PubMed] [Google Scholar]
- 2.Jackson, I.J. (1991) A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res., 19, 3795–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thanaraj, T.A. (1999) A clean data set of EST-confirmed splice sites from Homo sapiens and canonicals for clean-up procedures. Nucleic Acids Res., 27, 2627–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burset, M., Seledtsov,I.A. and Solovyev,V.V. (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res., 28, 4364–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solovyev, V.V. and Salamov,A.A. (1999) INFOGENE: a database of known gene structures and predicted genes and proteins in sequences of genome sequencing projects. Nucleic Acids Res., 27, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, D.A., Boguski,M.S., Lipman,D.J., Ostell,J., Ouellette,B.F., Rapp,B.A. and Wheeler,D.L. (1999) GenBank. Nucleic Acids Res., 27, 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salamov, A. and Solovyev,V. (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res., 10, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]